Abstract
Chemokines refer to a rapidly expanding family of small cytokines whose primary function is recruitment of leukocytes to inflammatory sites. These are known to bind to seven-transmembrane-domain containing receptors. A cDNA clone, CHEMR1, resembling the typical G protein-coupled receptor, was isolated from a mouse cytotoxic T-lymphocyte (CTL) library. Northern blot analysis in mouse cell lines suggests that its expression is found in a variety of cells, including T cells, B cells, and macrophages. The CHEMR1 gene Scya3r2 is a single-copy gene whose open reading frame may be in a single exon and maps to the distal region of mouse Chr 9 where the mouse macrophage inflammatory protein-1α (MIP-1α) receptor gene Scya3r and two related C-C chemokine receptor-like genes reside. Amino acid sequence comparison shows that CHEMR1 is 84% identical to human CCR-4, indicating that CHEMR1 is likely to be a mouse CCR-4. Binding assays using 125I-labeled C-C chemokines in mammalian cells indicated that CHEMR1 did not bind MIP-1α, RANTES, or MIP-1β, whereas CCR-1 binds MIP-1α and RANTES. Our result is different from the reported properties of human CCR-4. This suggests that CHEMR1 may be a receptor for unidentified C-C chemokine or a low-affinity receptor for MIP-1α.
THE CHEMOKINES COMPRISE a group of small cytokines that range from 8 to 12 kD in size and are involved in inflammatory processes, immunomodulation, and inhibition of proliferation of hematopoietic stem cells and T cells.1-3 These chemokines can be divided into two families, depending on whether the first two cysteines are adjacent (C-C chemokine or β) or are separated by an intervening amino acid (C-X-C chemokine or α). The former family includes monocyte chemotactic protein-1 (MCP-1), MCP-2, MCP-3, macrophage inflammatory protein-1α (MIP-1α), MIP-1β, I309, RANTES, eotaxin, macrophage inflammatory protein-related protein-1 (MRP-1; C10), and MRP-2; and the latter includes interleukin-8 (IL-8), interferon-γ–induced protein (IP)-10, neutrophil-activating peptide-2 (NAP-2), GRO, stromal cell differentiation factor (SDF ), and ENA-78.4-8 It has been shown that the C-C chemokines exhibit a preferential chemo-attractant potential for monocytes, whereas the C-X-C chemokines are chemo-attractants for neutrophils. It has been documented that IL-8, RANTES, MCP-1, MCP-2, MCP-3, SDF, MIP-1α, and MIP-1β are able to chemo-attract T lymphocytes.9-12 Recent studies showed that MIP-1α, MIP-1β, and RANTES inhibited human immunodeficiency virus–type 1 (HIV-1) viral replication and that HIV-1 used chemokine receptors as cofactors for the viral entry into CD4 T cells or macrophages.13-16
At present, four types of C-X-C chemokine receptors have been identified17-20: the human IL-8RA, which is selectively activated by IL-8; human IL-8RB, which is activated by IL-8, GROα, and NAP-214; CX CR3, which binds IP-10 and Mig; and fusin (or CX CR4) binding SDF. A murine homologue of the human IL-8RB was recently isolated21 and the presence of another member of IL-8R, probably IL-8RA, was proposed.22
Six members of human C-C chemokine receptor have been isolated and characterized: CCR-1, CCR-2A, CCR-2B, CCR-3, CCR-4, and CCR-5.23-27 These have overlapping ligand specificities, but the precise expression of these receptors is not yet known. In addition, isolation of several orphan receptors resembling the standard frame of C-C or C-X-C chemokine receptors makes correlation between biologic or immunologic activities of chemokines and the receptors mediating activities complicated. Especially with regards to HIV pathogenicity, therapeutic implication of the orphan receptors becomes increasingly of interest because various HIV strains seem to use different chemokine receptors as cofactors.28 29
We previously showed that CTLL-R8 cells (a mouse cytotoxic T-cell line) expressed MIP-1α receptor and that their proliferation was suppressed by MIP-1α.2 We found CTLL-2, an IL-2–dependent cytotoxic T-lymphocyte (CTL) line, also expressed the MIP-1α receptor (our unpublished observation). We and others have shown that recombinant MIP-1α suppresses hematopoietic stem and progenitor cell growth1-3,30,31 and prohibits proliferation signal through T-cell receptor (TCR) in primary T cells stimulated by immobilized anti-CD3.2
In an effort to understand the underlying mechanisms of the immunomodulatory function of chemokines on T cells and bone marrow stem and progenitor cells, we have attempted to clone an MIP-1α receptor. As a result, we discovered a putative G-protein–coupled receptor that has striking homology to CCR-4. We report here the identification and characterization of another potential member of the C-C chemokine receptor family.
MATERIALS AND METHODS
Cell lines.The IL-2–dependent CTLL-2 cells (a mouse cytolytic T-cell clone, ATCC TIB214) were maintained in RPMI 1640 (GIBCO Laboratories, Life Technology Inc, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; GIBCO), 100 U/mL penicillin (GIBCO), 100 μg/mL streptomycin (GIBCO), and 10 U/mL of IL-2 (Boehringer Mannheim, Indianapolis, IN). The B-cell lymphoma A20 and 2PK-3 cells were maintained in RPMI 1640 supplemented with 10% (vol/vol) FBS, antibiotics, nonessential amino acids (GIBCO), and 1 mmol/L sodium pyruvate (GIBCO). The D10 mouse T-helper cell clone was maintained in RPMI 1640 supplemented with 10% FBS, antibiotics, 10 μmol/L β-2 mercaptoethanol (2-ME; Sigma, St Louis, MO), and 100 U/mL of IL-2. The CTLL-R8, p388D1, RAW 264.7, and WEHI3 cells were maintained in RPMI 1640 supplemented with 10% FBS and antibiotics. Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and antibiotics. Mo7e cells are a human leukemic cell line and were routinely cultured in RPMI 1640 supplemented with 20% FBS plus 100 U/mL recombinant human granulocyte-macrophage colony-stimulating factor (rhuGM-CSF; Immunex, Seattle, WA).32
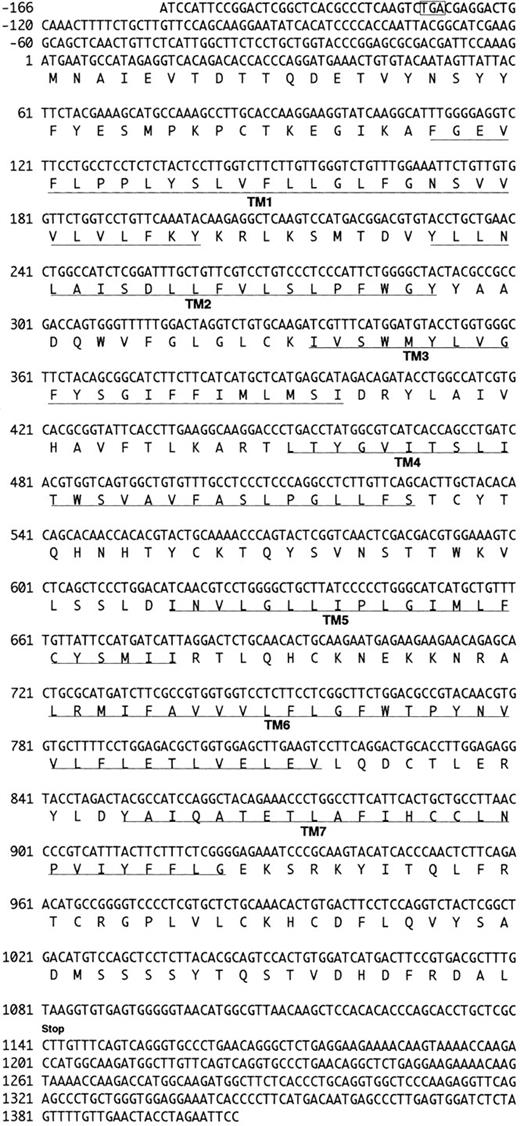
Reverse transcriptase-polymerase chain reaction (RT-PCR).The RT-PCR method was used to obtain cDNAs for potential G-protein–coupled receptors from CTLL-2. One microgram of cytoplasmic RNA was reverse transcribed by superscript (GIBCO), followed by PCR with the set of primers under the following conditions: for the first round of PCR in three cycles, denaturation was at 94°C for 1 minute, annealing at 37°C for 1.5 minutes, and extension at 72°C for 2 minutes; and for the second round of PCR in the next 30 cycles, denaturation was at 94°C for 1 minute, annealing at 48°C for 1.5 minutes, and extension at 72°C for 2 minutes. A pair of degenerate PCR primers was designed from the deduced amino acid sequence representing the sixth and seventh transmembrane domains of the published human IL-8 and FMLP receptors22: for the forward primer, 5′-TT(T/C)(T/C)TITG(T/C)TGGI(T/C/G)CCITA-3′ deduced from FLLCWLPY; and for the reverse primer, 5′IGG(A/G)TTIA(A/G)(A/G)CAI(G/C) (A/T)(A/G)T(G/T)IA-3′ deduced from HSCLNP. These primers are expected to produce an approximately 150-bp fragment from the members of the chemokine receptor family. After PCR, a 150-bp product was gel-purified, repaired by Klenow and T4 polynucleotide kinase, and ligated to the Sma I site in phosphatased pGEM3 (Promega, Madison, WI). Because we expected that the 150-bp fragments contained IL-8 receptor sequence, the plasmid library containing the PCR product was screened by IL-8 receptor probe to eliminate sequences corresponding to IL-8 receptor. Consequently, the cDNA clones that did not hybridize to IL-8 receptor probe were collected and analyzed. The nucleotide sequence of a cDNA insert was similar to IL-8 receptor sequences but sufficiently different to be a new member of receptor. The cDNA was named as CHEMR1, which stands for chemokine receptor 1.
Library screening.A λgt11 cDNA library prepared from CTLL-A11 cells was screened using the CHEMR1 probe to obtain a full-length version using standard procedures.33 Three overlapping cDNA clones were isolated. cDNA insert was prepared and subcloned into pBluscript SK+ (Stratagene, La Jolla, CA). The entire nucleotide sequences were determined using the Erase-A base system (Promega). Two of those (6-11 and 2-7) contained an overlapping 5′ untranslated region (UTR) with different length and different truncation of CHEMR1 open reading frame (ORF ), but lacked putative translational stop codon. The remaining cDNA clone (3-2) represented most of ORF (from TM1 to the putative stop codon) and 3′ UTR. To generate the longest CHEMR1 cDNA, the cDNA 6-11 was double-digested with Sac I and Stu I, which are located at the multicloning site of the vector and TM4 (around +513), respectively. The resulting fragments contained 5′ UTR to TM4 and were gel-purified with Gene-clean (BIO 101, La Jolla, CA) and ligated into the cDNA 3-2 that had been double-digested with Sac I and Stu I. Recombinant clones were confirmed by restriction analysis and sequencing of 5′ UTR, the junctional region around Stu I, and 3′ UTR. To obtain the human homologue of CHEMR1, a phytohemagglutinin (PHA)-stimulated human peripheral blood T-cell cDNA library from Clonetech (Palo Alto, CA) was screened with the CHEMR1 probe at low stringency; hybridization was conducted at 55°C in 6× SSC and the final wash was performed at 55°C in 2× SSC. The cDNA insert was prepared as mentioned above and its entire sequence was determined.
Northern blot.Poly (A+) RNA was prepared from mouse CTLL-R8, CTLL-2, D10, A20, 2PK3, p388D1, RAW 264.7, and WEH13 cells using oligo (dT) cellulose column chromatography (Boehringer Mannheim). Three micrograms of Poly (A+) RNA was electrophoresed on a 1% formaldehyde gel, transferrred to the Qiabrene (Qiagen, Santa Clarita, CA), and hybridized to CHEMR1 probe labeled by random priming. The filter was washed at 65°C for 30 minutes in 0.5× SSC and exposed to x-ray film (Eastman Kodak, Rochester, NY). The hybridized filters were stripped and rehybridized to the mouse β actin probe.34
Mouse genetic mapping.The chromosomal location of the gene encoding CHEMR1 was determined by analysis of two multilocus crosses: (NFS/N or C58/J X Mus musculus musculus ) X Mus musculus musculus (Kozak et al35 ) and (NFS/N X Mus spretus ) X M. spretus or C58/J (Adamson et al36 ). DNAs from the progeny of these crosses have been typed for more than 700 markers distributed over the mouse genome, including the chromosome (Chr) 9 markers Gnat1 (guanine nucleotide binding protein, α transducing subunit), Gnai2 (guanine nucleotide binding protein, α inhibiting activity polypeptide 2), Cck (cholecystokinin), and the microsatellite markers D9Mit16 and D9Mit20.D9Mit16 and D9Mit20 were typed using the primers and conditions described by Deitrich et al.37Cck was typed using as probe a 2-kb BamHI-EcoRI fragment of the CCK plasmid B5 obtained from Dr J. Friedman (The Rockefeller University, New York, NY).38 The progeny of both crosses were typed after digestion with EcoRI polymorphisms of Cck. The probe for Gnat1 was described previously39 and identified an HindIII restriction fragment length polymorphism in the spretus cross. Gnai2 was identified as a BamHI polymorphism in the spretus cross using as probe the 1.5-kb insert of the clone HHCPC5740 obtained from ATCC (Rockville, MD).
Linkage was determined using the program LOCUS designed by C.E. Buckler (NIAID, Bethesda, MD). The percentage of recombination and standard errors were determined according to Green41 and the gene order was determined by minimizing double recombinants.
Genomic Southern blot and genomic PCR.Twenty micrograms of mouse genomic DNA from 129/J or human genomic DNA prepared from Mo7e cells was cleaved with appropriate restriction endonucleases, electrophoresed in a 1% agarose gel, and transferred to a Quiabrene membrane. As a probe, CHEMR1 ORF was gel-purified, labeled to greater than 109 counts per minute (cpm)/μg of specific activity by the random primer method, and hybridized with the membrane in a solution containing 6× SSPE, 5× Denhardt solution, 1% sodium dodecyl sulfate (SDS), and 100 μg/mL sonicated salmon sperm DNA at 65°C overnight. The filter was washed with 0.1× SSC and 0.1% SDS at 65°C for 30 minutes, dried, and exposed to x-ray film (Eastman Kodak). A number of studies have shown that C-C chemokine receptor genes are intronless. To determine whether CHEMR1 gene constitutes intronless ORF, 1 μg of 129/J genomic DNA was amplified with the forward and reverse primers encompassing CHEMR1 ORF by using Pfu. As controls, CCR-4 and CCR-5 ORFs were amplified from OCM1 (a human melanoma cell line) genomic DNA.
The following primers were used for the amplification: 5′-GCC CAA GCT TGC GCG ACG ATT CCA AAG ATG-3′ (CHEMR1 forward primer); 5′-GCG CTC TAG AAT GTT ACC CCC ACT CAC ACC TTAC-3′ (CHEMR1 reverse primer); 5′-GCC CAA GCT TGC CTG TAG AGT TAA AAA ATG-3′ (CCR-4 forward primer); 5′-GCG CTC TAG ATT CTA CAG AGC ATC ATG GAG ATC-3′ (CCR-4 reverse primer); 5′-GCC CAA GCTT AAC CAG AGA GAA GCC GGG-3′ (CCR-1 forward primer); and 5′-GCG CTC TAGA GGC CTC CTA TGG TCT GAG-3′ (CCR-1 reverse primer). The amplified fragments were digested with Hind III/Xba I and clones into pRC/CMV.
Expression, transfection, and binding assay.A Kpn I/Hpa I fragment of CHEMR1 that included 26 bp of the 5′ UTR, the entire ORF, and 27 bp of the 3′ UTR was treated with T4 DNA polymerase and cloned into the BstXI site blunt-ended with T4 DNA polymerase of pRc/CMV (Invitrogen, San Diego, CA). To express CCR-1, the entire ORF was amplified from the λgt11 clone with pfu polymerase (Stratagene, La Jolla, CA). The sequences of the forward and reverse primers were as follows: 5′-GCCCAAGCTTAACCAGAGAGAAGCCGGG-3′ (forward) and 5′-GCGCTCTAGAGGCCTCCTATGGTCTGAG-3′ (reverse). The underlined bases represent HindIII and Xba I sites, respectively. The amplified fragment was gel-purified, cleaved with HindIII and Xba I, and cloned into pRc/CMV cleaved by HindIII and Xba I. Plasmids with insert were prepared according to standard procedures.33 In parallel with the above-mentioned expression strategy, CHEMR1 and CCR-4 ORF were directly amplified from human genomic DNA. After sequence verification, a FLAG epitope-tagged CHEMR1 CCR-4 and CCR-1 were created with appropriate primers that had nucleotide sequences encoding FLAG epitope. Reverse primers were the same as before. The sequences of forward primers were as follows: 5′-GCCCAAGCTT GCC GCC ATG GAC TAC AAG GAC GAC GAT GAC AAG AAT GCC ATA GAG GTC ACAG-3′ (CHEMR1 FLAG); 5′-GCCCAAGCTT GCC GCC ATG GAC TAC AAG GAC GAC GAT GAC AAG AAC CCC ACG GAT ATA GCAG-3′ (CCR-4 FLAG); and 5′-GCCCAAGCTT GCC GCC ATG GAC TAC AAG GAC GAC GAT GAC AAG GAA ACT CCA AAC ACC ACA GAG-3′ (CCR-1 FLAG). The underlining indicates the nucleotide sequence encoding FLAG epitope.
Five micrograms of each chemokine was lyophilized, resuspended in 10 μL of 100 mmol/L sodium borate buffer, pH 8.5, and added to 1 mCi of 125I-labeled Bolton-Hunter reagent (2200 Ci/mmol, monoiodinated; Dupont-NEN, Boston, MA). The reaction was continued on ice for 60 minutes. The reaction mixture was applied to a PD10 column (Pharmacia LKB, Piscataway, NJ) that had been pre-equilibrated with phosphate-buffered saline containing 0.1% gelatin. The iodinated protein was purified from free iodine, and bovine serum albumin and sodium azide were then added to the collected fractions to final concentrations of 1% and 0.025%, respectively. 125I-human MIP-1α (2,000 Ci/mmol) purchased from Amersham (Arlington Heights, IL) was used for receptor-binding assays.
Subconfluent HEK293 cells were transfected by the calcium phosphate method.42 After 24 hours, 2 × 106 cells were collected with the addition of 1 mmol/L EDTA in phosphate-buffered saline and resuspended in 200 μL of the binding medium (DMEM with 1% bovine serum albumin). Two nanomoles per liter each of iodinated MIP-1α, RANTES, and MIP-1β was used for the binding assay. The binding assay for 125I-labeled MIP-1α was performed at room temperature or at 37°C for 1 hour in the presence of 100-fold excess of cold MIP-1α, whereas the binding assay for RANTES and MIP-1β was performed at room temperature for 1 hour. The entire mixture was layered on a 10% sucrose cushion, and the radioactivity of cell pellets was counted. The specific binding was calculated by the difference in counts due to cold ligand competition. For cold competition of MIP-1α, 0.1 nmol/L 125I–hMIP-1α was used in the presence of varying concentration of cold hMIP-1α.
Fluorescence-activated cell sorting analysis of transfected HEK 293 cells expressing FLAG epitope-tagged chemokine receptors.Transfected HEK 293 cells (2 × 106) were washed once with 5 mL cold 1× Hanks' balanced salt solution (HBSS) containing 5% FBS and then resuspended in 2 mL of the same buffer. One microgram of anti-FLAG M2 monoclonal antibody (MoAb; mouse IgG1 ) was added to 100 μL of cell suspension and left on ice for 30 minutes. The treated cells were then washed with 5 mL HBSS and resuspended in 100 μL HBSS. A total of 0.5 μg of goat antimouse IgG-fluorescein isothiocyanate (FITC) (Zymed Laboratories, South San Francisco, CA) was added and incubated on ice for 30 minutes. The cells were washed the same as before and finally resuspended in 300 μL 2% paraformaldehyde. Receptor expression was measured by flow cytometer (Becton Dickinson, Mountain View, CA).
RESULTS
Molecular cloning of a potential chemokine receptor.We cloned genes encoding potential G-protein–coupled receptors by RT-PCR using primers based on the known sequence of human IL-8, N-formyl peptide, and rabbit N-formyl peptide receptors.17 The most conserved regions were chosen from transmembrane (TM) regions 6 and 7 to prepare degenerate oligonucleotides. The RT-PCR with the cytoplasmic RNA from CTLL-2 using these primer sets produced two bands with sizes of 700 and 150 bp (data not shown). Both bands were purified and cloned. The sequence of 700 bp did not match any gene sequence from GenBank.
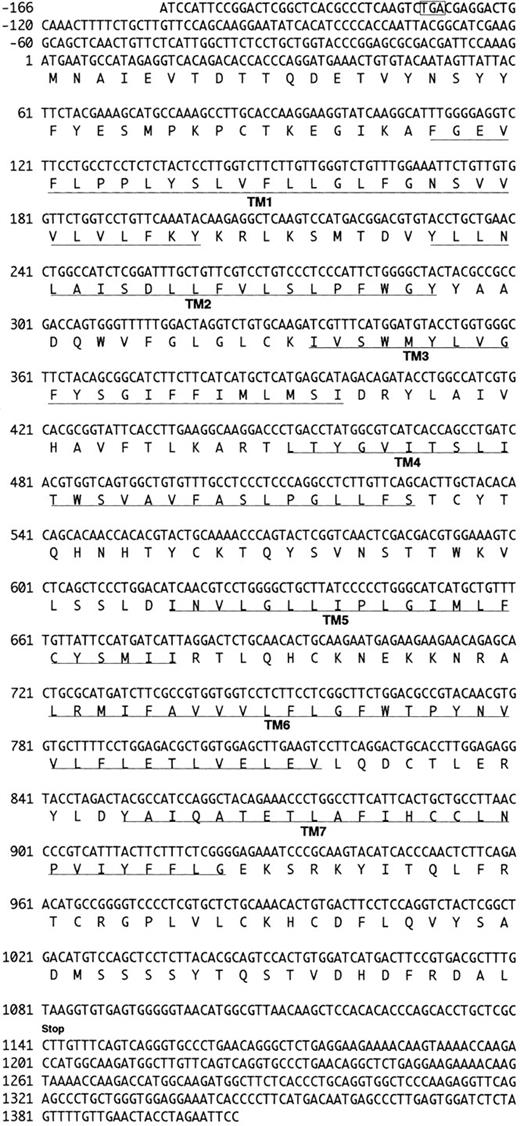
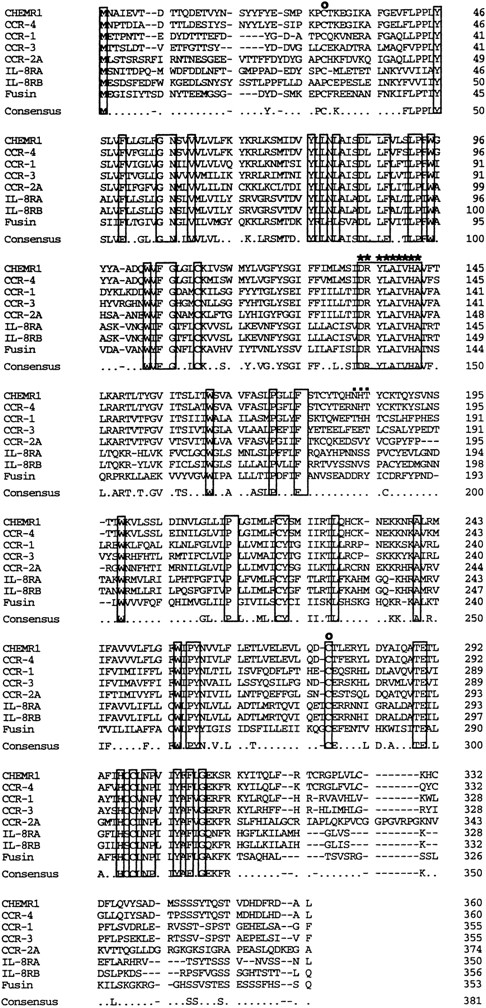
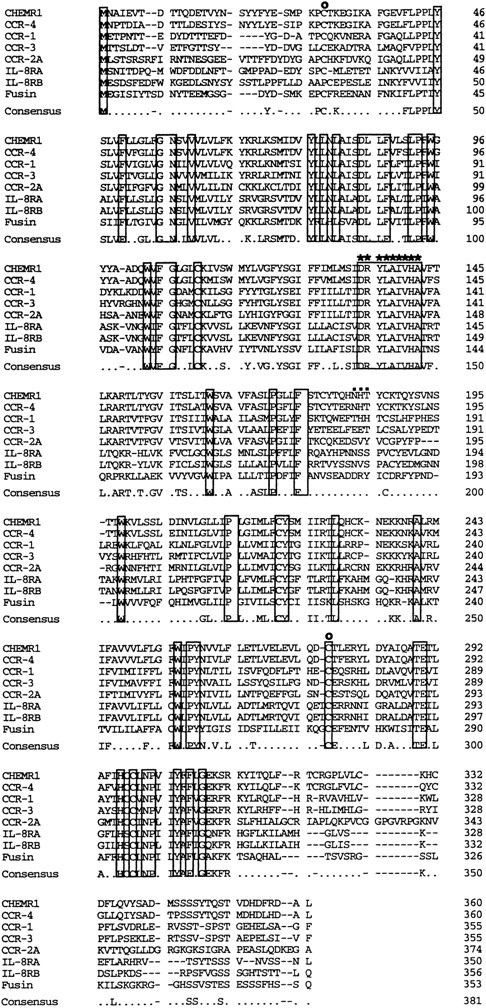
We analyzed approximately 200 150-bp band-derived cDNA clones. We first eliminated clones corresponding to the murine IL-8 receptor because the majority of the clones turned out to be the mouse homologue of the human type B IL-8 receptor (IL-8RB).21 We then grouped the rest of the clones by cross-hybridization and obtained two groups of cDNA clones. The first groups turned out to be a mouse homologue of fusin.43 The second group, termed CHEMR1, was most interesting to us because it exhibited some degree of homology to the human IL-8 receptor and MIP-1α/RANTES receptor (CCR-1) in the amplified region and seemed to be novel. Three overlapping cDNA clones were isolated by screening a cDNA library from CTLL-A11 with the radiolabeled CHEMR1 PCR fragment as a probe and were sequenced. One of the longest ORFs encoding 360 amino acids was started from the methionine located at 166 bp downstream of the cDNA. We found a putative translation stop codon located in frame in 5′ UTR, indicating that the initiation methionine we assigned may be a presumed initiator. As shown in Fig 1, the cDNA appeared to resemble a typical G-protein–coupled receptor, based on the presence of 7 putative membrane spanning domains. Moreover, as shown in Fig 2, it also contained a putative G-protein binding domain, DRYLAIVHA, which is represented by asterisks, a domain that is commonly found in other chemokine receptors.44 The putative amino-terminal ligand binding domain is composed of highly acidic amino acids. Compared with known chemokine receptors, a Pro28-Cys29 motif in the amino-terminal region and a Cys276 in the third predicted extracellular loop are conserved in CHEMR1. The third predicted intracellular loop is comprised of highly basic amino acids. These features suggest that CHEMR1 represents a potential chemokine receptor.45 Interestingly, the amino-terminal region lacks an N-glycosylation site. But the second predicted extracellular domain has a potential N-glycosylation site (Asn183-His184-Thr185 ) indicated by solid boxes. During preparation of this manuscript, eosinophil- and basophil-derived C-C chemokine receptors were isolated, CCR-3 and CCR-4, respectively.25,26 Macrophage tropic HIV cofactor, CCR-5, was recently isolated.27 The rank order of amino acid identity of known human chemokine receptors to CHEMR1 was CCR-4 (84%) >CCR-1 (47%) ∼CCR-5 (47%) >CCR-3 (41%), ∼CCR-2A (41%), >IL-8RA (37%), ∼IL-8RB (37%).
Nucleotide and deduced amino acid sequence of the CHEMR1 cDNA clone. Nucleotide numbering in CHEMR1 is referred to the first nucleotide (position 1) in the translation initiation codon (ATG). The putative transmembrane domains (TM1-TM7) are underlined. The translation termination codon is indicated by “stop.” Indicated in an open box is the potential stop codon found in 5′ UTR in frame relative to the initiation codon.
Nucleotide and deduced amino acid sequence of the CHEMR1 cDNA clone. Nucleotide numbering in CHEMR1 is referred to the first nucleotide (position 1) in the translation initiation codon (ATG). The putative transmembrane domains (TM1-TM7) are underlined. The translation termination codon is indicated by “stop.” Indicated in an open box is the potential stop codon found in 5′ UTR in frame relative to the initiation codon.
Comparison of the primary structure of CHEMR1 with other chemokine receptors. Shown is an alignment of CHEMR1 with CCR-4, the MIP-1α/RANTES receptor (CCR-1), eosinophil-derived C-C chemokine receptor (CCR-3), the MCP-1 receptor (CCR-2A), the two IL-8 receptors, and fusin (HUMSTSR), the T-tropic HIV-1 cofactor. The conserved residues are indicated in a box. A potential G-protein binding domain is indicated by asterisks.45 (○) A putative disulfide bridge that forms a ligand binding domain.45 (▪) A potential N-glycosylation site.
Comparison of the primary structure of CHEMR1 with other chemokine receptors. Shown is an alignment of CHEMR1 with CCR-4, the MIP-1α/RANTES receptor (CCR-1), eosinophil-derived C-C chemokine receptor (CCR-3), the MCP-1 receptor (CCR-2A), the two IL-8 receptors, and fusin (HUMSTSR), the T-tropic HIV-1 cofactor. The conserved residues are indicated in a box. A potential G-protein binding domain is indicated by asterisks.45 (○) A putative disulfide bridge that forms a ligand binding domain.45 (▪) A potential N-glycosylation site.
CHEMR1 expression.Northern blot analysis was performed using poly (A+) RNA from various murine cell lines. As shown in Fig 3, CTLL-2 (CD8+ T-cell line), D10 (CD4+, Th2 cell line), 2PK3 (B-cell lymphoma), and WEHI3 (macrophage) expressed CHEMR1-related mRNA. A CHEMR1 message was detected in CTLL-R8 cells at prolonged exposure. In all cases, a single mRNA of about 4.0 kb was detected, suggesting that the native CHEMRI message may include longer 5′ or 3′ UTR than the CHEMR1 cDNA presented here.
Northern analysis of CHEMR1 mRNA expression. Poly (A+) RNA was isolated from each cell line indicated. Three micrograms of the RNA were electrophoresed on a 1.2% formaldehyde gel, transferred to Quiabrane (Quiagen), and hybridized sequentially with [32P] CHEMR1 and β-actin cDNA.
Northern analysis of CHEMR1 mRNA expression. Poly (A+) RNA was isolated from each cell line indicated. Three micrograms of the RNA were electrophoresed on a 1.2% formaldehyde gel, transferred to Quiabrane (Quiagen), and hybridized sequentially with [32P] CHEMR1 and β-actin cDNA.
The CHEMR1 is a single copy gene that maps to mouse Chr 9.Genomic southern hybridization was performed at high stringency. CHEMR1 cDNA representing its ORF was used as a probe. Results suggest that the CHEMR1 gene is a single copy gene. As shown in Fig 4A, HincII or Kpn I digestion produced bands of 531 bp and 867 bp, respectively, which are the same size as expected from CHEMR1 cDNA. The result suggests that CHEMR1 ORF may be intronless. HincII digestion produced a faint band around 1.9 kb, which was also observed in C57Bl/6. The nature of the band is currently unknown. We performed a human genomic Southern blot using the same probe at high stringency. This showed that human genomic DNA possessed the human homologue of CHEMR1.
Genomic analysis of CHEMR1 gene. Evidence for existence of the human homologue of CHEMR1 and for CHEMR1 being intronless. (A) 129/J genomic DNA (20 μg) and human genomic DNA prepared from Mo7e cells were digested with the restriction enzymes indicated, run on a 1% agarose gel, transferred to Quiabrane, and hybridized with [32P] CHEMR1 ORF. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blots were exposed to XAR-5 film in cassettes at −70°C for 6 days. DNA molecular standards from BRL were run in parallel. The CHEMR1 cDNA has two Kpn I (−26 and 840) and HincII (580 and 1,110) sites, which release around 867 and 531 bp, respectively. These bands are indicated by arrows. (B) One microgram of 129/J genomic DNA or OCM1 genomic DNA was amplified with Pfu polymerase in the presence of 50 pmol of each of the corresponding primers as indicated in the Materials and Methods. After 30 cycles of amplification, one-tenth volume of PCR reaction mixture was resolved on a 1% agarose gel. Indicated with arrows are amplified ORFs of chemokine receptors. The same molecular standards were used as in (A).
Genomic analysis of CHEMR1 gene. Evidence for existence of the human homologue of CHEMR1 and for CHEMR1 being intronless. (A) 129/J genomic DNA (20 μg) and human genomic DNA prepared from Mo7e cells were digested with the restriction enzymes indicated, run on a 1% agarose gel, transferred to Quiabrane, and hybridized with [32P] CHEMR1 ORF. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blots were exposed to XAR-5 film in cassettes at −70°C for 6 days. DNA molecular standards from BRL were run in parallel. The CHEMR1 cDNA has two Kpn I (−26 and 840) and HincII (580 and 1,110) sites, which release around 867 and 531 bp, respectively. These bands are indicated by arrows. (B) One microgram of 129/J genomic DNA or OCM1 genomic DNA was amplified with Pfu polymerase in the presence of 50 pmol of each of the corresponding primers as indicated in the Materials and Methods. After 30 cycles of amplification, one-tenth volume of PCR reaction mixture was resolved on a 1% agarose gel. Indicated with arrows are amplified ORFs of chemokine receptors. The same molecular standards were used as in (A).
To determine whether the ORF portion of CHEMR1 gene is intronless, the CHEMR1 ORF was directly amplified from 129/J mouse genomic DNA. As controls, CCR-4 and CCR-5 were also amplified from human genomic DNA. CCR-5 has been shown to be an intronless gene46 and CCR-4 has been shown to be intronless as well (our unpublished observation). Figure 4B clearly shows that CHEMR1 ORF could be directly amplified from mouse genomic DNA as were CCR-5 and CCR-4. This suggests that CHEMR1 is intronless and could be classified as a member of C-C chemokine receptor supergene family.
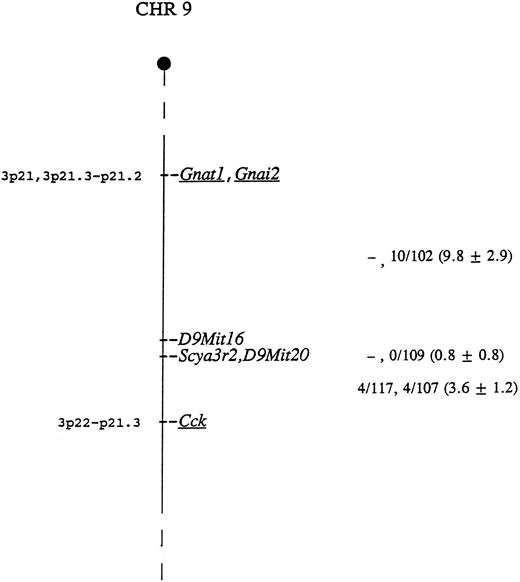
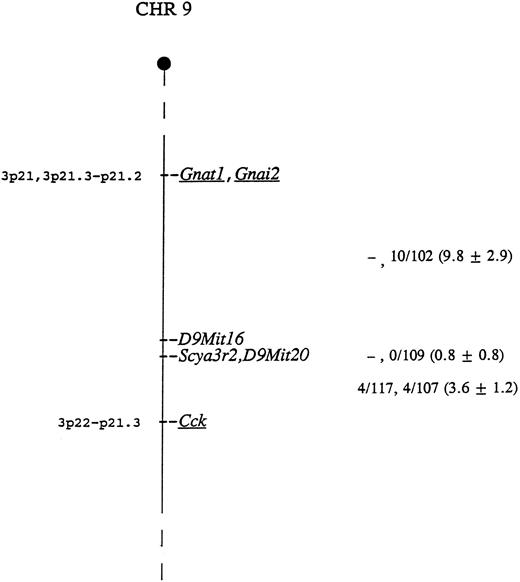
The mouse locus encoding CHEMR1 is termed Scya3r2, for small inducible cytokine 3 receptor 2. This locus was mapped using CHEMR1 cDNA probe as hybridization probe to type DNAs from two genetic crosses. BamHI digestion generated fragments of 3.7 and 2.5 kb in NFS/N and 6.4 kb in spretus, and Pvu II digestion produced fragments of 5.8 and 2.9 kb in NFS/N and 8.4 kb in musculus, respectively. Inheritance of the polymorphic fragments was followed in the progeny of both crosses and compared with that of more than 700 markers previously typed and mapped. Scya3r2 was mapped to the distal end of Chr 9 based on an analysis of both crosses (Fig 5). Closest linkage was observed with D9Mit20, with no recombinants identified in 121 mice, indicating that, at the 95% confidence level, these genes are within 2.4 centimorgans.
Genetic map location of Scya3r2 on mouse Chr 9. To the right of the map are given recombination fractions between adjacent loci with the first fraction representing data from the musculus cross and the second representing data from the spretus cross. The numbers in parentheses represent the percentage of recombination and the standard error. In the musculus cross, D9Mit16 and D9Mit20 were typed only in the progeny identified as recombinant between Gnat1 and Cck. Of the 12 recombinants in the interval Gnat1 - Scya3r2 in the two crosses, 11 were proximal to D9Mit16, indicating that D9Mit16 is just proximal to Scya3r2 and D9Mit20. The map locations for the human homologs of the underlined genes are given to the left of the map.
Genetic map location of Scya3r2 on mouse Chr 9. To the right of the map are given recombination fractions between adjacent loci with the first fraction representing data from the musculus cross and the second representing data from the spretus cross. The numbers in parentheses represent the percentage of recombination and the standard error. In the musculus cross, D9Mit16 and D9Mit20 were typed only in the progeny identified as recombinant between Gnat1 and Cck. Of the 12 recombinants in the interval Gnat1 - Scya3r2 in the two crosses, 11 were proximal to D9Mit16, indicating that D9Mit16 is just proximal to Scya3r2 and D9Mit20. The map locations for the human homologs of the underlined genes are given to the left of the map.
As indicated in Fig 5, the most telomeric region of Chr 9 shows conserved linkage with human chromosome 3, suggesting that the human homolog of this gene maps to chromosome 3p22-21. This is interesting because the human homolog of the MIP-1α/RANTES receptor gene, termed CMKBR1, has been mapped to 3p21. This suggests the possibility that these genes may be clustered.
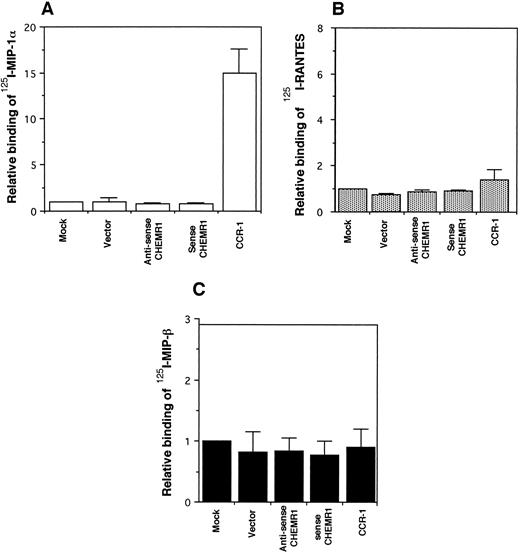
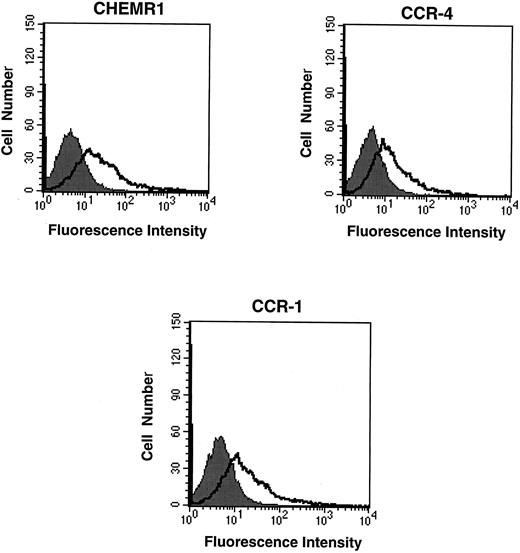
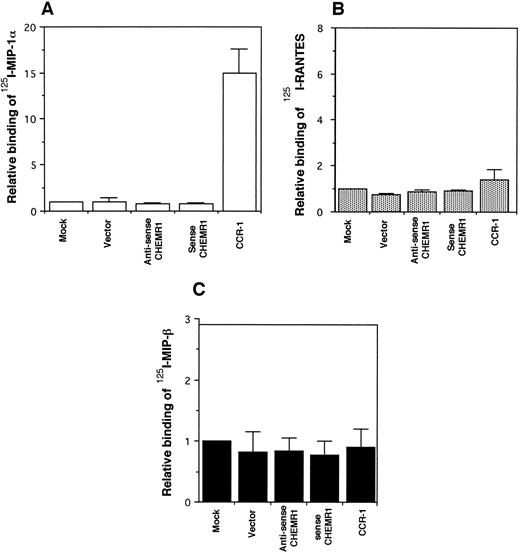
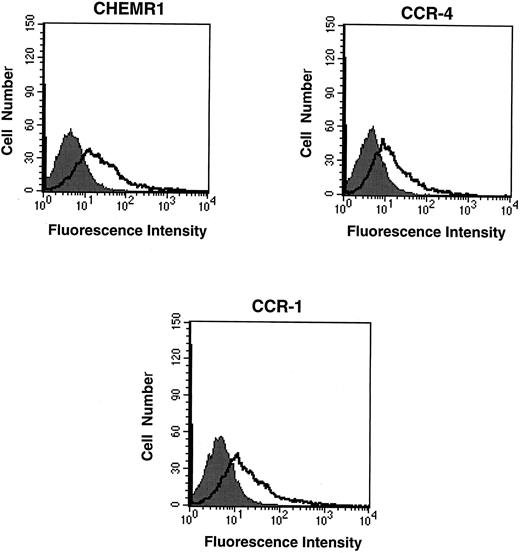
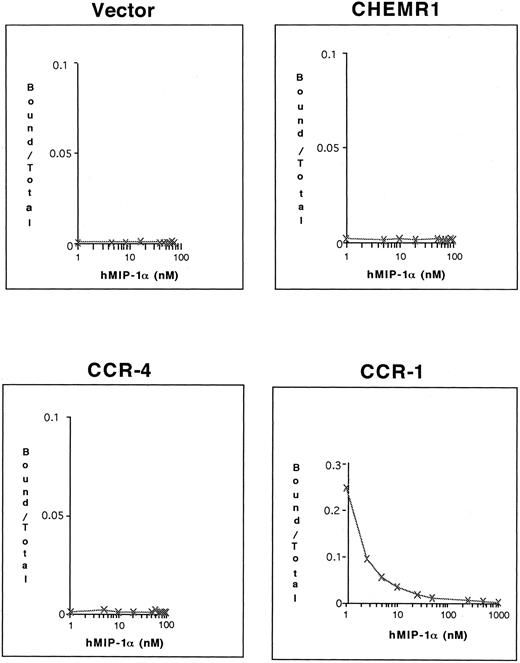
CHEMR1 may not be a MIP-1α, MIP-1β, or RANTES receptor.The CHEMR1 and CCR-1 ORFs were inserted downstream of the CMV promoter in both sense and antisense orientations. These receptors were then expressed transiently in HEK 293 cells. A single species of mRNA was detected below 18S for both receptors (Fig 6). In chemokine binding assays with transfected HEK 293 cells, 125I–MIP-1α, 125I-RANTES, and 125I–MIP-1β did not bind to CHEMR1, whereas 125I–MIP-1α and 125I-RANTES did bind to CCR-1 (Fig 7A, B, and C). 125I–MIP-1α efficiently bound to CCR-1 at 37°C (Fig 7A). 125I-RANTES bound slightly to CCR-1 at room temperature (Fig 7B). MIP-1β did not bind to CCR-1 or CHEMR1 at room temperature (Fig 7C) or at 37°C (data not shown). One possible reason for our inability to detect appreciable binding of such chemokines to CHEMR1 is the lack of appropriate surface expression in the transfected cells. To determine whether our transfection system allows chemokine receptors to be extracellularly expressed, FLAG epitope was introduced to CHEMR1, CCR-1, and CCR-4. CCR-4 was used for comparison with CHEMR1 because of its sequence similarity. After transfection, extracellular presentation of the receptors was detected by flow cytometry with anti-FLAG MoAb. As shown in Fig 8, all three receptors were expressed on the HEK 293 cell surface.
Northern blot analysis of HEK293 cells transfected with vector (pRc/CMV), the antisense CHEMR1, sense CHEMR1, and CCR-1 in pRc/CMV. HEK293 cells were transfected with 20 μg of each DNA using the modified calcium phosphate method. After 24 hours, total RNA was isolated with Tri-reagent (Molecular Research Center, Cincinnati, OH). Twenty micrograms of the RNA was run on a 1.2% formaldehyde gel, blotted to Quiabrane, and hybridized with [32P] CHEMR1 cDNA. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blot was exposed to XAR-5 film in a cassette at −70°C for 2 days. The same filter was stripped and subjected to sequential hybridization with [32P] CCR-1 and [32P] rat GAPDH probe, respectively.
Northern blot analysis of HEK293 cells transfected with vector (pRc/CMV), the antisense CHEMR1, sense CHEMR1, and CCR-1 in pRc/CMV. HEK293 cells were transfected with 20 μg of each DNA using the modified calcium phosphate method. After 24 hours, total RNA was isolated with Tri-reagent (Molecular Research Center, Cincinnati, OH). Twenty micrograms of the RNA was run on a 1.2% formaldehyde gel, blotted to Quiabrane, and hybridized with [32P] CHEMR1 cDNA. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blot was exposed to XAR-5 film in a cassette at −70°C for 2 days. The same filter was stripped and subjected to sequential hybridization with [32P] CCR-1 and [32P] rat GAPDH probe, respectively.
Binding assay of radiolabeled human MIP-1α (A), RANTES (B), and MIP-1β (C) to CHEMR1. HEK293 cells were transfected with vector only, the CHEMR1 expression construct, or CCR-1 expression construct. After 24 hours, cells were harvested (about 2 × 106 cells) and incubated with 2 nmol/L 125I–MIP-1α, RANTES, and MIP-1α. The binding temperature for the MIP-1α was 37°C, whereas those for RANTES and MIP-1β were room temperature. The relative binding was obtained by dividing specific binding (bound cpm) of the radiolabeled chemokines to transfectants with the expression constructs by the specific-binding (bound cpm) to the mock-transfected cells. The average specific binding of 125I–MIP-1α to CCR-1 at 37°C was 14,000 cpm. The average bound cpm of 125I-RANTES to CCR-1 was 1,500 cpm. The average nonspecific binding of 125I–MIP-1α, RANTES, and MIP-1β to the mock-transfected cells was less than 800 cpm. Standard deviations were calculated by using three independent transfections and binding assays.
Binding assay of radiolabeled human MIP-1α (A), RANTES (B), and MIP-1β (C) to CHEMR1. HEK293 cells were transfected with vector only, the CHEMR1 expression construct, or CCR-1 expression construct. After 24 hours, cells were harvested (about 2 × 106 cells) and incubated with 2 nmol/L 125I–MIP-1α, RANTES, and MIP-1α. The binding temperature for the MIP-1α was 37°C, whereas those for RANTES and MIP-1β were room temperature. The relative binding was obtained by dividing specific binding (bound cpm) of the radiolabeled chemokines to transfectants with the expression constructs by the specific-binding (bound cpm) to the mock-transfected cells. The average specific binding of 125I–MIP-1α to CCR-1 at 37°C was 14,000 cpm. The average bound cpm of 125I-RANTES to CCR-1 was 1,500 cpm. The average nonspecific binding of 125I–MIP-1α, RANTES, and MIP-1β to the mock-transfected cells was less than 800 cpm. Standard deviations were calculated by using three independent transfections and binding assays.
Flow cytometric analysis of the expression of chemokine receptors in transfected HEK 293 cells. Twenty micrograms of FLAG epitope-tagged CHEMR1, CCR-4, and CCR-1 and 0.5 μg of CMV-luciferase gene were transfected into HEK 293 cells. After 12 hours, 2 × 105 cells were washed, resuspended in HBSS, and mixed with 1 μg of either anti-FLAG MoAb (M2) or mouse IgG1 isotype control. After washing the cells, the cells were incubated with goat antimouse IgG-FITC, washed, and subjected to flow cytometric analysis. The shaded areas indicate the isotype control, whereas the lines denote fluorescence reflecting expression of the chemokine receptors. Each 2 × 105 transfected cells produced approximately 10,000 light units of luciferase activity; therefore, the fluorescence intensity shown above represents a quantitative difference of the surface expression of each receptor due to compensating the difference of transfection efficiency. At this condition, the mean fluorescence intensities (MFI) for CHEMR1, CCR-4, and CCR-1 were 127.7, 24.9, and 91.2, respectively.
Flow cytometric analysis of the expression of chemokine receptors in transfected HEK 293 cells. Twenty micrograms of FLAG epitope-tagged CHEMR1, CCR-4, and CCR-1 and 0.5 μg of CMV-luciferase gene were transfected into HEK 293 cells. After 12 hours, 2 × 105 cells were washed, resuspended in HBSS, and mixed with 1 μg of either anti-FLAG MoAb (M2) or mouse IgG1 isotype control. After washing the cells, the cells were incubated with goat antimouse IgG-FITC, washed, and subjected to flow cytometric analysis. The shaded areas indicate the isotype control, whereas the lines denote fluorescence reflecting expression of the chemokine receptors. Each 2 × 105 transfected cells produced approximately 10,000 light units of luciferase activity; therefore, the fluorescence intensity shown above represents a quantitative difference of the surface expression of each receptor due to compensating the difference of transfection efficiency. At this condition, the mean fluorescence intensities (MFI) for CHEMR1, CCR-4, and CCR-1 were 127.7, 24.9, and 91.2, respectively.
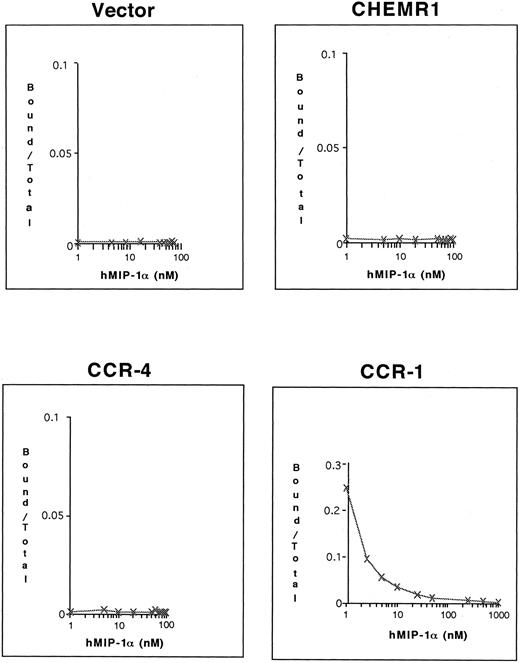
To determine whether 125I–MIP-1α binds to these receptors, competition binding assays were conducted. As shown in Fig 9, only CCR-1 bound 125I–MIP-1α, whereas neither CHEMR1 nor CCR-4 bound 125I–MIP-1α. These results again suggest that CHEMR1 may not be functionally identical to MIP-1α, RANTES, or a MIP-1β receptor and may be a novel chemokine receptor for a yet unidentified C-C chemokine.
Competition of 125I–MIP-1α binding to HEK 293 cells transfected with CHEMR1, CCR-4, and CCR-1. Transfected cells (2 × 105 cells) were incubated with 0.1 nmol/L 125I–MIP-1α and varying concentrations of unlabeled MIP-1α at 37°C for 1 hour. The incubation mixtures were layered on 10% sucrose gradients, and labeled cells were recovered from pellets. In the case of CCR-1, 25% of the total input 125I–MIP-1α (∼105 cpm) bound to transfected cells in the absence of cold hMIP-1α, whereas the background level was less than 200 cpm.
Competition of 125I–MIP-1α binding to HEK 293 cells transfected with CHEMR1, CCR-4, and CCR-1. Transfected cells (2 × 105 cells) were incubated with 0.1 nmol/L 125I–MIP-1α and varying concentrations of unlabeled MIP-1α at 37°C for 1 hour. The incubation mixtures were layered on 10% sucrose gradients, and labeled cells were recovered from pellets. In the case of CCR-1, 25% of the total input 125I–MIP-1α (∼105 cpm) bound to transfected cells in the absence of cold hMIP-1α, whereas the background level was less than 200 cpm.
DISCUSSION
In the present study, we report the isolation and characterization of a novel mouse cDNA encoding a potential chemokine receptor. CHEMR1 contained the predicted G-protein binding domain, DRYLAIVHA,45 a pair of cysteines that form an interdisulfide bond between Pro28-Cys29 and Cys276-Thr277, which form a putative ligand-binding domain.45 The C-terminal tail is comprised of serine- and threonine-rich motifs that are known to be substrates for Ser/Thr kinases such as casein kinase, protein kinase C, and some G-protein receptor kinases.47 This region is believed to be involved in receptor desensitization.48
Previous studies by our laboratory have shown that mouse CTL cell lines express MIP-1α receptors. Several lines of evidence strongly indicate that T cells have receptors for MIP-1α, MIP-1β, and RANTES.49 Because CHEMR1 originated from a CTL cDNA library, we initially tested chemokine binding of CHEMR1 using radiolabeled C-C chemokines. Our results showed that CHEMR1 did not bind MIP-1α, MIP-1β, or RANTES (Figs 7, 8, and 9). Preliminary data suggest that the stable transfectants expressing CHEMR1 did not seem to mobilize Ca2+, responding to each 100 nmol/L of recombinant MIP-1α, IL-8, MCP-1, and IP-10, whereas the stable ones expressing CCR-1 exhibited small, transient elevation of Ca2+ upon stimulation with 100 nmol/L of MIP-1α (unpublished observation). Based on a significant identity of amino acid between CHEMR1 and CCR-4 (84% amino acid identity), we believe that CHEMR1 would be most related to CCR-4. However, our binding results established a conflict with the report by Power et al.26 CCR-4 was able to stimulate a Ca2+-activated chloride channel in Xenopus oocytes upon stimulation with 1 μmol/L of MIP-1α, RANTES, and MCP-1 using the voltage clamp technique. Because a very high concentration of chemokines was used and more sensitive measurements than equilibrium binding assay or conventional Ca2+ mobilization assays were used in the studies of Power et al,26 the chemokines could still trigger CCR-4 if it were a low-affinity receptor for MIP-1α, RANTES, or MCP-1. During preparation of this report, Hoogewerf et al50 reported the murine CCR-4 and showed that the murine and human CCR-4 were high-affinity receptors for MIP-1α and RANTES in transfected HL60 cells. By contrast to their previous report, MCP-1 did not bind to these receptors.50 The murine CCR-4 has a 97% amino acid identity with CHEMR1, but the nucleotide sequences of 5′ and 3′ UTR are somewhat different between the two receptor genes. Therefore, CHEMR1 could be a receptor different from the murine CCR-4. This could explain the difference in binding activities between CHEMR1 and the murine CCR-4. If CHEMR1 and the murine CCR-4 are the same gene product and the differences in amino acid sequence and UTR sequence are simply due to an allelic variation, the conflict simply may reflect the difference in experimental approach. For example, Hoogewerf et al50 used HL60 cells to express recombinant CCR-4. Because it is known that HL60 cells express endogenous CCR-5,51 the high-affinity binding of MIP-1α and RANTES seen by Hoogewerf et al50 might be due to endogenous CCR-5. Because our data clearly showed that CHEMR1 and the human CCR-4 were extracellularly expressed, the negative binding of MIP-1α to these receptors may not be at least due to an experimental flaw. Therefore, our data do not support the binding results of Hoogewerf et al.50 Alternatively, it is possible that MIP-1α did not bind to CHEMR1 or CCR-4 because MIP-1α is a low-affinity ligand to the receptors.
Although CHEMR1 may not bind MIP-1α, RANTES, or MIP-1β, it may still be an important chemokine receptor, because it is expressed in different hematopoietic cell lines. The CHEMR1 mRNA was detected in CTL lines (CTLL-2 and CTLL-R8), a B-cell lymphoma cell line (2PK3), a macrophage cell line (WHE13), and a T-helper cell clone (D10). Hoogewerf et al50 also described that they detected a significant expression of the mCCR-4 in a CTL line. It is worthwhile to note that CHEMR1 cDNA was derived from a CTL cDNA library.
It has been shown that CCR-3 was specifically expressed in eosinophils, whereas the rest of known C-C chemokine receptors were expressed in a rather wide range of tissues. Because CHEMR1 was detected in different hematopoietic cell lines, we believe that CHEMR1 would be expressed in lymphoid organs.
Kozak et al52 reported the chromosomal location of one mouse MIP-1α receptor and two C-C chemokine receptor-like genes. The gene symbols for the mouse genes are Scy3ar, Scy3ar-rs1, and Scy3ar-rs2. The genes for these three receptors, Scy3ar, Scy3ar-rs1, and Scy3ar-rs2, all map to a cluster on distal Chr 9, but not at the same position as the CHEMR1 gene, which is about 5 centimorgans proximal to this cluster. This region of Chr 9 is homologous to human 3p21-22, which contains the human CCR-1 gene.
The chromosomal locations of the two IL-8 receptors (IL-8RA and IL-8RB) as well as that of an IL-8 receptor pseudogene were found to reside in human chromosome 2q35.53 This region shares linkage homology with a region in mouse Chr 1 that encodes the Ity-Lsh-Bcg and Vil loci.21 Using the human IL-8RB cDNA probe, Cerretti et al21 isolated the mouse homologue of the human IL-8RB. As predicted, the mouse IL-8RB gene maps between the Ity-Lsh-Bcg and Vil loci in mouse Chr 1. These observations suggest the intriguing possibility that the genes for mouse and human C-C chemokine receptors are clustered in mouse Chr 9 and human chromosome 3 in homologous regions, respectively, whereas the genes for mouse and human C-X-C chemokine receptors are clustered in regions of conserved synteny of mouse Chr 1 and human chromosome 2, respectively. This is reminiscent of the finding that a cluster of mouse C-C chemokine loci Scya (small inducible cytokine) has been mapped to Chr 11, and the human corresponding region has been mapped to chromosome 17q-11-21.3, where seven known C-C chemokine genes reside.11 Even though the chromosomal location of mouse C-X-C chemokines is not currently known, human C-X-C chemokine is known to be on human chromosome 4.11
Because the numbers of chemokines are still increasing, further characterization of CHEMR1 or related members of this family of chemokine receptors may provide useful information on the specific function of this rapidly expanding family of molecules. The identification of specific C-C chemokines that bind to CHEMR1 is currently under investigation.
NOTE ADDED IN PROOF
CCR4 is reported to be a specific receptor for TARC (thymus and activation-regulated chemokine; Keystone Symposia, Copper Mountain, CO, March 31 to April 5, 1997: The role of chemokine in leukocyte trafficking and disease. [abstr 227]).
ACKNOWLEDGMENT
The authors gratefully acknowledge the valuable comments on this manuscript of Drs Young-June Kim, Beth Ann Garni-Wagner, and Hal E. Broxmeyer. We thank Seung Kim, Ihn-K. Jang, Jon McMahel, and Susan Grigsby for their excellent technical support in the measurement of Ca2+ mobilization using FACS. We thank Dr Philip M. Murphy for sharing information on characteristics of three murine-related C-C chemokine receptors before publication. We also thank Audrey Carson for typing this manuscript.
Supported by National Institutes of Health Grants No. AI-28175, AR-40248, and R03 DE12156 (to B.S.K.).
The nucleotide sequences of the CHEMR1 cDNA have been submitted to the GenBank/EMBL Data Bank with Accession No. U15208.
Address reprint requests to Byoung S. Kwon, PhD, Indiana University School of Medicine, Department of Microbiology and Immunology, 635 Barnhill Dr, Indianapolis, IN 46202-5120.

![Fig. 3. Northern analysis of CHEMR1 mRNA expression. Poly (A+) RNA was isolated from each cell line indicated. Three micrograms of the RNA were electrophoresed on a 1.2% formaldehyde gel, transferred to Quiabrane (Quiagen), and hybridized sequentially with [32P] CHEMR1 and β-actin cDNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/12/10.1182_blood.v89.12.4448/3/m_bl_0032f3.jpeg?Expires=1769105725&Signature=S353~AmDgk~~Fq7PmKUqPicz7O0mmLisp3pYm4eNOvmzfgRqtPJzUoRPewYBqLYbf4z-6v4ZRTsYprIPEvSD4cyTSp59MJp-jy6FDp17euJSrHz5o41~gyql~u6pF1aYcCTxWElfzJ4P4ZyWFueYCiLeFsn4qAgVK9Zz2fib8K77TTZOteCu3B9c2awtdUMP4JpwySdUv2YPyltQ56om-Uf6nSqlDmfHIBkSDtMjtlFSFxoKGsgi5mwo0QBPxLjLeCjwQqmfrtlkXHQNsvsfnM4WJrc6mtUjG2f3G-avmOm384ACz18i9D4KXvjNnx0PjXegZvsJm15AQToHwl7PLQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Genomic analysis of CHEMR1 gene. Evidence for existence of the human homologue of CHEMR1 and for CHEMR1 being intronless. (A) 129/J genomic DNA (20 μg) and human genomic DNA prepared from Mo7e cells were digested with the restriction enzymes indicated, run on a 1% agarose gel, transferred to Quiabrane, and hybridized with [32P] CHEMR1 ORF. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blots were exposed to XAR-5 film in cassettes at −70°C for 6 days. DNA molecular standards from BRL were run in parallel. The CHEMR1 cDNA has two Kpn I (−26 and 840) and HincII (580 and 1,110) sites, which release around 867 and 531 bp, respectively. These bands are indicated by arrows. (B) One microgram of 129/J genomic DNA or OCM1 genomic DNA was amplified with Pfu polymerase in the presence of 50 pmol of each of the corresponding primers as indicated in the Materials and Methods. After 30 cycles of amplification, one-tenth volume of PCR reaction mixture was resolved on a 1% agarose gel. Indicated with arrows are amplified ORFs of chemokine receptors. The same molecular standards were used as in (A).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/12/10.1182_blood.v89.12.4448/3/m_bl_0032f4.jpeg?Expires=1769105725&Signature=38p1T5wq0ur3d~v4sukxkDtcQh7kWRVVMHqvPmRu6SIW7cKKwoOSwPzF2RTFmmU0crVZB2hYoKgFsfVH~T5xz2lVQk6tGiC~~BQzTxuZ3IwTA77omESYp9n4w0owcfDOG~gbxRKkQi8l0QgQn7kvQpHRlrB4UGQtkN7IJhBDj0JqkweD2PF9bX7kjzAB92yJnslS4xy31YLfkwZ8jO85Bnsu3WS1waEYx-hsBIjhRCx0udWSzQg18wfYyrYW7ZEkMEqqIXsxJOMljnpcN1z92cnr1vDfEQJRA5FH1UQpuqsehfbhM9oB-tBK3BzbFWMy0zDlSt3b-2hHyBMPBNpCog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Northern blot analysis of HEK293 cells transfected with vector (pRc/CMV), the antisense CHEMR1, sense CHEMR1, and CCR-1 in pRc/CMV. HEK293 cells were transfected with 20 μg of each DNA using the modified calcium phosphate method. After 24 hours, total RNA was isolated with Tri-reagent (Molecular Research Center, Cincinnati, OH). Twenty micrograms of the RNA was run on a 1.2% formaldehyde gel, blotted to Quiabrane, and hybridized with [32P] CHEMR1 cDNA. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blot was exposed to XAR-5 film in a cassette at −70°C for 2 days. The same filter was stripped and subjected to sequential hybridization with [32P] CCR-1 and [32P] rat GAPDH probe, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/12/10.1182_blood.v89.12.4448/3/m_bl_0032f6.jpeg?Expires=1769105725&Signature=SL2tx1rImTdLfdC9IU0oHDq3hUD6e63SwZtv2ba8BzrNAR0e0v0DLKGEhMigOloDc4bqHyDEXCtgJC1jWrhx5bB-tZq4Fo0JfC99W8MNIou~CudEMENA3apqyNhGuKAfw6yRne~nMUtU8TkFE7gWaOmIKT3AB95rl6Gv~bb4H-paR~KyObJxD8WaTWBXIoXAqTJ5RZ6U68vI~SkDPtAfl5HicGYq4fMGtcumWC62FSpka0Lo0YTfV6GnH-VpvKjRkuU2~arfu5iQ~jSr4m76ckT8VWeHmQdTHhbUP76ZsZ6W5S6xSsQH1X8qqxE0WTOEZyITgPSORWBv5DSVV5K5aA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 3. Northern analysis of CHEMR1 mRNA expression. Poly (A+) RNA was isolated from each cell line indicated. Three micrograms of the RNA were electrophoresed on a 1.2% formaldehyde gel, transferred to Quiabrane (Quiagen), and hybridized sequentially with [32P] CHEMR1 and β-actin cDNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/12/10.1182_blood.v89.12.4448/3/m_bl_0032f3.jpeg?Expires=1769195806&Signature=PLhUSSnLp~Es4J~gLoUlBYi3bMGekkHAGAybPdFOtK08gP5CZn4K5yHrqsTYAvWfTKtoEmALT7EFujuXs9vf~E1djndetDG9JwPzb53goG81BAqYfdPgPu1tNhRcEcS-MfybisXKAK-pmqbqWSVvP6Xqtls07AfuOTfd4CMO4uaBHg1Lz~RP09Lut4Dd69y4JOM0PgMa6HOJ7q4qzZFpO7OomeFL~9SyODVpSYgc18e2Q169pP0GcH~DXVR7TGnYQdD5ABKp2BTbFRiJp0By1Pevo9ZYrJY7TpK7FJznAKLjKNNPFvHcTgS~hICPrSD-vokwJFm54D44sHxv5739Ng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Genomic analysis of CHEMR1 gene. Evidence for existence of the human homologue of CHEMR1 and for CHEMR1 being intronless. (A) 129/J genomic DNA (20 μg) and human genomic DNA prepared from Mo7e cells were digested with the restriction enzymes indicated, run on a 1% agarose gel, transferred to Quiabrane, and hybridized with [32P] CHEMR1 ORF. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blots were exposed to XAR-5 film in cassettes at −70°C for 6 days. DNA molecular standards from BRL were run in parallel. The CHEMR1 cDNA has two Kpn I (−26 and 840) and HincII (580 and 1,110) sites, which release around 867 and 531 bp, respectively. These bands are indicated by arrows. (B) One microgram of 129/J genomic DNA or OCM1 genomic DNA was amplified with Pfu polymerase in the presence of 50 pmol of each of the corresponding primers as indicated in the Materials and Methods. After 30 cycles of amplification, one-tenth volume of PCR reaction mixture was resolved on a 1% agarose gel. Indicated with arrows are amplified ORFs of chemokine receptors. The same molecular standards were used as in (A).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/12/10.1182_blood.v89.12.4448/3/m_bl_0032f4.jpeg?Expires=1769195806&Signature=kN7lKFtsmwbdqb1erodwvhhKYAGE83M6acOTBWfjjyniQuosxbY0wSH5QI9yic1VX4UM~X7tzPXB7qgCS49T7WX-YdXLY~VyhW0bbnPUKE-vkgDhzg0Vj9EZLGkPTdYpjebVft~lKelXNRfTFCMgT~EmbbTRyht64~~hL55IeqCQ8WYIH8OkXj5xn9IUATVP1Y2~xR8ZceaNC4vxTXgj8y68XYB-WiHLlIzRa6LySalQpa3~i8wNBM8FjBbcXdRfoHziF29Xff2ncEBLFFOZCh6g-gklVYOu44AjuIP0S5Lo5VCVy0btw88X8Gex4X32JsVI9s6VSkghHDTnl2NcYw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Northern blot analysis of HEK293 cells transfected with vector (pRc/CMV), the antisense CHEMR1, sense CHEMR1, and CCR-1 in pRc/CMV. HEK293 cells were transfected with 20 μg of each DNA using the modified calcium phosphate method. After 24 hours, total RNA was isolated with Tri-reagent (Molecular Research Center, Cincinnati, OH). Twenty micrograms of the RNA was run on a 1.2% formaldehyde gel, blotted to Quiabrane, and hybridized with [32P] CHEMR1 cDNA. The filter was washed finally with 0.5× SSC, 0.1% SDS at 65°C, and the blot was exposed to XAR-5 film in a cassette at −70°C for 2 days. The same filter was stripped and subjected to sequential hybridization with [32P] CCR-1 and [32P] rat GAPDH probe, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/12/10.1182_blood.v89.12.4448/3/m_bl_0032f6.jpeg?Expires=1769195806&Signature=aR3reXG3if~8SbbTELWpDdMUb1HC4GiJ5qeaQpNJ3fRzc7TGgmgZdEMbtuOVL3mbIa99pHhFCiUEeshXsJ3h02PrutNMIbbZZsyLfpzh4LPW4tSX76sXB45Di~OPUy-ouWzUm0VBuhEsdigCft6rbShVbfMPgY2Yx4r9bwFjEZsPECdz4x2mEQPi5xoaJkKH-7boDc8o-s9dKQiC58WZBbN4Evtap7PC8LVu4ruU5HyWpfqWWerJWge2Px3iwtURgCu~jwLod2QGNKqhcszyZlHFsi59EMzQ7jAluVhM6ayybPS6ACG5v-OMxsr~TDFMR3txnW7Z-R8dZDJsHc9GtA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)