Abstract
B-cell chronic lymphocytic leukemia (B-CLL) is a slowly progressive disease characterized by the clonal expansion of CD5+/CD23+ B lymphocytes. The malignant transformation is assumed to occur at the level of mature B lymphocytes. We asked whether CD34+ progenitor cells are involved in the malignant process in B-CLL. Furthermore, we investigated the possibility of aberrant CD34 expression by the malignant B-cell clone. Bone marrow and peripheral blood samples from 75 patients with B-CLL were tested for the presence of trisomy 12 and deletion of the retinoblastoma gene (Rb) by fluorescence in situ hybridization. CD34+ subpopulations were isolated by fluorescence-activated cell sorting and analyzed for the presence of the informative genetic marker. Bone marrow and peripheral blood samples of 10 B-CLL patients were analyzed for coexpression of CD34/CD5/CD20. Trisomy 12 was detected in 15 of 75 (20%) and Rb-deletion was detected in 6 of 30 patients (20%). In 7 patients with trisomy 12, hematopoietic progenitor cells were sorted, with the sort purity being between 85% and 99.8%. The genetic marker was detected in the CD34+/CD38+ cells as well as in the CD34+/38− subsets in 3 patients. Progenitor cells were also sorted in 2 patients with Rb-deletion. In 1 patient, Rb-deletion was present in 10% of CD34+/38+ cells. In the other patient, Rb-deletion was neither detected in the CD34+/38+ nor in the CD34+/CD38− subsets. In all 10 patients investigated for coexpression of CD34/CD5/CD20, we could not find a subpopulation coexpressing these markers. We conclude that trisomy 12 and Rb-deletion are present in a considerable subset of patients with B-CLL. In part of these patients, the genetic marker was detected at the level of CD34+ stem cells. CD34 expression is not related to an aberrant phenotype of the malignant B-cell clone. These results suggest that the malignant transformation in B-CLL may involve early hematopoietic stem cells and place a note of caution on future strategies using autologous stem cell transplantation.
B-CELL CHRONIC lymphocytic leukemia (B-CLL) is generally believed to result from the clonal expansion of mature B cells. B-CLL cells are immunophenotypically characterized by the coexpression of CD19, CD22, and CD23; low expression of CD20; absence or low expression of surface Ig; and high expression of CD5.1,2 CD5 is found on all T cells, but is absent from most B cells from adult blood and tissues, with the exception of a minority of B cells localized in the follicles of lymphoid tissues.3 These normal CD5+ B cells (B1 cells)4 share a number of cellular and molecular properties with B-CLL cells, such as production of monoclonal polyreactive IgM autoantibodies in the absence of somatic hypermutations,5 expression of cross-reactive idiotypes,6 and the expression of myelomonocytic antigens.7 Based on these observations, B1 cells have been proposed as the normal counterpart of B-CLL cells and as the cells from which the disease originates.2 In contrast, Estrov et al8 were able to generate B-lymphoid colonies in vitro from CD34+ progenitor cells. This observation points to pluripotent stem cells as the target of malignant transformation and places B-CLL in the same group with other hematological malignancies, including acute myeloid leukemia,9-11 chronic myeloid leukemia,12 and myelodysplastic syndrome.10 13
Normal hematopoietic stem cells are immunophenotypically characterized by the expression of CD34, a 100- to 115-kD cell surface molecule. During early differentiation, CD34+ cells sequentially acquire lineage-nonrestricted antigens such as CD38, HLA-DR14-17 and more lineage-restricted antigens such as stem cell factor receptor (SCF-R; CD117)18 and CD19.19 Operationally, the most primitive CD34+/CD38−/HLA-DR− cells have been defined by their ability to form mixed colonies,20,21 to induce long-term bone marrow culture,22,23 and to reconstitute hematopoiesis in animal models.24 25
In the current study, we asked whether in B-CLL CD34+ progenitor cells are also involved in the malignant process. Aberrant expression of CD34 on the malignant clone is well known from acute myeloid leukemia.26 To exclude the possibility of aberrant coexpression of CD34 on the malignant B-cell clone, we have analyzed patients with B-CLL for aberrant expression of progenitor cell markers. To test the hypothesis of progenitor cell involvement in B-CLL, we selected genetic markers and decided to use trisomy 12 and deletion of the retinoblastoma gene (Rb) on chromosome 13q14, which comprise the most frequent cytogenetic abnormalities in B-CLL. Trisomy 12 occurs with an incidence of 15% to 35%27-32 and abnormalities of 13q14 in 16% to 31%.33,34 They are more frequently detected when fluorescence in situ hybridization (FISH) is performed on interphase cells using either α-centromeric probe specific for chromosome 12 or a probe for the retinoblastoma gene.29,35 This higher incidence is due to the low numbers of metaphases of the malignant B-cell clone.36 Studies on the correlation between FISH data and laboratory features and clinical course have found an association of trisomy 12 with atypical lymphocyte morphology, advanced disease, and an aggressive clinical course, whereas deletions of 13q14 are common in stage A disease, are associated with typical morphology, and do not carry an adverse prognostic significance.27,33 37 In the present study, we have investigated peripheral blood and bone marrow samples from patients with B-CLL for the presence of trisomy 12 and Rb-deletion using FISH. In patients with trisomy 12 or Rb-deletion, highly purified CD34+ subpopulations were isolated by fluorescence-activated cell sorting (FACS) and analyzed by FISH for the presence of the informative genetic marker.
PATIENTS AND METHODS
Patients.Diagnostic and follow-up bone marrow or peripheral blood samples of 75 unselected patients with B-CLL were used for interphase FISH analysis and sorting procedure. Patients were admitted between January 1992 and October 1996 to the Department of Hematology and Oncology of the University of Göttingen, Germany. Forty-seven patients were men and 28 were women, with a median age of 63 years. Diagnosis of CLL was based on light microscopy of Pappenheim-stained slides and on immunophenotypical expression of CD19, CD20, and CD23 together with CD5 and weak or absent surface Ig.
FACS.For isolation of subpopulations by cell sorting, fresh and cryopreserved bone marrow aspirates or peripheral blood samples were used. Mononuclear cells were isolated by Ficoll gradient centrifugation, washed twice in RPMI 1640 (GIBCO, Karlsruhe, Germany) and 20% fetal calf serum (Hyclone, Logan, UT), and counted. Cells (2 × 107) were double-stained with anti-CD34 directly conjugated to fluorescein isothiocyanate (FITC; HPCA-2; Becton Dickinson Immunocytometry Systems [BDIS], San José, CA) and phycoerythrin (PE)-conjugated anti-CD38 (Leu 17; BDIS) monoclonal antibodies. All steps were performed under sterile conditions. Analysis and isolation were performed with an FACS (FACS Vantage; BDIS) equipped with a 488-nm laser using phosphate-buffered saline as sheath fluid. Cells were sorted directly on slides as described previously.38 Two different subsets were defined on the distinction of different maturational subgroups and in agreement with characteristic light scatter properties defined by low forward and low to intermediate side scatter profile: CD34+/38+ and CD34+/38−. Sort purity was determined by immunophenotypical reanalysis of sorted subpopulations by using a FACScan (BDIS). In samples with small numbers of CD34+ cells, analysis of sort purity was performed on CD38+ subpopulations.
FISH.Sorted as well as unsorted cells were fixed in methanol/acetic acid 3:1 and air-dried. The slides were then denatured in 70% formamide 2× SSC solution at 72°C for 2 minutes. After this, they were dehydrated for 2 minutes in 70%, 85%, and 100% ethanol and air-dried. One microliter of an α-satellite DNA-probe D12Z3 for chromosome 12, or 1 μL of a locus-specific DNA-probe for 13q14, directly conjugated to spectrum orange (Vysis, Stuttgart, Germany) was diluted in 7 μL of hybridization mixture (Vysis) and 2 μL ddW, then denatured at 75°C for 5 minutes and placed on ice. Directly before hybridization, slides were prewarmed at 45°C for 2 minutes and DNA-probe mixture was incubated at 37°C for 20 minutes. The hybridization mixture (10 μL) was dropped on the slide, cover-slipped, sealed with rubber cement, and incubated at 37°C overnight. The slides were washed in 42°C 50% formamide in 2× SSC for 30 minutes, followed by 10 minutes of washing in 47°C 2× SSC and 5 minutes of washing in 47°C 2× SSC/0.1% NP-40. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and signals were visualized using a Zeiss Axiophot dual filter fluorescence microscope (Zeiss, Göttingen, Germany) under 1,000× magnification. Two hundred single interphase cells with well-delineated fluorescent spots were examined. Control slides with mononuclear cells from 10 normal donors were used as negative controls.
Flow cytometry.The cell concentration of bone marrow aspirates and peripheral blood samples of 10 patients with B-CLL was adjusted to 1 × 106 cells/mL with phosphate-buffered saline containing 1% bovine serum albumin (pH 7.3; PBS). Twenty microliters each of monoclonal antibody, CD34 FITC (HPCA-2 FITC; BDIS), CD20 PerCP (L27 PerCP; BDIS), and CD5 PE (L17F12 PE; BDIS) was added to 100 μL of cell suspension. Cells were lysed for 10 minutes at room temerature in FACS lysing solution (BDIS) and centrifuged at 335g for 5 minutes. The pellet was washed twice in PBS and finally resuspended in 0.5% paraformaldehyde. In the control experiments, cells were incubated with fluorescently labeled isotype controls. Instrument set-up included an unstained sample and samples stained with CD3 PE (SK7 PE; BDIS), CD45 FITC (2D1 FITC; BDIS), and CD45 PerCP (2D1 PerCP; BDIS). Flow cytometric analysis was performed on a FACScan (BDIS). Data of 50,000 events were stored in listmode data files and the analysis was performed using the PAINT-A-Gate software (BDIS).
RESULTS
Incidence of trisomy 12 and Rb deletion.In situ hybridization with a DNA-probe for chromosome 12 was performed on specimens of 75 patients. Slides from 10 normal patients served as controls. Three intracellular signals were observed on average in 0.4% (SD, 0% to 0.9%; double SD, 0% to 1.4%). Fifteen patients showed trisomy 12 in 22% to 83% of interphase nuclei (mean, 51%), ie, an incidence of trisomy 12 of 20%. Thirty patients were analyzed for deletion of the Rb gene. In 2.1% (mean) of the control nuclei, only one hybridization signal could be identified (SD, 0.6% to 3.6%; double SD, 0% to 5.1%). Rb deletion was found in 6 patients, ie, an incidence of 20%. The frequency of cells with a single hybridization signal ranged from 12% to 95%, with a mean of 51% (Table 1).
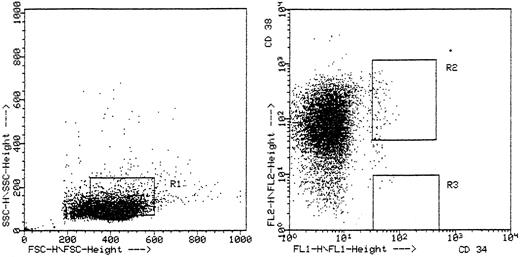
Isolation of CD34+ cells.Progenitor cell compartments were defined by the immunophenotypical expression of CD34 and CD38 and in addition by forward and side scatter profile (Fig 1). In 9 patients we found a sufficient number of CD34+ cells and sorted two different bone marrow progenitor cell compartments: CD34+/CD38− and the more mature CD34+/38− CD34+/38+ progenitor cells. The number of sorted CD34+/38+ cells was between 9 and 500, with a median of 107 cells. In the CD34+/38− subset, it was possible to sort between 5 and 164 cells, resulting in a median of 42 cells. In patient no. 5 it was not possible to detect any CD34+/38− cells, resulting in 8 patients with sufficient numbers of sorted CD34+/38− cells. The sort purity was between 85% and 99.8% (Table 2).
Representative flow cytometric analysis of patient no. 1. Ten thousand cells were aquired in listmode on a FACScan with two gates showing the sorted two different maturational subpopulations: CD34+/38+ cells in fluorescence gate R2 with light scatter gate R1 and CD34+/38− cells in fluorescence gate R3 with light scatter gate R1.
Representative flow cytometric analysis of patient no. 1. Ten thousand cells were aquired in listmode on a FACScan with two gates showing the sorted two different maturational subpopulations: CD34+/38+ cells in fluorescence gate R2 with light scatter gate R1 and CD34+/38− cells in fluorescence gate R3 with light scatter gate R1.
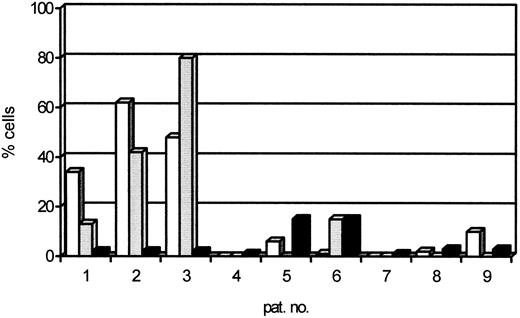
Genetic markers in CD34+ cells.In situ hybridization with a DNA-probe for chromosome 12 was performed on sorted cells of 7 patients. CD34+ cells from 4 normal patients served as controls. Three intracellular signals were observed on average in 0.6% (SD, 0.2% to 1.0%; double SD, 0% to 1.4%). In 3 of 7 patients with trisomy 12, the genetic marker was found in sorted CD34+/38+ cells in 34%, 62%, and 48%, respectively, and in the CD34+/38− compartment in 13%, 42%, and 80% (Table 2 and Figs 2 and 3). These percentages were significantly greater than the percentage of sort unpurity of 2% in these samples. In contrast, in the 4 other patients with trisomy 12, the progenitor cell compartments, both the CD34+/38+ and the CD34+/38− cells, were negative for the genetic marker. Trisomy 12 was found in 0%, 6.3%, 1%, and 0% of CD34+/38+ cells and in 0%, 15%, and 0% of CD34+/38− cells. These percentages were less than the sort unpurity that ranged from 0.02% to 15%.
The percentage of cells within sorted CD34+/38+ (□) and CD34+/38− () fractions that contain the genetic marker detected by FISH. (▪) The percentage of sort unpurity.
The percentage of cells within sorted CD34+/38+ (□) and CD34+/38− () fractions that contain the genetic marker detected by FISH. (▪) The percentage of sort unpurity.
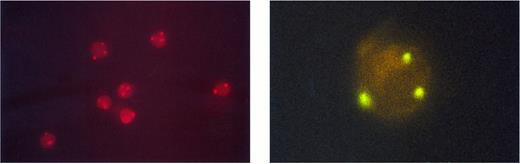
Unsorted peripheral blood cells after FISH with a chromosome 12-specific α satellite DNA probe conjugated to spectrum orange. Three fluorescent hybridization signals indicate trisomy 12; two signals represent diploid cells (left). The right side shows a sorted CD34+/38+ cell hybridized to a spectrum green labeled a chromosome 12-specific α satellite DNA probe. The three hybridization spots indicate trisomy 12 (right).
Unsorted peripheral blood cells after FISH with a chromosome 12-specific α satellite DNA probe conjugated to spectrum orange. Three fluorescent hybridization signals indicate trisomy 12; two signals represent diploid cells (left). The right side shows a sorted CD34+/38+ cell hybridized to a spectrum green labeled a chromosome 12-specific α satellite DNA probe. The three hybridization spots indicate trisomy 12 (right).
Rb-deletions were analyzed in peripheral blood samples of 2 patients. In 2.2% (mean) of the control nuclei, only one hybridization signal could be identified (SD, 0.5% to 3.9%; double SD, 0% to 5.6%). In 1 patient with the genetic marker, no cell with the genetic abnormality was found in neither the CD34+/38+ progenitor cell compartment nor in the more immature CD34+/38− cell subset. In the other patient with Rb deletion, the genetic abnormality was detected in 2 of 20 CD34+/38+ cells and in none of 4 CD34+/38− cells. Sort purity in these two investigations was 97%.
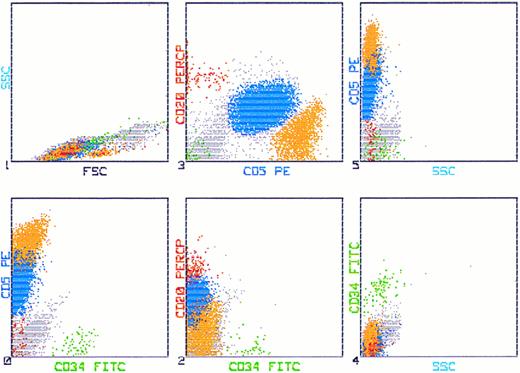
Coexpression of CD34 on CD5+/CD20+ cells.We have investigated 10 patients for aberrant expression of CD34 on CD5+/CD20+ cells. In all investigated samples we did not find a subpopulation coepressing these markers. A typical example of bone marrow cells that express CD5/20 and are negative for CD34 is shown in Fig 4.
Flow cytometric analysis of bone marrow of a CLL patient stained with CD34 FITC (HPCA-2), CD20 PerCP (L27), and CD5 PE (L17F12). Fifty thousand cells were aquired in listmode on a FACScan. The leukemic cells are depicted blue by simultaneous analysis of CD5 PE and CD20 PerCP, whereas the CD34+ cells appear as green. There is no population detectable coexpressing CD34, CD5, and CD20.
Flow cytometric analysis of bone marrow of a CLL patient stained with CD34 FITC (HPCA-2), CD20 PerCP (L27), and CD5 PE (L17F12). Fifty thousand cells were aquired in listmode on a FACScan. The leukemic cells are depicted blue by simultaneous analysis of CD5 PE and CD20 PerCP, whereas the CD34+ cells appear as green. There is no population detectable coexpressing CD34, CD5, and CD20.
DISCUSSION
Immunologic characterization and isolation of hematologic progenitor cells allow the assignment of chromosomal abnormalities to specific progenitor cell populations. Recent studies have shown clonal chromosomal abnormalities in progenitor cells of acute myeloid leukemia10,11 by simultaneous analysis of cell surface immunophenotypic markers and chromosome analysis and in progenitor cells of acute lymphoblastic leukemia by the detection of leukemic clone specific T-cell-receptor delta rearrangements in the CD34+CD38− cells.39 To assess the involvement of early hematopoietic progenitor cells in B-CLL, we have investigated whether genetic markers are detectable in CD34+ subpopulations.
To identify patients with genetic abnormalities, we performed FISH analysis for trisomy 12 and Rb-deletion on bone marrow and peripheral blood samples. In this screening, trisomy 12 and also the Rb-deletion were found each with an incidence of 20%. These data are consistent with reports of other groups who found trisomy 12 in B-CLL with an incidence between 11.5% and 35% by conventional cytogenetics or FISH.27-32 The incidence of Rb-deletion in B-CLL is reported to be between 16% and 31%.33,34 This variable incidence of trisomy 12 and Rb-deletion in B-CLL still remains to be explained. Most probably, heterogeneity of patient population in different series may be responsible. Another explanation might be that patients with splenic lymphoma with villous lymphocytes (SLVL) and lymphoplasmocytic lymphoma were included.29 In our series, only B-CLL cases with typical morphology and immunophenotype were analyzed.40
In patients with trisomy 12 or Rb-deletion, we have investigated progenitor cells for the presence of the genetic marker. Trisomy 12 was found in the CD34+ cells of 3 of 7 patients, both in the 38+ as well as in the 38− compartment. Rb-deletion was not detectable in the progenitor cells of one patient. In contrast, the other one had the genetic marker in the CD34+/38+ compartment but only in a small percentage; the CD34+/38− cells of this patient did not show the Rb-deletion.
Our results suggesting pluripotent stem cells as target of malignant transformation in B-CLL are confirmed by Estrov et al,8 who generated B-lymphoid colonies in vitro from CD34+ progenitor cells. Furthermore, B-CLL cells show germline encoded VH region Ig genes that reflect the absence of germinal center (GC)-based process of V region gene hypermutation and suggest the origination of B-CLL from an immature pre-GC cell.41
Other studies using immunocytochemical techniques (alkaline phosphatase antialkaline phosphatase [APAAP]) or a combining FISH/FACS approach have shown that mature clonal B cells with and without trisomy 12 coexist in some patients with B-CLL.30 42 The demonstration of such mosaicism taken together with our finding of trisomy 12 in hematopoietic progenitor cells suggests that the acquisition of an extra copy of chromosome 12 is an early but not the first event in the pathogenesis of B-CLL. Trisomy 12 in early progenitor cells may result from genetic instability of already transformed more immature stem cells.
A different explanation for trisomy 12 and Rb-deletion in CD34+ cells could be aberrant antigen expression in B-CLL cells. This phenomenon of aberrant antigen expression is well known in acute leukemias.26 To rule out this phenomenon in chronic lymphocytic leukemia, we have investigated 10 patients with B-CLL for the coexpression of CD34 and CD5/CD20, but we could not find any subpopulation showing this marker combination. In addition, the coordinated expression of CD38 in all samples argues against this explanation. If the assumption of aberrant antigen expression was true, it would implicate an important immunophenotypical bias regarding stem cell involvement that can only be excluded by functional analysis of CD34+ progenitor cells in B-CLL.
The finding of few progenitor cells with the genetic marker, as in patient no. 9, in quantitative small sort products can be explained most likely by sort contamination. One or two aberrant cells of 20 sorted cells results in a significant percentage of aberrant progenitor cells, above the cut-off level of sort unpurity. In addition, especially in Rb-deletions, one signal may result in three-dimensional perspective from overlaying of one hybridization signal above the other. To minimize the problem of sort contamination and to ensure the quantitative significance of our results, we have reanalyzed the sort purity in each of our sorting experiments.
The interpretation of normal diploid progenitor cells is less clear-cut, because retention of two signals using a locus-specific probe or an α-centromeric probe, respectively, does not exclude a microdeletion or a point mutation within the region encompassed by the locus-specific probe nor a genetic abnormality in another region of 13q14 or chromosome 12. Submicroscopic changes may result in the same pathogenetical course as deletions or duplications that are detectable by conventional molecular or cytogenetic methods. Another explanation could be the restricted number of detectable and analyzable progenitor cells in B-CLL. In our experiments, the range of sorted progenitor cells was between 4 and 500, implicating low statistical significance in small numbers of sorted cells. This may limit the interpretation of the data on CD34+/38− cells, but not on the total population of CD34+ cells in the informative patients, in whom the number of positive cells was significantly above the level of background hybridization and sort purity.
These findings of genetically altered progenitor cells in B-CLL is particularly important for actual curative concepts in B-CLL using myeloablative chemotherapy and radiotherapy with autologous stem cell transplantation.43-45
In summary, we have found that, in a small subset of B-CLL patients, the CD34+ progenitor cells carry the genetic markers. These data suggest that the malignant transformation in B-CLL may involve early hematopoietic stem cells. Subsequent immunophenotypical and functional studies are necessary to confirm these data using additional markers and in vitro and xenotransplant models.
Address reprint requests to B. Gahn, MD, Department of Hematology and Oncology, Robert-Koch-Straβe 40, 37 075 Göttingen, Germany.