Abstract
The human gene for γ-glutamyl carboxylase is 13 kb in length and contains 15 exons. Transcription starts at a cytosine 217 base pair upstream of the first codon. There are two major transcripts in all tissues examined. They are distinguished by the presence of an Alu sequence in the 3′ nontranslated end of the longer species. Relative mRNA levels for 12 bovine tissues are presented.
γ-GLUTAMYL carboxylation, accomplished by the integral membrane microsomal enzyme γ-glutamyl carboxylase, is a posttranslational modification essential for the biological activities of the vitamin K-dependent proteins. These include coagulation factors II, VII, IX, X, and proteins C and S. In addition, osteocalcin, matrix Gla protein1 and a recently discovered growth arrest specific protein, gas6,2 are carboxylated. The recognition of the vitamin K-dependent proteins by the γ-glutamyl carboxylase is dependent on a short peptide sequence.3 With the exception of the matrix Gla protein, where the recognition sequence resides within the mature protein, this recognition sequence is an approximately 18 amino acid propeptide, which is removed before secretion.
In addition to its protein substrates, the vitamin K-dependent carboxylase requires oxygen, carbon dioxide, and the cofactor vitamin K hydroquinone to accomplish carboxylation. Concomitant with the conversion of each glutamate residue to a Gla residue a vitamin K hydroquinone is oxidized to its vitamin K 2,3-epoxide. To accomplish complete carboxylation at the normally low physiological concentration of vitamin K, it is necessary to continuously regenerate vitamin K hydroquinone from vitamin K 2,3-epoxide by the DTT-dependent, warfarin sensitive, microsomal enzyme, vitamin K epoxide reductase. However, conversion of vitamin K to vitamin K hydroquinone can also be accomplished by a warfarin insensitive, NADPH dependent-vitamin K reductase.1 4 Although incapable of using vitamin K epoxide, the warfarin insensitive vitamin K reductase can be used, at least in liver, to generate reduced vitamin K from administered vitamin K in cases of epoxide reductase deficiencies or of warfarin poisoning.
Because vitamin K deficiency was first associated with a bleeding disorder and because synthesis of coagulation proteins occurs predominantly in the liver, liver has been considered the primary organ where carboxylation occurs. However, as the number of Gla-containing proteins continues to grow, it is becoming apparent that carboxylase is present in most tissues.
We have previously published the cDNA sequences for the human and most of the bovine carboxylase cDNA.5 In addition, we have mapped the chromosomal location of the gene for the human carboxylase to chromosome 2 at position p12.6
It is important to characterize the entire carboxylase gene for two reasons. The first is to facilitate the genetic analysis of the combined vitamin K-dependent coagulation protein deficiencies, which could result from loss of either carboxylase or epoxide reductase activities. The second is to allow studies to be done to understand the regulation of carboxylase synthesis in different tissues. In this paper we report the complete genomic sequence of the human carboxylase, its transcription start site, the relative level of carboxylase mRNA in 12 human tissues and the characterization of two major transcripts, which appear in all tissues examined. In addition, we define 4 polymorphisms within the human γ-glutamyl carboxylase gene.
MATERIALS AND METHODS
Isolation of the Human γ-Glutamyl Carboxylase Genomic DNA
A human cosmid genomic library from Stratagene (La Jolla, CA) was used for isolation of the carboxylase genomic DNA. The complete human γ-carboxylase cDNA5 was used as a probe to screen the library. Two independent clones of about 38 kb were identified; each contained the complete carboxylase gene.
Transcription Start Site
RNA preparation.Total human liver RNA was isolated using TRIZOL reagent (BRL). Liver poly(A)+ RNA was prepared using a PolyATract mRNA isolation system II from Promega (Madison, WI).
Primer extension assay. Primer extension was performed using two different primers. One, a 22-mer, was complementary to residues 753-774 and the other, a 21 mer was complementary to residues 695-715 (see Fig 1). Each primer was end labeled with 32P using the enzyme T4 polynucleotide kinase (New England Biolabs, Beverly, MA) and purified by centrifuging through a G-25 quick spin column (Boehringer, Indianapolis, IN). About 0.1 μg (1 × 106 counts per minute) primer was added to 0.5 μg human liver poly(A)+ RNA and annealed at 65°C for 90 minutes in 150 mmol/L KCl, 10 mmol/L Tris-HCl (pH 8.3) and 1 mmol/L EDTA (pH 8.0). Primer extension was accomplished with 200 U of Superscript II (BRL) at 45°C for 60 minutes. The reaction was stopped by adding RNase A (20 μg/mL) and salmon sperm DNA (100 μg/mL) as a carrier. The reaction mixture was incubated at 37°C for an additional 15 minutes. The product was then phenol-chloroform extracted, precipitated with 2 volumes of ethanol, and washed with 70% ethanol. The pellet was resuspended in 80% formamide, 1 mmol/L EDTA (pH 8.0), 0.1% bromophenol blue, 0.1% xylene cyanol and analyzed on a 6% polyacrylamide sequencing gel. A reference sequence was generated using the same primer and a template containing the genomic sequence of the γ-carboxylase gene.
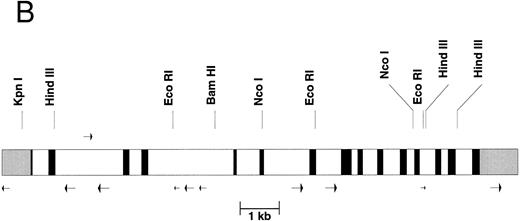
The gene organization of the γ-glutamyl carboxylase. (A) The nucleotide numbers are based on the numbers submitted to Genbank. The transcription initiation site at nucleotide number 515 is labeled with an overhead “➭”. The coding sequence is in uppercase boldfaced. The 5′- and 3′-untranslated regions are in lowercase italics. The intron sequences are shown in lowercase. The potential polyadenylation sites are underlined and capitalized. Each transcription stop site (identified by cDNA cloning) is double underlined and capitalized. (B) The intron-exon organization is shown. 5′ and 3′ untranslated regions are shown in gray scale, the exons are black, and the introns are represented by clear rectangles. Alu sequences are represented by arrows below the figure whereas the mer 20 sequence is represented by an arrow above the figure. Some restriction sites are shown above the figure.
The gene organization of the γ-glutamyl carboxylase. (A) The nucleotide numbers are based on the numbers submitted to Genbank. The transcription initiation site at nucleotide number 515 is labeled with an overhead “➭”. The coding sequence is in uppercase boldfaced. The 5′- and 3′-untranslated regions are in lowercase italics. The intron sequences are shown in lowercase. The potential polyadenylation sites are underlined and capitalized. Each transcription stop site (identified by cDNA cloning) is double underlined and capitalized. (B) The intron-exon organization is shown. 5′ and 3′ untranslated regions are shown in gray scale, the exons are black, and the introns are represented by clear rectangles. Alu sequences are represented by arrows below the figure whereas the mer 20 sequence is represented by an arrow above the figure. Some restriction sites are shown above the figure.
DNA Sequencing
DNA was sequenced at the UNC-CH automated facility on a model 373A DNA sequencer (Applied Biosystems, Foster City, CA) using the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems). Both strands of the intron regions was sequenced and both strands of the exons and exon-intron borders were sequenced at least 5 times.
Northern Blot Analysis
Northern blots (Human Multiple Tissue Northern Blot and Human Endocrine System Multiple Tissue Northern Blot) were purchased from Clontec Laboratories Inc (Palo Alto, CA). The probe was prepared by random priming using the carboxylase cDNA.5 An internal standard of β-actin was used with Clontech's ExpressHyb solution according to the manufacturer's protocol. The results were imaged with Molecular Dynamics Storm 840 Phosphorimager (Sunnyvale, CA) and quantitation was achieved with ImageQuant software.
Analysis of Polymorphisms
For all polymerase chain reaction (PCR) analyses the following buffer was used: 2.5 mmol/L MgCl2 , 10 mmol/L Tris-HCl, 50 mmol/L KCL (pH 8.3). All reactions were amplified for 30 cycles but the temperature and times varied slightly for each analysis and are given for each of the 4 described polymorphisms
(CAA)n repeat in intron 6.Nested PCR analysis was used. For the first PCR we used the following pair of oligonucleotides: 5′ TGTAACTCAGGAGCATGGATT C and 5′ TGGCTAGTCCCTTCCTGCAAAACTG. The PCR conditions were: 94°C, 1 minute; 57°C, 40 seconds; 72°C, 3 minutes. The first 3,099 base pairs (bp) amplified fragment was used as a template for a second PCR analysis using the following pair of oligonucleotides: 5′ 32P GCCCAGGAGTTTAGCTAC and 5′ ATCACAGCACGAATGTGCTT. The PCR conditions were: 94°C, 1 minute; 56°C, 40 seconds; 72°C, 1 minute. The 5 amplified DNA fragments, which ranged in size from 180 to 192 bp were analyzed by denaturing gel electrophoresis.
EcoR1 polymorphism.The oligonucleotide pair 5′ CCCCTCAATGTTTACCT and 5′ CAGCTTTTCAGCATTGGT were used to amplify a 2,142-bp PCR fragment including exons 5, 6, and 7. The PCR conditions were: 94°C, 1 minute; 49°C, 40 seconds; 72°C, 3 minutes. The amplified fragment was subjected to EcoRI digestion and the fragments were analyzed in 0.8% agarose gels.
Coding sequence polymorphism.The following pairs of oligonucleotides were used to amplify a 252-bp fragment that introduces a Taq 1 site by primer template mismatch in the G allele but not the A allele: 5′ AGCTGGTGTCCTACTGCCCTC and 5′ CGTTCCTTTCTAAGTCCAG. The PCR conditions were: 94°C, 1 minute; 57°C, 40 seconds; 72°C, 3 minutes. The amplified fragment was digested with Taq 1 and analyzed on 4% agarose gels.
Silent polymorphism in exon 9 of the coding region.The following oligonucleotides were used to generate a 90-bp fragment with a primer/template mismatch that introduces an Alu I site in the C but not the T allele: 5′ GGACATGATGGTGCACTCCAG and 5′GGCTACCTTAACCCTGGG. The PCR conditions were: 94°C, 1 minute; 57°C, 40 seconds; 72°C, 3 minutes. The amplified fragment was subjected to Alu I digestion and the resulting fragments were analyzed on 4% agarose.
RESULTS
DNA Sequence
It was necessary to determine the genomic sequence of the normal carboxylase to compare it to the genomic sequence of patients with a suspected defect in the carboxylase gene. Therefore, we have determined the entire 13-kb sequence of the human γ-glutamyl carboxylase. Part of the sequence and the intron-exon boundaries are shown in Fig 1A while the overall exon structure is shown in Fig 1B. The complete genomic sequence is available under Genbank accession number U65896. The carboxylase gene has 15 exons, and each exon has a canonical splicing sequence at its boundary. Within the introns of the gene there are 10 Alu sequences, represented by arrows under Fig 1B, and one middle repetitive mer 20 sequence, represented by an arrow above Fig 1B. Mer 20 is a middle repetitive element, which is found in the haploid human genome with a frequency of 200 to 400 copies.7 The Alu sequences include both the Alu J and S types with the Alu S being divided into Sq, Sx, and Sp subtypes.8 The density of Alu repeats, one per 1.3 kb, is about the same as has been reported for other regions of the human genome sequence.9
Interestingly, a gene for methionine adenosyltransferase, which is localized to chromosome 2 region p11.2,10 is found in the same cosmid that contains the complete carboxylase gene. This gene is found at a distance of 6 kb from the carboxylase gene but is transcribed in the opposite direction.
Transcription Start Site
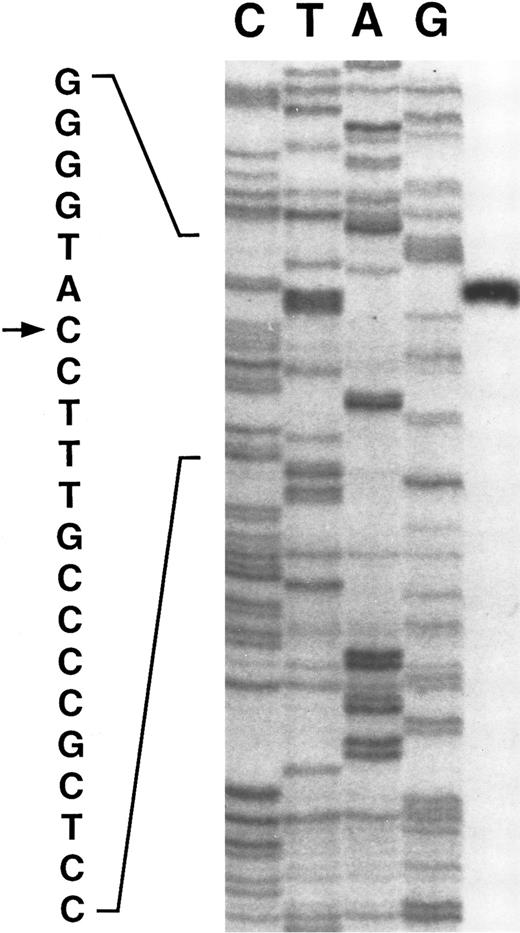
Primer extension of poly A+ RNA isolated from human liver indicates a single transcription start site at a C residue 217 bases upstream of the first codon, residue 515 on Fig 1 (see Fig 2). The same site is found with either of two primers. Although most mRNAs initiate at an A or G, initiation at C is not uncommon.11
The transcription initiation site (indicated by →) for the γ-glutamyl carboxylase determined by primer extension. The control sequence on the left panel was determined using a genomic clone and primer 1 (residues 753-774 of Fig 1).
The transcription initiation site (indicated by →) for the γ-glutamyl carboxylase determined by primer extension. The control sequence on the left panel was determined using a genomic clone and primer 1 (residues 753-774 of Fig 1).
Characterization of Transcripts
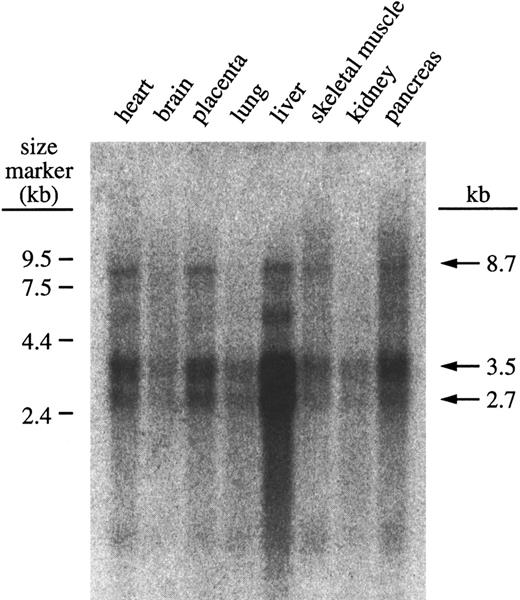
Northern blotting experiments revealed that there are 2 predominant carboxylase mRNA transcripts, one of about 2.7 kb and the other of about 3.6 kb (Fig 3). These 2 mRNA species are found both in human and bovine tissues in all organs that have detectable levels of carboxylase mRNA. These two species of mRNA probably arise from termination at each of the two consensus poly A+ addition signals shown in Fig 1. The second consensus sequence is actually repeated three times in an overlapping pattern. In addition to these consensus poly A+ addition signals there is an ATTAAA sequence beginning at nucleotide 13085, which is the most common alternative poly A+ addition signal.12 13 We have examined more than 30 cDNA clones and there are clearly mRNAs that terminate at a position consistent with this alternative signal being functional. The difference between the two major forms of the carboxylase mRNA is that the longer has an Alu Sx sequence interposed between the first and second consensus poly A addition sequences.
Representative Northern blot probed with γ-glutamyl carboxylase cDNA show mRNA present in 8 human tissues. The probe was prepared by random priming from the carboxylase cDNA. Hybridization was with Clontech's ExpressHyb solution according to the manufacturer's protocol. The results were imaged with Molecular Dynamics Storm 840 Phosphorimager and quantitation was achieved with ImageQuant software.
Representative Northern blot probed with γ-glutamyl carboxylase cDNA show mRNA present in 8 human tissues. The probe was prepared by random priming from the carboxylase cDNA. Hybridization was with Clontech's ExpressHyb solution according to the manufacturer's protocol. The results were imaged with Molecular Dynamics Storm 840 Phosphorimager and quantitation was achieved with ImageQuant software.
The significance of the Alu sequences in the 3′ end of the longer transcript is still unknown. However, there are numerous reports of Alu sequences affecting gene expression by different mechanisms. For example, Almenoff et al14 have shown that a transcript of an Alu Sx sequence can induce the expression of a receptor for bacterial heat stable enterotoxins in intestinal cell lines.
There is also evidence that an Alu sequence can function as a binding site for a retinoic acid receptor and increase transcription up to 35-fold.15 Because Alu sequences can have a number of different effects on gene expression, it is probably important to further characterize the longer Alu containing carboxylase transcript by expressing it in cells producing factor IX to determine if it has a role in increasing carboxylation.
The relative amounts of carboxylase mRNA from 12 tissues were examined. As expected, it was found that the level of carboxylase mRNA was highest in the liver where the vitamin K-dependent coagulation factors are synthesized. All tissues examined contain detectable amounts of carboxylase mRNA but none contained more than 20% of the levels found in liver. Figure 4 shows the mRNA levels relative to liver in brain, placenta, mammary gland, thyroid gland, and adrenal gland.
Northern blot analysis of various human organs probed with γ-glutamyl carboxylase cDNA. mRNA levels are shown as percentages relative to the mRNA level in liver. Quantitiation was performed with the ImageQuant software.
Northern blot analysis of various human organs probed with γ-glutamyl carboxylase cDNA. mRNA levels are shown as percentages relative to the mRNA level in liver. Quantitiation was performed with the ImageQuant software.
Polymorphisms
Microsatellite.A trinucleotide repeat sequence (CAA)n was identified in intron 6 of the carboxylase gene. This microsatellite can be analyzed by PCR amplification as described in Materials and Methods. Five different alleles of 192, 189, 186, 183, and 180 bp were observed in 132 unrelated normal chromosomes. The heterozygosity was 0.63 with 44.5% having the (CAA)9, 12.1% the (CAA)10, 22% the (CAA)11, 19.7% the (CAA)12, and 1.3% the (CAA)13 genotype.
EcoRI polymorphism.An EcoRI polymorphism was identified at intron 5, 230 bp from the end of exon 5 where a nucleotide substitution of C for T generates an EcoRI restriction site. Alleles of 124 unrelated normal chromosomes were analyzed. The heterozygosity was 0.49 and 70% of the individuals examined have an EcoR1 site in one or both alleles.
Coding sequence polymorphism.This coding sequence polymorphism is produced by a G to A transition at nucleotide 8762. The consequence of this polymorphism is the substitution of Gln for Arg at position 325. Forty-two percent of the individuals examined were heterozygous at this position and 94% had arginine at one or both alleles.
Silent Polymorphism in Exon 9 Coding Region
There is a silent polymorphism within the coding region created by a T to C transition at nucleotide 9167. The observed heterozygosity was 0.37 and 92% of the individuals tested (124) had the C allele in one or both chromosomes.
We used the information obtained from our genomic sequence analysis to design primers for sequencing the carboxylase genes in 5 patients who had combined factor II, VII, IX, and X deficiencies. Clinically, it is difficult to predict whether patients with combined vitamin K-dependent coagulation deficiencies bear mutations in the carboxylase gene or the vitamin K epoxide reductase gene. Of the 5 patients we examined, only one had a naturally-occurring mutation in the carboxylase gene (in preparation). It is likely that the others have mutations in their epoxide reductase gene. However, other unknown genes may also be essential for carboxylation.
The data presented in this report should allow the facile examination of presumed carboxylase deficient patients. It also presents the background needed for analysis of the transcriptional control of this essential gene.
Supported by National Institutes of Health Grants No. HL48318 to D.W.S. and HL48322 to K.A.H.
Address reprint requests to Darrel W. Stafford, PhD, Department of Biology, University of North Carolina-Chapel Hill, Chapel Hill, NC 27599-3280.