In this issue of Blood, Kwon et al1 provide evidence that posttranslational modification of the transcriptional factor RUNX1, through serine residue phosphorylation by CDK9, facilitates megakaryocyte lineage specification from bipotential megakaryocyte-erythroid progenitors (MEPs).
In adult hematopoiesis, RUNX1 has previously been shown to be of importance in megakaryocyte lineage specification through conditional murine Runx1 knockout models,2 including in megakaryocyte specification from platelet/megakaryocyte-primed hematopoietic stem/progenitor cell populations.3 RUNX1 forms a sequence-specific core-binding factor transcription factor complex with its heterodimeric partner CBFβ. Earlier work has suggested posttranslational modification of RUNX1 via serine residue phosphorylation by members of several kinase families, including cyclin-dependent kinases, can increase RUNX1 transcriptional activity and ubiquitin-mediated degradation through several potential molecular mechanisms (reviewed by Goyama et al4).
Interestingly, CDK9,5 the serine/threonine kinase component of the positive transcriptional elongation factor complex (P-TEFb) that modulates transcriptional elongation and termination via RNA polymerase II component phosphorylation, may also modulate cell-differentiation programs. Indeed, CDK9 and P-TEFb have been implicated in megakaryocyte-erythroid specification in vitro and in vivo. At the molecular level, P-TEFb activity has been shown to contribute to the transcriptional cooperation of GATA-1 and RUNX1 in the activation of the megakaryocytic IIb promoter,6 with flavopiridol, a relatively selective CDK9 inhibitor, impairing megakaryocyte differentiation from human CD34+ cells.
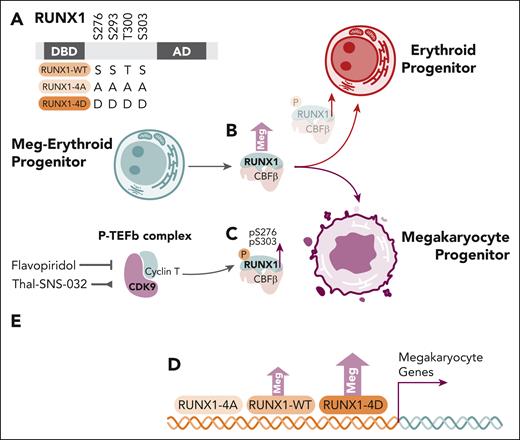
Kwon and colleagues expand on earlier works by exploring the role of RUNX1 and RUNX1 serine phosphorylation in primary human MEPs and the bipotential HEL cell line (see figure panel A). The authors confirmed RUNX1 overexpression in primary MEPs favored megakaryocyte colony formation (see figure panel B), whereas Ro5-3335, a putative but unconfirmed inhibitor of RUNX1/CBFβ,7 favored the erythroid lineage specification. Further in vivo analysis of pS276P and pS303P RUNX1 residues revealed increased RUNX1 phosphorylation with megakaryocyte progenitor formation from MEPs, more so than in erythroid progenitors (see figure panel C).
The role of RUNX1 serine residue phosphorylation and CDK9 in megakaryocyte specification from megakaryocyte-erythroid progenitors. (A) RUNX1 serine/threonine residues, with schemata of wild-type, alanine-mutated, phosphorylation-resistant (4A) and aspartate-mutated phosphomimetic (4D) constructs. (B) Overexpression of RUNX1-WT in transduced MEPs results in increased megakaryocyte colony formation. (C) Phosphoserine residues pS276P and pS303P are relatively increased megakaryocyte progenitors. (D) Overexpression of RUNX1-WT and RUNX1-4D constructs, but not RUNX1-4A, in HEL cells induces increased megakaryocyte colony formation and expression signatures enriched for megakaryocyte genes. (E) Inhibition (flavopiridol) or degradation (Thal-SNS-032) of CDK9 inhibits RUNX1 pS276P and pS303P, whereas degradation (Thal-SNS-032) also inhibits megakaryocyte colony formation from MEPs. AD, activation domain; DBD, DNA binding domain; Meg, megakaryocyte; MEP, megakaryocyte-erythroid progenitor.
The role of RUNX1 serine residue phosphorylation and CDK9 in megakaryocyte specification from megakaryocyte-erythroid progenitors. (A) RUNX1 serine/threonine residues, with schemata of wild-type, alanine-mutated, phosphorylation-resistant (4A) and aspartate-mutated phosphomimetic (4D) constructs. (B) Overexpression of RUNX1-WT in transduced MEPs results in increased megakaryocyte colony formation. (C) Phosphoserine residues pS276P and pS303P are relatively increased megakaryocyte progenitors. (D) Overexpression of RUNX1-WT and RUNX1-4D constructs, but not RUNX1-4A, in HEL cells induces increased megakaryocyte colony formation and expression signatures enriched for megakaryocyte genes. (E) Inhibition (flavopiridol) or degradation (Thal-SNS-032) of CDK9 inhibits RUNX1 pS276P and pS303P, whereas degradation (Thal-SNS-032) also inhibits megakaryocyte colony formation from MEPs. AD, activation domain; DBD, DNA binding domain; Meg, megakaryocyte; MEP, megakaryocyte-erythroid progenitor.
Transduction of the HEL bipotential cell line with wild-type (RUNX1-WT), alanine-mutated, phosphorylation-resistant (RUNX1-4A), or aspartate-mutated phosphomimetic (RUNX1-4D), constructs demonstrated megakaryocyte-lineage specification with RUNX1-WT was further increased with the RUNX1-4D construct, whereas the RUNX1-4A construct did not demonstrate activity greater than the empty vector control (see figure panel D). Interestingly, distinct gene expression changes induced by RUNX1-4A, RUNX1-WT, and RUNX1-4D in HEL cells compared with empty vector control were noted, but cut-and-run analysis, including at megakaryocyte and erythroid-specific gene loci, did not identify clearly different binding patterns between the various RUNX1 constructs, despite clear differences in gene expression by RNAseq.
Flavopiridol and the Thal-SNS-032 degrader used to inhibit CDK9 activity and reduce CDK9 levels, respectively (see figure panel E), resulted in reduction of RUNX1 phosphorylation. Consistent with earlier data, Thal-SNS-032 treatment of MEPs also increased erythroid colony formation, yet this effect was mitigated by overexpression of RUNX1-WT and RUNX1-4D.
Although providing incremental advances, Kwon et al lend support to earlier data that CDK9 and the P-TEFb complex have a role in hematopoietic lineage specification, with their evidence suggesting that CDK9 facilitates phosphorylation of important serine residues of RUNX1 that favor megakaryocyte lineage specification.
Important and intriguing questions, however, remain to be explored. The authors have duly acknowledged that their overexpression systems may not precisely represent the molecular interactions during normal megakaryocyte development from MEPs. Future work must consider validation of their model using gene editing of endogenous gene loci. Such approaches may also help elucidate the relative contribution of a complex array of potential kinases8 and phosphorylation targets of CDK9 and the P-TEFb complex. This may help identify those factors, the phosphorylation of which most contribute to megakaryocyte lineage specification. Similarly, demonstrating direct interaction between CDK9/P-TEFb and RUNX1 (and other factors, such as CBFβ and GATA-19) in MEPs in vivo would certainly strengthen their model. The molecular mechanisms by which RUNX1 phosphorylation increases transcriptional activity, particularly at megakaryocyte gene loci, also remains to be explored.
Overall, this and other studies highlight an important layer of gene expression regulation, where posttranslational modification of transcription factor complexes can have major effects on transcriptional activity with physiological consequences. The complex interplay of RUNX1, CBFβ, GATA-1 transcription factors, and phosphorylation by CDK9/P-TEFb during megakaryocyte lineage specification is a rich field for further investigation. Understanding how these players could integrate cell-signaling pathways with transcription factor complex function not only provides important biochemical and biologic insights but could also illuminate molecular targets for potential therapeutic approaches to human disease.
Conflict-of-interest disclosure: A.P.N. declares no competing financial interests.