Abstract
The inv(11)(p15q22) is a recurrent chromosomal abnormality associated with de novo and therapy-related myeloid malignancies. Here we report the molecular definition of this chromosomal aberration in four patients. Positional cloning showed the consistent rearrangement of the DDX10 gene on chromosome 11q22, which encodes a putative RNA helicase. The translocation targets the NUP98 gene on 11p15, a member of the FG peptide repeat nucleoporin family. In DDX10 and NUP98, the inv(11) breakpoints occurred within two introns of each gene and the two genes merged in-frame to produce the chimeric transcripts characteristic of this translocation. Although two reciprocal chimeric products, NUP98-DDX10 and DDX10-NUP98, were predicted, only NUP98-DDX10 appears to be implicated in tumorigenesis. DDX10 is predicted to be involved in ribosome assembly. NUP98 has been identified as a nuclear pore complex protein and a target of chromosomal translocation in acute myeloid leukemia through the t(7; 11)(p15; p15) translocation. The predicted NUP98-DDX10 fusion protein may promote leukemogenesis through aberrant nucleoplasmic transport of mRNA or alterations in ribosome assembly.
SPECIFIC CHROMOSOMAL translocations are frequently found in hematopoietic malignant tumors and in some types of solid tumors.1 Molecular analyses of the chromosomal translocations in leukemia have shown rearrangements of genes involved in the programmed regulation of proliferation and differentiation in the hematopoietic system. In many myeloid leukemias, chromosomal alterations have been shown to result in the production of unusual chimeric proteins.2 3 Some of the structures of the abnormal transcripts have been clarified and these transcripts have been proposed to play an important role in malignant transformation.
A number of different recurring aberrations involving chromosome band 11q22-23 are observed in human de novo acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), and myelodysplastic syndrome (MDS). The band 11q22-23 has also been reported to be involved in therapy-related AML (t-AML) or MDS (t-MDS) that occasionally develops after cytotoxic chemotherapy including the use of topoisomerase II (topo II) inhibitors. Several groups of investigators have cloned the MLL/ALL1/HRX/HTRX14-7 gene on 11q23 and shown it to be disrupted and fused with up to 40 different partner genes in many aberrations of 11q22-23, including those resulting from therapy-related secondary leukemia.8,9 However, translocations or inversions in the 11q22-23 region that do not involve the MLL locus have been reported.10 11 Recent studies in hematopoietic neoplasms identified the additional genes, PLZF,12RCK,13 and LPC14 in 11q23. PLZF was identified in acute promyelocytic leukemia and found to be associated with the t(11; 17)(q23; q21) translocation. The RCK and LPC genes are involved in the t(11; 14)(q23; q32) translocation found in lymphomas.
The inversion 11 (p15q22) has been observed rarely in de novo AML and MDS, but is found more frequently in t-AML and t-MDS.15,16 We previously reported detection of the inv(11)(p15q22) rearrangement by fluorescence in situ hybridization (FISH) with a yeast artificial chromosome (YAC) clone, y981g12, which mapped to a region distinct from the MLL locus.17 From this YAC, we constructed a P1 contig and identified a P1 clone which spanned the breakpoints. Here we report that a DEAD-box putative RNA helicase gene, DDX10,18 is rearranged. Subsequent analysis shows that DDX10 is fused in-frame to a nucleoporin gene NUP98,19 which is found associated with t(7; 11)(p15; p15) AML.20 21 This rearrangement was observed in four inv(11) patients with de novo AML, de novo MDS, t-AML, and t-MDS. Thus, we suggest that the production of a chimeric protein between DDX10 and NUP98 is involved in the pathogenesis of inv(11) AML and MDS.
MATERIALS AND METHODS
Patient samples.We studied leukemic cells obtained from four patients with inv(11)(p15q22) associated with malignant myeloid diseases. The clinical and cytogenetic data of the patients have been described previously.17 Two of the four patients had non-Hodgkin's lymphoma (no. 1) or ALL (no. 2) as the primary malignancy and had received cytotoxic chemotherapy, including the use of topo II inhibitors. They developed AML (M4) (no. 1) and MDS (no. 2) 12 and 36 months, respectively, after the initiation of chemotherapy. The other two had de novo AML (M1) (no. 3) or MDS (no. 4).
Genomic cloning.A P1 library of the total human genome was screened by the polymerase chain reaction (PCR) method as described.22 Seven anonymous DNA markers (D11S1897, D11S1818, D11S2180, D11S1294, D11S535, D11S2179, and D11S384) were mapped above a YAC clone, y981g12, and were then used as probes for the initial screening of the P1 library. The P1 contig was constructed by chromosome walking with sequence-tagged sites (STS) from end fragments of the P1 DNAs. One P1 clone, P189, containing a part of the NUP98 gene, was isolated using PCR primers NF1 (CACAAATACCAGTGGTGGGAATA) and NR1 (GTTCCAAATCCTGCACCAAG) developed from the 93-bp exon A sequence.20 End fragments from P1s were isolated by ligation-mediated PCR23 with minor modifications. In brief, P1 clones were digested with Alu I, Rsa I, or HaeIII and ligated with pre-annealed linker. The ligated mixtures were amplified by PCR using the linker primer (LM1, GCGGTGACCCGGGAGATCTGAATTC) and a P1 vector end-specific primer (PSup1, TACGTAGGCCTAATTGGCCGTCGAC or PTup1, TCGAGCTTGACATTGTAGGACTATA). PCR amplification was performed for 35 cycles (denaturation at 94°C for 30 seconds, annealing at 60°C for 45 seconds, and extension at 72°C for 90 seconds) in a GeneAmp PCR system 9600 (Perkin Elmer, Foster City, CA). Secondary PCR was performed using LM1 and an nested P1 vector end-specific primer (PSup2, GGCCGTCGACATTTAGGTGACACTA or PTup2, CGGCCGCTAATACGACTCACTATAG). The amplified fragments were used as probes for Southern blot analysis and sequenced by an ALF DNA sequencer (Pharmacia, Uppsala, Sweden) or 373S DNA sequencer (Perkin Elmer). All STSs were mapped back to human chromosome 11 by PCR analysis or Southern blot analysis of YAC y981g12 and a human-mouse somatic hybrid cell line, A9neo11, which contains chromosome 11 as the only human chromosome. Restriction mapping of the genomic clones was performed by standard techniques using single and double digestions of genomic DNA as described.22
FISH.P1 clones were labeled with biotin-16-dUTP and/or digoxigenin-11-dUTP by nick translation. Hybridization to metaphase cells was done as described previously.17
Southern blot analysis.Genomic DNA (5 μg) was isolated from patient samples and two cell lines, Namalwa and C498. DNA was digested with appropriate restriction enzymes, separated by electrophoresis in agarose gels, and transferred to Hybond-N membrane (Amersham, Bucks, UK). Hybridization with a random-primer-labeled probe was performed at 42°C in 6× standard saline citrate (SSC)/10% dextran sulfate/1% sodium dodecyl sulfate (SDS)/1× Denhart's solution/50% formamide containing denatured salmon sperm DNA (100 μg/mL). If probes containing repetitive sequences were used, human placenta DNA was added to the hybridization mixture. The final washing was in 0.1× SSC/0.1% SDS at 65°C. Autoradiography was performed using a bioimage analyzer, Fujix BAS2000 (Fujifilm, Tokyo, Japan).
Sequencing of the exon-intron boundaries.The location of exons was determined by restriction mapping and Southern blot analysis of genomic clones using various parts of DDX10 and NUP98 cDNAs as probes. The exon-intron boundaries of the genes were determined by two methods. One was the ligation mediated PCR23 method and the other was the direct sequencing method. The ligation mediated PCR was performed as described above. The ligated mixtures were amplified by PCR using the linker primer LM1 and primers corresponding to the DDX10 and NUP98 cDNAs. Secondary PCR was performed with the same primer pair. The amplified fragments were used as probes for Southern blot analysis and were sequenced. When direct sequencing was applied, P1 DNA was purified through polyethylene glycol precipitation and phenol-chloroform extraction. One microgram of purified P1 DNA was sequenced with a PRISM dye-terminator FS cycle sequencing kit (Perkin Elmer) and cDNA specific primers.
Reverse transcription (RT)-PCR and primers.Total RNA of peripheral blood or bone marrow cells was isolated by the acid guanidinium thiocyanate/phenol/chloroform method.24 From 1 μg of this RNA, cDNA was synthesized in 20 μL of the reaction mixture with random nonamer primers and reverse transcriptase XL using a RNA PCR kit (Takara, Tokyo, Japan). The reaction was diluted to 40 μL and 1 or 2 μL was used for PCR. PCR amplification was performed for 35 cycles (denaturation at 94°C for 25 seconds, annealing at 55 or 60°C for 60 seconds, and extension at 72°C for 60 seconds), followed by denaturation at 94°C for 3 minutes and extension at 72°C for 10 minutes. PCR products were separated by electrophoresis through a 1.5% GTG agarose gel (FMC Bioproducts, Rockland, ME) in 1× TAE. Fragments were excised from the gel and sequenced directly. PCR primers for the DDX10 and NUP98 were designed according to the known cDNA sequences as follows: DF26, AAATATACTCGTGTGCACACCAGG; DR27, TTTGCTGCAGCTCACAGACTATGT; NF2, GCTGGACAGGCATCTTTGTT; NR14, TCGTCATCCAGCCCATCAAAG; NR17, GGAGAAAAGAGATTGCTATTA.
Northern blot analysis.Membranes of human multiple tissue Northern (MTN) blot II and human fetal MTN blot II were purchased from Clontech (Palo Alto, CA). Hybridization and autoradiography were performed as for Southern blot analysis.
RESULTS
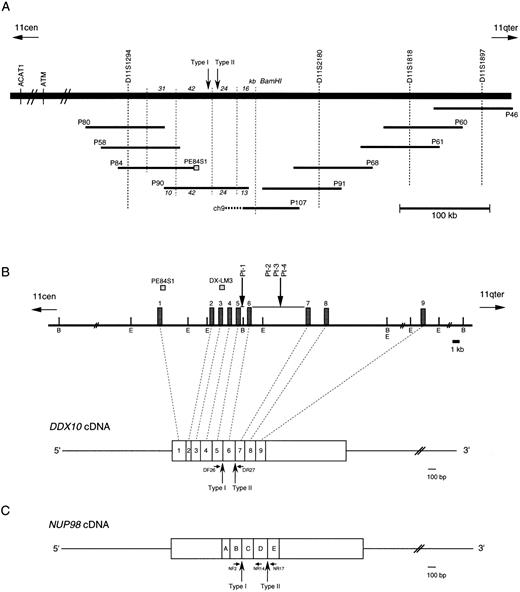
The rearranged gene at 11q22 is DDX10.We previously reported that it was possible to detect the rearrangement, 11q22.3-q23.1, in leukemic cell DNAs from inv(11) AML and MDS patients (patients 1, 2, and 4) using a YAC clone, y981g12.17 The YAC contains about 2,000 kb of a genomic insert without chimerism, and two important house keeping-genes, ACAT1,25 which encodes the acetyl-Coenzyme A acetyltransferase 1 gene, and ATM,26 which encodes the ataxia-telangiectasia mutated gene.27 FISH analyses using P1 clones containing these genes showed that both ACAT1 and ATM were centromeric to the inv(11) breakpoint (data not shown). To localize the breakpoint more precisely, we constructed P1-based contig maps using STS markers that had been mapped on y981g12 (Fig 1A). FISH analyses with P1 clones on the contig indicated that P90 was split in leukemic cells from patients 1 and 2, whereas the overlapping P1 clones, P84 and P107, were not (data not shown). From the overlapping BamHI fragments of the three P1 clones, the breakpoint was predicted to be located within either the 42-kb, 24-kb, or 16-kb genomic BamHI fragment (Fig 1A). However, Southern blot analysis using the 42-kb, 24-kb, and 13-kb BamHI fragments of P90 as probes did not allow the detection of any rearranged band, probably because of high background signals, originating from highly repetitive sequences, and nonchromosome 11 signals.
Physical map of the 11q22 region and the structure of the DDX10 gene and the NUP98 gene. Vertical arrows indicate breakpoints of both DDX10 and NUP98. Small horizontal arrows below the transcripts indicate primers used for RT-PCR. (A) A schematic outline of the P1 contig in 11q22. The BamHI restriction map around P90 is also shown. P107 is chimeric and its chimeric portion originates from chromosome 9. (B) Restriction map around the inv(11) breakpoints on 11q22 and its relationship to the DDX10 cDNA. B and E indicate the BamHI and EcoRI sites, respectively. Exons of the DDX10 gene are represented by boxes (not shown to scale) on the map. Two probes used for Southern blot analysis are shown above the map. (C) cDNA structure of NUP98.
Physical map of the 11q22 region and the structure of the DDX10 gene and the NUP98 gene. Vertical arrows indicate breakpoints of both DDX10 and NUP98. Small horizontal arrows below the transcripts indicate primers used for RT-PCR. (A) A schematic outline of the P1 contig in 11q22. The BamHI restriction map around P90 is also shown. P107 is chimeric and its chimeric portion originates from chromosome 9. (B) Restriction map around the inv(11) breakpoints on 11q22 and its relationship to the DDX10 cDNA. B and E indicate the BamHI and EcoRI sites, respectively. Exons of the DDX10 gene are represented by boxes (not shown to scale) on the map. Two probes used for Southern blot analysis are shown above the map. (C) cDNA structure of NUP98.
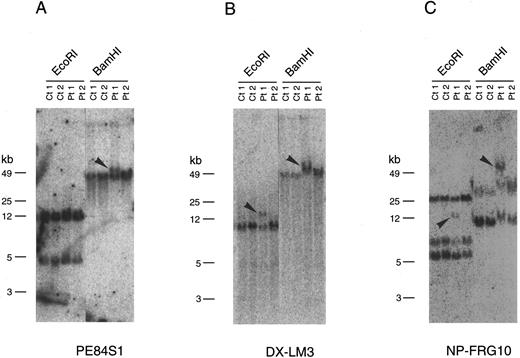
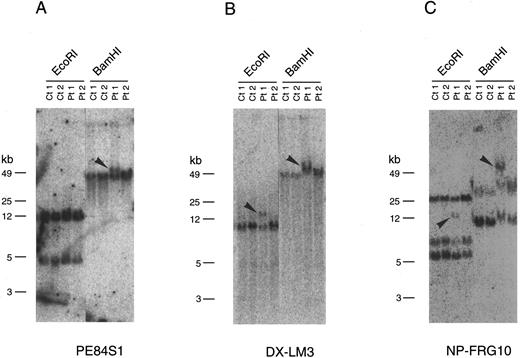
Southern blot analysis with the end sequences of the P1 clones including those of P84, P90, and P107 indicated that PE84S1, a fragment from P84, could detect a rearranged band in patient 1 (Fig 2A). BLAST database28 searching revealed that the sequence of PE84S1 contained an exon-intron junction of a putative RNA helicase gene, DDX10. A 3.2-kb cDNA sequence18 that encompasses the entire predicted coding frame was then amplified by RT-PCR and used for Southern blot analysis. The DDX10 gene is composed of at least 12 exons and spans more than a 200-kb region on P84, P90, P107, P91, P68, P61, and P60. Exon-intron boundary sequences were then determined by ligation-mediated PCR and by direct sequencing using P90 DNA and primers of DDX10 cDNA sequences. This demonstrated the existence in P90 of at least 9 exons starting from the 5′ terminus (Fig 1B). Sequence comparison with the cDNA sequence18 indicated the following exon sequences: exon 1 from the nucleotide sequence 1 to 251, exon 2 from 252 to 312, exon 3 from 313 to 443, exon 4 from 444 to 602, exon 5 from 603 to 723, exon 6 from 724 to 913, exon 7 from 914 to 1040, exon 8 from 1041 to 1203, and exon 9 from 1204 to 1387. Probe PE84S1, which detected the rearranged band in patient 1, was found to contain the boundary sequence between exon 1 and intron 1. Analysis using the probe DX-LM3, a 241-bp ligation-mediated PCR product from the exon 3-intron 3 junction, showed that the breakpoint mapped within an 8.5-kb EcoRI fragment in patient 1 (Fig 2B).
Southern blot analysis of genomic DNA from two patients carrying the therapy-related inv(11). Genomic DNA samples from two inv(11) patients (Pt1, patient 1; Pt2, patient 2), as well as control cell lines (Ct1, Namalwa; Ct2, C498) were digested with EcoRI or BamHI. Rearranged bands are indicated by arrowheads. Hybridization of three probes, PE84S1 (A), DX-LM3 (B), and NP-FRG10 (C), is shown. kb, kilobases.
Southern blot analysis of genomic DNA from two patients carrying the therapy-related inv(11). Genomic DNA samples from two inv(11) patients (Pt1, patient 1; Pt2, patient 2), as well as control cell lines (Ct1, Namalwa; Ct2, C498) were digested with EcoRI or BamHI. Rearranged bands are indicated by arrowheads. Hybridization of three probes, PE84S1 (A), DX-LM3 (B), and NP-FRG10 (C), is shown. kb, kilobases.
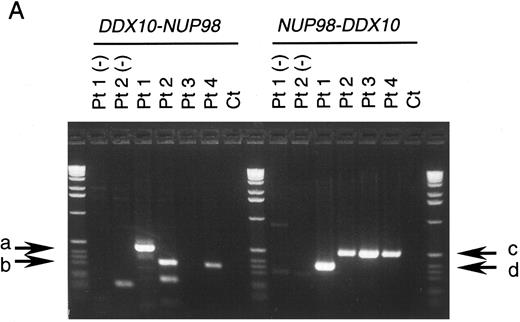
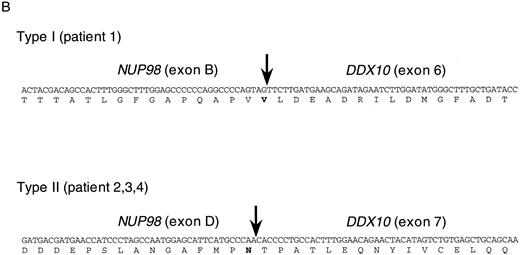
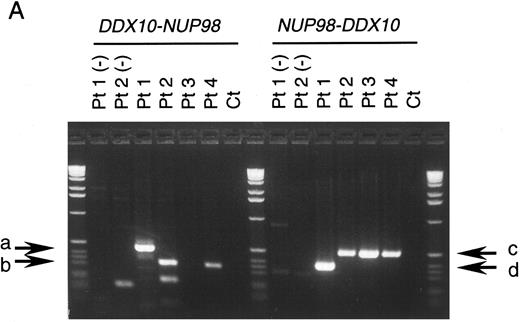
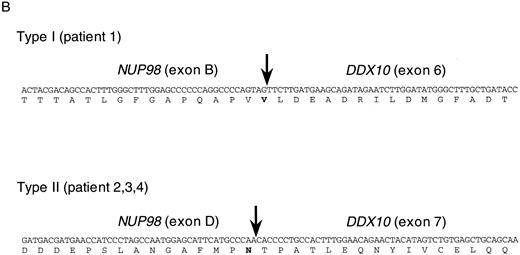
NUP98 is fused with DDX10 in de novo and therapy-related inv(11) patients. NUP98 was recently identified as a gene located on 11p15 that becomes rearranged and fused to the HOXA9 gene at 7p15 following the t(7; 11) translocation associated with AML (M2 and M4 subtypes).20,21 The breakpoint of inv(11) maps cytogenetically to the same 11p15 band as the t(7; 11) translocation. This suggested that the NUP98 gene might also be implicated in the myeloid malignancy associated with inv(11)(p15q22). Thus, we prepared several cDNA probes for NUP98 by RT-PCR and performed Southern blot analysis on DNA from patients 1 and 2. Rearranged bands were detected as 14-kb EcoRI and 50-kb BamHI bands in patient 1 using the probe NP-FRG10, which was amplified with primers NF2 and NR14 (Fig 2C). The rearranged bands were the same size as those detected using a fragment of DDX10 as a probe (Fig 2B). We isolated a P1 clone, P189, containing a part of the NUP98 gene, and determined the exon-intron junctions. We tentatively named the 5 exons, exons A-E, as indicated in Fig 1C. Sequence comparison with the cDNA sequence21 indicated the following exon sequences: exon A from the nucleotide sequence 1319 to 1411, exon B from 1412 to 1552, exon C from 1553 to 1687, exon D from 1688 to 1874, exon E from 1875 to 1991. Probe NP-LM12, a 221-bp ligation-mediated PCR product from the intron C-exon D junction of NUP98, detected rearranged bands in patient 2 (data not shown). These results indicate that NUP98 is rearranged and juxtapositioned to DDX10 in at least patient 1 and that a fusion gene(s) is formed between NUP98 and DDX10. To confirm the existence of the NUP98-DDX10 or DDX10-NUP98 fusion transcript, RT-PCR amplification using NUP98 and DDX10 sequence-specific primers was performed on RNA from four patients including an additional two cases with de novo myeloid disorder. Although the NUP98-DDX10 fusion transcript was detected in all four patients, the DDX10-NUP98 fusion transcript was not detected in patient 3 (Fig 3A). The absence of the DDX10-NUP98 transcript in patient 3 suggested that the DDX10-NUP98 fusion gene might not be involved in the pathogenesis of inv(11) malignancy. In both NUP98-DDX10 and DDX10-NUP98 fusion messages, two PCR products of different sizes were generated as shown in Fig 3A. In the case of NUP98-DDX10, 373-bp and 505-bp products were generated using primers NF2 and DR27. In the case of DDX10-NUP98, 333-bp and 465-bp products were generated using DF26 and NR17. Sequence analysis of these PCR products showed that in both NUP98-DDX10 and DDX10-NUP98,DDX10 and NUP98 were fused in-frame to produce chimeric proteins (Fig 3B, and the sequence data have been registered to DDBJ/EMBL/GenBank as AB000267, AB000268, AB001342, and AB001343). Thus, the NUP98-DDX10 fusion transcripts are presumed to encode two types of chimeric protein (Fig 4). As shown in Figs 1B, 1C, and 4, exon B of NUP98 was fused in-frame to exon 6 of DDX10 in patient 1. On the other hand, exon D of NUP98 was fused in-frame to exon 7 of DDX10 in the other three patients. The breakpoint of NUP98 in patient 1 is the same as those in t(7; 11) AML.20 21
RT-PCR analysis of the DDX10-NUP98 and NUP98-DDX10 chimeric transcripts in four inv(11) patients with de novo or secondary myeloid malignancies. (A) Detection of RT-PCR products by electrophoresis. The DDX10-NUP98 and NUP98-DDX10 chimeric transcripts were amplified with primers DF26 and NR17, and primers NF2 and DR27, respectively (see Fig 1C and D). Pt1, Pt2, Pt3, Pt4: RT-PCR amplified patient RNA samples. Pt1 (−), Pt2 (−): patient 1 and 2 PCR product without reverse transcription. Ct: RT-PCR amplified normal peripheral blood RNA. Arrows indicate the detected fusion transcripts. (B) Sequences of the junctions of the NUP98-DDX10 chimeric transcripts (DDBJ/EMBL/GenBank accession nos. AB000267 and AB000268). Two RT-PCR products (c and d) were sequenced. Nucleotide sequences and predicted amino acid sequences around the junctions are shown. The vertical arrows indicate the junction points of the two genes.
RT-PCR analysis of the DDX10-NUP98 and NUP98-DDX10 chimeric transcripts in four inv(11) patients with de novo or secondary myeloid malignancies. (A) Detection of RT-PCR products by electrophoresis. The DDX10-NUP98 and NUP98-DDX10 chimeric transcripts were amplified with primers DF26 and NR17, and primers NF2 and DR27, respectively (see Fig 1C and D). Pt1, Pt2, Pt3, Pt4: RT-PCR amplified patient RNA samples. Pt1 (−), Pt2 (−): patient 1 and 2 PCR product without reverse transcription. Ct: RT-PCR amplified normal peripheral blood RNA. Arrows indicate the detected fusion transcripts. (B) Sequences of the junctions of the NUP98-DDX10 chimeric transcripts (DDBJ/EMBL/GenBank accession nos. AB000267 and AB000268). Two RT-PCR products (c and d) were sequenced. Nucleotide sequences and predicted amino acid sequences around the junctions are shown. The vertical arrows indicate the junction points of the two genes.
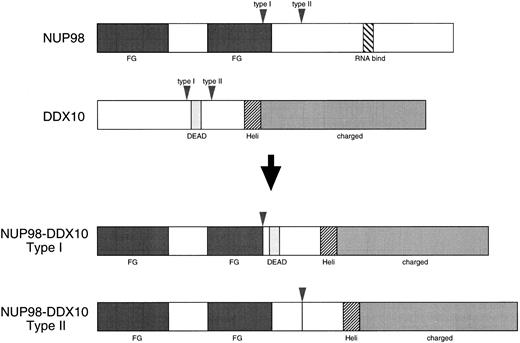
Schematic diagram of the wild-type proteins and deduced chimeric proteins. Vertical arrowheads indicate breakpoints of both DDX10 and NUP98. Regions that correspond to the structural or functional domains of the proteins are also shown: DEAD, DEAD motif; Heli, putative RNA helicase domain; charged, charged amino acids motif; FG, FG peptide repeat motif; RNA bind, putative RNA binding domain.
Schematic diagram of the wild-type proteins and deduced chimeric proteins. Vertical arrowheads indicate breakpoints of both DDX10 and NUP98. Regions that correspond to the structural or functional domains of the proteins are also shown: DEAD, DEAD motif; Heli, putative RNA helicase domain; charged, charged amino acids motif; FG, FG peptide repeat motif; RNA bind, putative RNA binding domain.
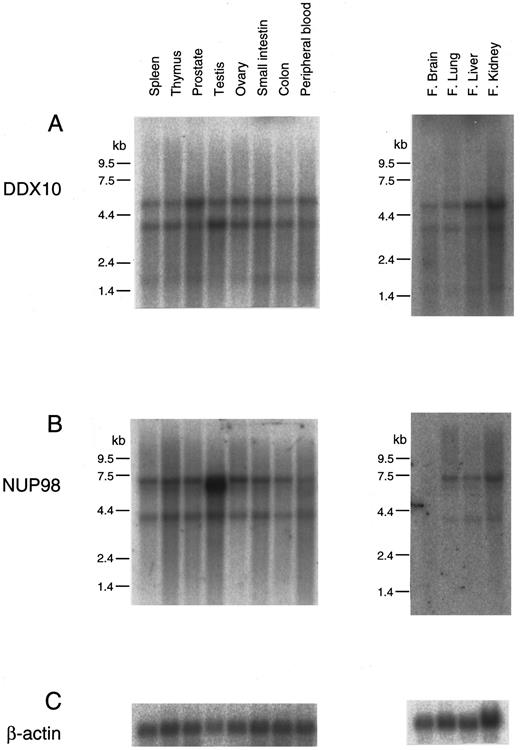
The expression of DDX10 and NUP98 was assessed in adult and fetal tissues. Both genes were ubiquitously expressed in all the tissues examined (Fig 5). DDX10 expressed 3.2-kb and 5.0-kb transcripts, and NUP98 expressed 3.6-kb and 7.0-kb transcripts. In adult testes, the 6.5-kb form of NUP98 was additionally expressed at a high level. NUP98 was rarely expressed in fetal brain. The structural features of the NUP98-DDX10 fusion protein are displayed in Fig 4. In all the four patients, the N terminal portion containing the FG peptide repeat motif of NUP98 was fused to the C terminal portion of DDX10. Both type I and II fusion proteins possessed the RNA helicase domain29 HRAGRTAR, but type I fusion protein did not retain the DEAD motif.30
Northern blot analysis of the expression of DDX10 and NUP98 genes in human tissues. Northern blot filters from human adult and fetal tissues (Clontech) were used for analysis. Probes: (A), DX-FRG116 (nucleotides 21 to 1347 of DDX10 cDNA18 ); (B), NP-FRG123 (nucleotides 93 to 2655 of NUP98 cDNA21 ); (C), β-actin. kb, kilobases.
Northern blot analysis of the expression of DDX10 and NUP98 genes in human tissues. Northern blot filters from human adult and fetal tissues (Clontech) were used for analysis. Probes: (A), DX-FRG116 (nucleotides 21 to 1347 of DDX10 cDNA18 ); (B), NP-FRG123 (nucleotides 93 to 2655 of NUP98 cDNA21 ); (C), β-actin. kb, kilobases.
DISCUSSION
In this study we analyzed four patients with malignant myeloid disease that had the associated inv(11)(p15q22) chromosome translocation. Two had t-AML and t-MDS, after receiving chemotherapy for the primary neoplasm which included treatment with topo II inhibitors, such as etoposide. The other two had de novo AML or MDS. The results showed that the translocation breakpoints in the four patients are located in the DDX10 gene on chromosome 11q22 and the NUP98 gene on chromosome 11p15. As a result of this translocation, the DDX10 gene is fused with the NUP98 gene to form two types of chimeric genes, DDX10-NUP98 and NUP98-DDX10. Expression of both DDX10 and NUP98 is ubiquitous in fetal and adult tissues. Both types of fusion genes were expected to be expressed in leukemic cells obtained from inv(11) patients. However, as indicated in Fig 3A, only the NUP98-DDX10 fusion transcript was seen in patient 3. Using FISH analysis, we previously reported that the genomic region covered by P84 was deleted in the inv(11) chromosome of patient 3 and that P90 hybridized only to the short arm of this inv(11) chromosome.17 This suggests that a region containing the promotor of DDX10 was deleted and that DDX10-NUP98 fusion transcript is not seen in patient 3 because it is not expressed.
Formerly, another nucleoporin gene NUP214/CAN was identified as being implicated in AML associated with t(6; 9)(p23; q34).31 In that case, the t(6; 9) translocation produced a DEK-NUP214/CAN fusion protein consisting of most of the entire DEK polypeptide and the C-terminal portion of NUP214/CAN. The t(7; 11) translocation generates a fusion protein containing the N-terminal portion of NUP98 and most of the coding region of HOXA9.20,21 In both translocations, the chimeric products containing the FG peptide repeat nucleoporin motif, the C-terminal portion of NUP214, and the N-terminal portion of NUP98 were thought to be implicated in leukemogenesis.20,21 32 Both these results and ours relating to patient 3 suggest that the NUP98-DDX10 chimeric product may function in inv(11) malignancies.
The DDX10 gene at 11q22 was recently identified by cDNA screening within the A-T (ataxia telangiectasia) locus.18 Although it encodes a putative adenosine triphosphate (ATP)-dependent DEAD box RNA helicase, its normal function has not yet been identified. The DEAD peptide motif30 is implicated in the binding of Mg2+, which is required for the hydrolysis of ATP by both RNA and DNA helicase.33-35 Many kinds of DEAD box RNA helicases have been found in prokaryotes and eukaryotes. The proteins have been shown to be involved in many aspects of RNA metabolism, including splicing, translation, and ribosome assembly. In Saccharomyces cerevisiae, genetic and biochemical studies have shown that some of the DEAD box proteins can be divided into subgroups: PRP2,36 PRP5,37 PRP16,38 PRP22,39 and PRP2840 for pre-mRNA processing; TIF141 and TIF241 for translation initiation; and SPB442 and DRS143 for ribosome assembly. DDX10 shows high similarity to yeast SPB4 and DRS1, which have been shown to be involved in the processing of yeast 27S into mature 25S rRNA.18 Ribosome biogenesis occurs primarily in the nucleolus of eukaryotes, where rRNA is transcribed, processed, and associated with ribosomal proteins imported from the cytoplasm.44 In the spb4 and drs1 mutant, ribosome subunit assembly is defective as a result of inhibition of ribosomal RNA processing.42,43 One of the nuclear localization signals (NLS) in humans has been identified as consisting of two interdependent basic domains.45 In the DDX10 protein, we found two portions which potentially function as NLSs (amino acids 24-39; KKKHSHRQNKKKQLRK and amino acids 665-680; KKKMTKVAEAKKVMKR). Hence, DDX10 may normally be transported into the nucleus and function therein. The ubiquitous expression and the putative nuclear localization of DDX10 are in agreement with a possible role in ribosome assembly through rRNA processing.
The NUP98 gene on 11p15 was initially identified by cloning the t(7; 11)(p15; p15) translocation breakpoint associated with AML.20,21 It encodes a member of the nucleoporin family.19 Nucleoporins are major constitutes of the nuclear pore complex (NPC) in the nuclear envelope.46 Active transport of proteins and RNAs between the nucleus and cytoplasm proceeds via the NPC.46,47 The FG peptide repeat motif of NUP98 and NUP214/CAN function as the docking sites for protein import at the nuclear pore in vitro.19,48 In nuclear import, NLS containing proteins bind to the nuclear envelope via a nuclear import receptor, the karyopherin α-β heterodimer. This NLS protein-receptor complex docks to the NPC via karyopherin β and is subsequently translocated from the cytoplasm to the nucleoplasm through the pore by an energy-dependent mechanism.46,47 By immunogold electron microscopy, NUP98 and NUP214/CAN have been localized to the nucleoplasmic side and the cytoplasmic side of the NPC, respectively.19,49 The FG repeat motif of the nucleoporins has been suggested to form an array of sites to mediate the docking of transport proteins, and to proceed transport by repeated docking and undocking reactions.19 46
In the chromosomal translocations associated with myeloid lineage malignancies, at least one of the rearranged genes is ordinarily a transcription factor.2,3NUP98-DDX10 may be a new type of fusion gene associated with myeloid malignancies. One of the specific structural features of DDX10 is two acidic regions and very highly charged amino acids at the C-terminus. The acidic motif could serve as a transcription activation domain like those of VP16 and GAL4.50 Therefore, there exists the possibility that DDX10 may be a transcriptional activation factor. However, the information on normal functions of nucleoporins and putative functions of DDX10 suggest that the fusion gene might interfere with the synthesis of protein(s) involved in normal process of myeloid lineage differentiation in hematopoiesis by aberrant necleoplasmic transport of mRNA or alteration of ribosome assembly.
ACKNOWLEDGMENT
We are grateful to Dr H. Eguchi of Kurume University and Dr H. Nakadate of Sapporo National Hospital for providing patient samples; Dr M. Oshimura of Tottori University for providing the cell line A9neo11; and Dr H. Ichikawa for critical reading of the manuscript. We also thank M. Fukushima, M. Mori, and C. Hatanaka for their technical assistance.
Supported in part by a Grant-in-Aid for the special promotion of Basic Medical Science from the Drug Organization; a grant from the Special Coordination Funds for the Promotion of Science and Technology from the Science and Technology Agency; and a Grant-in-Aid for the Second Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare of Japan.
Address reprint requests to Yasuhito Arai, MS, Radiobiology Division, National Cancer Center Research Institute, 5-1-1, Tsukiji, Chuo-ku, Tokyo 104, Japan.