Abstract
Polymorphic structures of the neutrophil Fcγreceptor IIIb (FcγRIIIb) result in alloantibody formation that causes alloimmune neonatal neutropenia and transfusion reactions. Alloantigens located on FcγRIIIb include the antigens NA1 and NA2. In four cases of alloimmune neonatal neutropenia, granulocyte-specific alloantibodies directed against a thus far unknown antigen were detected by granulocyte agglutination and immunofluorescence tests in the maternal sera. By the use of the monoclonal antibody–specific immobilization of granulocyte antigens (MAIGA) assay, the new antigen, termed SH, was located on the FcγRIIIb. Nucleotide sequence analysis of the FcγRIIIb coding region from a SH(+) individual showed a single-base C→A mutation at position 266, which results in an Ala78Asp amino acid substitution. A family study confirmed that this nucleotide difference is inherited, and corresponds to the SH phenotype. Serologic typing of 309 randomly selected individuals showed an antigen frequency of 5% in the white population. The same frequency was found by genotyping, for which a technique based on polymerase chain reaction (PCR) using sequence-specific primers (PCR-SSP) was developed. Typing of all SH(+) individuals for NA1 and NA2, and PCR-restriction fragment length polymorphism analysis of the NA-specific PCR products from five SH(+) individuals using the SH-specific endonuclease SfaN I showed that SH antigen is very probably the result of an additional mutational event in the NA2 form of the FcγRIIIB gene. Immunochemical studies also demonstrated that the SH determinants reside on the 65- to 80-kD NA2 isoform of the FcγRIIIb. Our findings show the existence of an additional polymorphism of the FcγRIIIb, which can result in alloantibody formation causing alloimmune neonatal neutropenia.
THE NEUTROPHIL Fcγreceptor IIIb (FcγRIIIb = CD16) is a low-affinity receptor for the Fc region of complexed, but not monomeric IgG antibodies. FcγRIIIb preferentially removes small immune complexes from the circulation.1 The FcγRIIIb is released from the neutrophil membrane during apoptosis, ie, programmed cell death,2 and soluble FcγRIIIb can inhibit proliferation and immunoglobulin production of stimulated B lymphocytes.3
Biochemical characterization has identified the FcγRIIIb as an extensively glycosylated glycosylphosphatidylinositol (GPI)-anchored glycoprotein with two extracellular regions composed of disulfide bonds.4,5 The gene for the FcγRIIIb is mapped to the long arm of chromosome 1 (band q22) and belongs to the immunoglobulin superfamily.6
In addition to the functional significance of the FcγRIIIb, immunologic studies have shown that FcγRIIIb is polymorphic, bearing the important neutrophil-specific NA1 and NA2 alloantigens.7 The NA antigens are involved in alloimmune neonatal neutropenia, autoimmune neutropenia, and transfusion-related acute lung injury.8-10 The NA isoforms of the FcγRIIIb differ in their relative molecular weight: 50 to 65 kD for FcγRIIIbNA1 versus 65 to 80 kD for FcγRIIIbNA2.11 The different molecular weights have been attributed to different glycosylation patterns12,13: two more N-linked glycosylation sites in the NA2 than in the NA1 isoform. The additional glycosylation sites result from two of the four amino acid differences between the NA isoforms. On the DNA level, the NA1 and NA2 alleles differ in five nucleotides, one of which is a silent mutation.12,13 Approximately 0.1% of the European population does not express the FcγRIIIb on their neutrophils, and as a consequence do not display the NA1 and NA2 antigens.14 This phenotype is called NA-null and is caused by a FcγRIIIB gene deficiency.15
In this study, we characterized the biochemical and molecular basis of a thus far unknown polymorphism of FcγRIIIb that has been identified by granulocyte-specific alloantibodies in the maternal sera from four cases of alloimmune neonatal neutropenia. This disease is caused by placental transfer of maternal granulocyte alloantibodies to the fetus during pregnancy.
CASE REPORTS
Case report no. 1.This case has been reported elsewhere,16 but in brief, a 34-year-old woman gave birth to a full-term baby boy. The mother had two prior pregnancies and one other child. The pregnancy and delivery were uncomplicated, but on the fourth day of life, the baby developed erythema around the umbilicus and at the site of circumcision. His white blood cell count was 6.9 × 109/L with 2% bands and 1% polymorphonuclear cells. The child was treated with antibiotics and he was given a single dose of intravenous immunoglobulin. His absolute neutrophil count increased to 2.72 × 109/L and the infection resolved. However, 2 weeks later his neutrophil count decreased to 0.92 × 109/L, but returned to normal 2 weeks later.
Case report no. 2.The healthy mother had eight previous pregnancies and three other children alive. At 31 weeks of gestation, she gave birth to a neonate with a body weight of 1,680 g. The neonate suffered from immediate severe respiratory distress. The neutrophil count at birth was 0.6 × 109/L (white blood cell count, 2.3 × 109/L); red blood cell and platelet counts were normal. Bone marrow examination showed no abnormalities. The neutrophil count decreased to less than 0.2 × 109/L and the neonate suffered from pulmonary infections. Severe neutropenia persisted until the child died as a result of a cerebral hemorrhage 19 days after birth.
Case report no. 3.The healthy mother had two previous in utero fetal deaths due to abruptio placentae and one spontaneous abortion. The child was delivered by cesarean section at the 34th week of pregnancy. Moderate respiratory distress made mechanical ventilation necessary for 4 days. Blood counts performed on days 1 and 3 of life showed severe neutropenia (<0.1 × 109/L). After treatment with high-dose intravenous immunoglobulin on the third day of life, the neutrophil count increased to 6 × 109/L and neutropenia did not reappear again.
Case report no. 4.The healthy mother had one other child who had no history of neonatal neutropenia or infection. Delivery of the baby was at term. On day 4 of life, the baby developed staphylodermia. The white blood cell count was 6 × 109/L, but no neutrophils were detectable. Red blood cell and platelet counts were normal. Bone marrow examination showed only a slight decrease of mature neutrophils. Despite prophylactic treatment with antibiotics, additional infections such as dermatitis, parotitis, lymphadenitis, and otitis media occurred, which resolved after 1 month, when the neutrophil count had increased.
MATERIALS AND METHODS
Sera.The sera analyzed in this study were from the four mothers described in the reported cases. The sera were numbered 1 to 4 according to the case report numbers. Antigen characterization was performed using serum no. 1.
Human NA-specific typing sera and FcγRIIIb-specific isoantibodies were obtained from immunized mothers of other cases of alloimmune neonatal neutropenia and from an immunized individual with FcγRIIIb deficiency (NA-null), respectively.
Granulocyte antibody screening.Granulocyte immunofluorescence (GIFT) and agglutination (GAT) tests were performed as follows. Granulocytes were isolated from heparin- (GIFT) or EDTA- (GAT) anticoagulated blood of healthy donors. After dextran sedimentation, granulocytes were isolated from the supernatant leukocyte-rich plasma by Ficoll-Hypaque gradient centrifugation and red blood cells were lysed with ammonium chloride solution. For GIFT, neutrophils were fixed with 1% paraformaldehyde and incubated for 30 minutes at 37°C with the sera to be tested in microtiter plates. After washing, the cells were incubated with fluorescein isothiocyanate (FITC)-labeled rabbit F(ab′)2-antihuman IgG (light plus heavy chain) (Dako, Hamburg, Germany), rewashed, and evaluated with a fluorescence microscope. The GAT assay was performed in Terasaki trays. Two microliters of granulocytes was incubated under oil with 2 μL of serum for 120 minutes at 37°C and then read microscopically.
To exclude HLA- or other lymphocyte-reactive antibodies, serum no. 1 was also tested against lymphocytes obtained from the granulocyte donors in a lymphocytotoxicity test (LCT) and lymphocyte immunofluorescence test (LIFT). The lymphocytes were isolated by density gradient centrifugation and the LCT was performed according to National Institutes of Health standards. The LIFT was performed according to the GIFT, but lymphocytes were used instead of granulocytes.
Phenotyping of granulocytes and antigen location.NA and SH phenoytpings were performed by GIFT and GAT and the antigen-specific monoclonal antibody (MoAb)-specific immobilization of granulocyte antigens (MAIGA) assay17 using human NA-specific typing sera and serum no. 1. The MAIGA assay was also used for antigen location. Briefly, 1 × 106 neutrophils from one individual fixed with 1% paraformaldehyde were incubated (30 minutes, 37°C) with human serum and MoAbs. In different reaction mixtures, we used the MoAbs 3G8 (Immunotech, Hamburg, Germany) and Bw 209/2 (Behringwerke, Marburg, Germany), which are specific for the FcγRIII (CD16), and the MoAbs 2E1, Bear 1, BL5, and J3D3 (Immunotech), which are specific for the FcγRII (CD32), the subunits of the leukocyte adhesion molecule CD11b/CD18 and the C3b complement receptor I (CR1, CD35). The cells were washed and solubilized by adding 100 μL of lysis buffer (1% Triton-X 100, 5 mmol/L EDTA, 2 mmol/L phenylmethylsulfonyl fluoride [PMSF], 0.5 μg/mL leupeptin, 500 kallikrein inactivating units (KIE)/mL aprotinin in 20 mmol/L Tris-buffered saline, pH 7.4) for 30 minutes at room temperature. After sonication (2 minutes) and centrifugation at 15,000g for 30 minutes, the supernatant of each reaction mixture was transferred to a separate tube coated with goat antimouse antibodies. Unattached antibodies were removed by washing, and goat antihuman IgG (heavy plus light chain) antibodies conjugated with peroxidase were added. After washing and subsequent addition of a substrate containing luminol, hydrogen peroxide, and 4-iodophenol, the emitted light (chemiluminescence) was measured over a period of 15 minutes in a luminometer (Lumat LB 9501, Berthold, Wildbad, Germany).
Immunochemical studies.These studies were performed by (lumino-)immunopreciptation and immunblot. For luminoimmunoprecipitation, unfixed granulocytes (1 × 108/mL in phosphate-buffered saline [PBS]) were biotinylated with 5 mmol/L of a water-soluble biotin analog with an extended spacer arm (NHS-LC-Biotin; Pierce, Rockford, IL) for 30 minutes on ice. After washing, to each 100 μL of cell suspension (1 × 107/mL in PBS), 100 μL of serum or MoAb solution (0.01 mg/mL) was added and incubated for 30 minutes at 37°C. The cells were washed and solubilized by adding 250 μL of lysis buffer (see MAIGA assay) for 30 minutes at room temperature. After sonication (3 minutes) and centrifugation at 15,000g for 30 minutes, the supernatants were incubated with rabbit antihuman IgG or antimouse immunoglobulin (Dako) antibodies coupled to Protein A-Sepharose 4B (Pharmacia, Freiburg, Germany) for 2 hours at room temperature. To couple the antibodies to the beads, 50 μL of Protein A-Sepharose CL-4B was preincubated with 50 μL rabbit antimouse immunoglobulin or antihuman IgG in 200 μL Tris-buffered saline, pH 8.0, for 45 minutes at room temperature. The Protein A-Sepharose CL-4B beads were then washed and resuspended in sodium dodecyl sulfate polyacrylamide gel electropheresis (SDS-PAGE) sample buffer, boiled for 3 minutes, and then subjected to 10% SDS-PAGE. After electropheresis, proteins were transferred onto nitrocellulose (Hibond C; Amersham, Little Chalfont, UK; 50 V for 6 hours using Tris/glycine transfer buffer). For visualization, the nitrocellulose was first blocked with 1.5% bovine serum albumin (BSA) and 0.05% Tween-20 in Tris-buffered saline (TBS) and then incubated with streptavidin conjugated to peroxidase. Unbound streptavidin was washed out, and the nitrocellulose was incubated with a chemiluminescent substrate (ECL Western Blotting Detection System; Amersham) and then exposed to x-ray films for 0.5 to 4 minutes.
For luminoimmunoblotting, 5 × 107 unfixed granulocytes were solubilized with 500 μL lysis buffer (see MAIGA assay). The lysate was then centrifuged (30 minutes, 15,000g) and the supernatant analyzed by SDS-PAGE and transferred to a nitrocellulose membrane. Membrane strips were blocked with 1.5% BSA and then incubated with diluted serum samples (1:20 to 1:200). The strips were washed with Tris-buffered saline pH 7.4 containing 0.05% Tween 20 and then incubated with peroxidase-conjugated rabbit antihuman antibodies (dilution, 1:200,000). After another washing step, a chemiluminescent substrate (ECL) was added for 0.5 to 4 minutes and the recognized antigens visualized on x-ray film.
Isolation and amplification of granulocyte mRNA.Granulocyte mRNA was isolated from EDTA-anticoagulated blood of NA- and SH-phenotyped donors as previously described.18 Ten-microliter aliquots of granulocyte mRNA were heated to 68°C for 10 minutes and quickly cooled on ice before reverse transcription. cDNA was then synthesized using 10 μmol/L oligo dT, 40 U RNAsin (Boehringer Mannheim, Mannheim, Germany), 2 mmol/L of each dNTP (Pharmacia), 500 U of cloned Moloney murine leukemia virus reverse transcriptase, and 5x enzyme buffer (GIBCO-BRL, Eggenstein, Germany) in a total volume of 30 μL. cDNA synthesis was performed at 40°C for 45 minutes and was stopped by chilling to 0°C.
The FcγRIIIB-specific primers used to amplify the entire coding region of the granulocyte FcγRIIIB based on the published sequence and numbering system of Ravetch and Perussia.13 Three microliters of cDNA was mixed with 5 μL of 10× polymerase chain reaction (PCR) buffer, 0.5 μmol/L of sense primer no. 1 (5′-1TCTTTGGTGACTTGTCCA18-3′) and 0.5 μmol/L of antisense primer no. 2 (5′-831GCCACTGCTCTTATTACT814-3′), 200 μmol/L of each dNTP, 0.2 mg/mL BSA, 4 mmol/L MgSO4, and 1 U of Deep-Vent DNA polymerase (New England Biolabs, Schwalbach, Germany), and were amplified on a DNA thermal cycler (Biometra, Göttingen, Germany) for 35 cycles. Each cycle consisted of denaturation at 97°C for 20 seconds, annealing at 52°C for 40 seconds, and primer extension at 72°C for 90 seconds. Five microliters of the PCR products (dilution 1:10) was amplified again for 39 cycles using nested primer pair no. 3 and no. 4 (5′-41AGCTGCTCCTCCCAACTG58-3′ and 5′-393CTCCTTGAACACCCACCG376-3′) or primer pair no. 5 and no. 6 (5′-309CCTCTCCACCCTCAGTGA326-3′ and 5′-655GGTACCCAGGTGGAGAGA638-3′) under the following conditions: denaturation at 95°C for 30 seconds, annealing at 57°C for 50 seconds, and primer extension at 73°C for 50 seconds. In the final cycle, the samples were kept at a temperature of 72°C for 10 minutes and then chilled to 4°C.
Analysis of PCR products.Five microliters of the PCR products was analyzed on 1.5% agarose gel containing ethidium bromide. The amplified DNA was isolated from the gel, purified by Geneclean (Dianova, Hamburg, Germany), flushed using Klenow DNA polymerase, and finally subcloned into the EcoRV site of the pGEM5 plasmid (GIBCO-BRL). Plasmid from positive clones was sequenced by the dideoxy chain termination reaction method using Sequenase 2.0 (United States Biochemical, Braunschweig, Germany).
In some cases, amplified cDNA was subjected to RFLP analysis using SfaN I endonuclease (New England Biolabs) and analyzed on Separide gel (GIBCO-BRL).
Genotyping by PCR with sequence-specific primers.For genotyping of NA antigens, a slightly modified PCR with sequence-specific primers (PCR-SSP) technique was used as previously described.19 Briefly, genomic DNA isolated from EDTA-anticoagulated blood was amplified by PCR using a thermal cycler with 2 U Taq DNA polymerase. The PCR reaction consists of 30 cycles (denaturation: 98°C/30 seconds; annealing: 57°C/1 minute; extension: 71°C/30 seconds; final extension: 71°C/5 minutes). The NA-specific primers were designed as sense primers and were situated at position 208 to 227 for NA1 and at position 130 to 147 for NA2. To enhance the specificity of the NA1 primer, at position 4 from the 3′ end the correct nucleotide A was substituted by a T. The nonspecific antisense primer is situated at position 331 to 348. As internal positive PCR control, two primers (HGH-I and HGH-II, see below) amplifying a 439-bp fragment of the human growth hormone gene (HGH) were used. After electropheresis in 1.6% agarose gel and staining with ethidium bromide, the PCR products were visualized by UV illumination and photographed.
Genotyping of the SH antigen.2.5 microliters of genomic DNA was amplified in a total volume of 22.5 μL using 0.5 μmol/L SH(+) sequence-specific antisense primer no. 7 (5′-285ACTGTCGTTGACTGTGTCAT 266-3′) or the antithetical SH(−) sequence-specific antisense primer no. 8 (5′-285ACTGTCGTTGACTGTGTCAG266-3′), 0.5 μmol/L sense primer no. 9 (5′-95AAGATCTCCCAAAGGCTGTG115-3′), 200 μmol/L of each dNTP, 2.25 μL 10× PCR buffer, and 0.8 U Taq polymerase on a DNA thermal cycler (Perkin Elmer, Weiterstadt, Germany) for 30 cycles. To enhance the specificity of the primers, the correct nucleotide G at position 269 was substituted by a T. Coamplification of the HGH gene using 0.125 μmol/L HGH I primer (5′-CAGTGCCTTCCCAACCATTCCCTTA-3′) and 0.125 μmol/L HGH II primer (5′-ATCCACTCACGGATTTCTGTTGTGTTTC-3′) was run as internal control. Each cycle consisted of denaturation at 95°C for 30 seconds, annealing at 60°C for 1 minute, and primer extension at 71°C for 30 seconds.
RESULTS
Antibody screening in serum no. 1.Serum no. 1 has been reported to contain an alloantibody that is exclusively detectable by GIFT and is directed against the leukocyte antigen SL, which was found to be expressed on lymphocytes and neutrophils with a frequency of 66%.16 Since positive reactions in the GAT with few donor neutrophils have also been reported, we again screened serum no. 1 in GAT against a panel of eight granulocytes typed for the alloantigens NA1, NA2, NB1, MART, and 5b. The serum reacted with the neutrophils from one (donor no. 1 in Table 1) of the eight donors. The pattern of reaction did not correlate with any antigen for which the panel cells were typed. Negative results of serum no. 1 in LIFT and LCT using the cells of donor no. 1 excluded that the positive reaction in GAT was due to HLA or other lymphocyte-reactive antibodies such as anti-SL. Testing of the serum with neutrophils from 12 additional individuals in GAT was also negative. These results indicated that the serum no. 1 contained in addition to anti-SL a granulocyte-specific alloantibody directed against a new low-frequency antigen, which was termed SH.
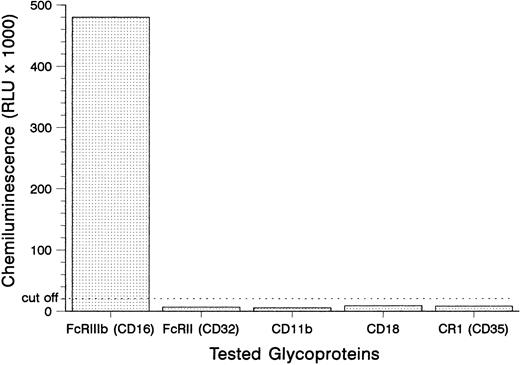
Antigen identification.To locate the antigen recognized by the SH antibody, we tested its binding to the isolated glycoproteins of the granulocytes from donor no. 1 in the MAIGA assay (Fig 1). The SH antibody only reacted with the FcγRIIIb (CD16), but not with the FcγRII (CD32), the subunits of the leukocyte adhesion molecule CD11b/CD18 and the C3b complement receptor CR1 (CD35). These results demonstrated that the SH alloantigen resides on the FcγRIIIb.
Detection of SH on the FcγRIIIb using the MAIGA assay. Neutrophils from a SH(+) donor were tested with the anti–SH-containing serum no. 1 for antibodies to FcγRIIIb, FcγRII, the subunits of the leukocyte adhesion molecule CD11b/CD18, and the C3b complement receptor CR1 (CD35).
Detection of SH on the FcγRIIIb using the MAIGA assay. Neutrophils from a SH(+) donor were tested with the anti–SH-containing serum no. 1 for antibodies to FcγRIIIb, FcγRII, the subunits of the leukocyte adhesion molecule CD11b/CD18, and the C3b complement receptor CR1 (CD35).
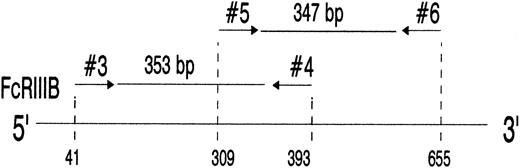
Analysis of the FcγRIIIB mRNA from SH(+) and SH(−) individuals.To elucidate the molecular basis of SH, we analyzed the nucleotide sequence of the entire coding region of the FcγRIIIB. Granulocyte mRNA was sequentially amplified by reverse-transcription PCR (RT-PCR) using three sets of primers. After FcγRIIIB cDNA amplification in the primary PCR with primers no. 1 and no. 2, secondary PCRs were performed (Fig 2) using either nested primers no. 3 and no. 4 or primers no. 5 and no. 6 to amplify overlapping regions encompassing nucleotides 41 to 393 (353 bp) and nucleotides 309 to 655 (347 bp), respectively. Both PCR products were subcloned and sequenced.
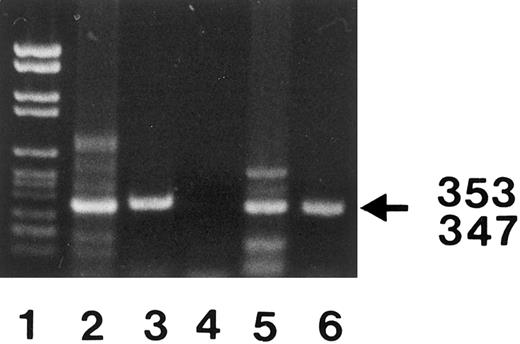
PCR amplification of nucleotides 41 to 393 (lanes 3 and 6) and 309 to 655 (lanes 2 and 5) of neutrophil FcγRIIIB mRNA. The locations of the 2 primer pairs (arrows) used for PCR amplification of the expected 353- and 347-bp products after nested PCR are illustrated above. cDNA derived from a SH(−) (lanes 2 and 3) and a SH(+) individual (lanes 5 and 6) were amplified with primer pair no. 3 and no. 4 (lanes 3 and 6) or primer pair no. 5 and no. 6 (lanes 2 and 5) and analyzed on 1.5% agarose gel stained with ethidium bromide. PCR without template was run as negative control (lane 4). DNA size standards (pBr 328 DNA.BglI + pBr 328 DNA.HinfI) are shown in lane 1. The resulting 353-bp and 347-bp products from the SH(+) individual (lanes 5 and 6) were isolated from preparative gels and subcloned for nucleotide sequence analysis.
PCR amplification of nucleotides 41 to 393 (lanes 3 and 6) and 309 to 655 (lanes 2 and 5) of neutrophil FcγRIIIB mRNA. The locations of the 2 primer pairs (arrows) used for PCR amplification of the expected 353- and 347-bp products after nested PCR are illustrated above. cDNA derived from a SH(−) (lanes 2 and 3) and a SH(+) individual (lanes 5 and 6) were amplified with primer pair no. 3 and no. 4 (lanes 3 and 6) or primer pair no. 5 and no. 6 (lanes 2 and 5) and analyzed on 1.5% agarose gel stained with ethidium bromide. PCR without template was run as negative control (lane 4). DNA size standards (pBr 328 DNA.BglI + pBr 328 DNA.HinfI) are shown in lane 1. The resulting 353-bp and 347-bp products from the SH(+) individual (lanes 5 and 6) were isolated from preparative gels and subcloned for nucleotide sequence analysis.
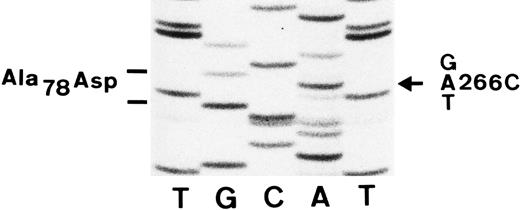
Comparison of the 353-bp nucleotide sequence of the mRNA derived from a SH(+) individual and a SH(−) individual (donors no. 1 and no. 6 in Table 1) showed a single nucleotide difference at base 266. The mRNA from the SH(+) individual had an A at this position (Fig 3), whereas that from the SH(−) individual had a C (data not shown). This change resulted in the substitution of an aspartic acid for an alanine at amino acid residue 78.
Nucleotide sequence analysis of amplified cDNA from an SH(+) individual. The region of the autoradiograph shown here includes the sequence from base 257 to 275. The nucleotide change from a C to an A (arrow) at position 266 in the SH(+) individual results in an alanine (GCT) to an aspartic acid (GAT) amino acid substitution at residue 78 of the FcγRIIIb polypeptide.
Nucleotide sequence analysis of amplified cDNA from an SH(+) individual. The region of the autoradiograph shown here includes the sequence from base 257 to 275. The nucleotide change from a C to an A (arrow) at position 266 in the SH(+) individual results in an alanine (GCT) to an aspartic acid (GAT) amino acid substitution at residue 78 of the FcγRIIIb polypeptide.
Analysis of the 353-bp insert obtained from four different clones from SH(+) donor no. 1 showed that the clones had either an A (two clones) or a C (two clones) at position 266, indicating that the SH-associated mutation was present only in one of the FcγRIIIB genes. In addition, all of these clones showed the nucleotides C, T, G, A, and A at the positions 141, 147, 227, 277, and 349, respectively corresponding to the NA1(−),NA2(+) phenotype of donor no. 1 (Table 1). No other nucleotide differences between the SH(+) versus SH(−) individuals were found in the 347-bp insert.
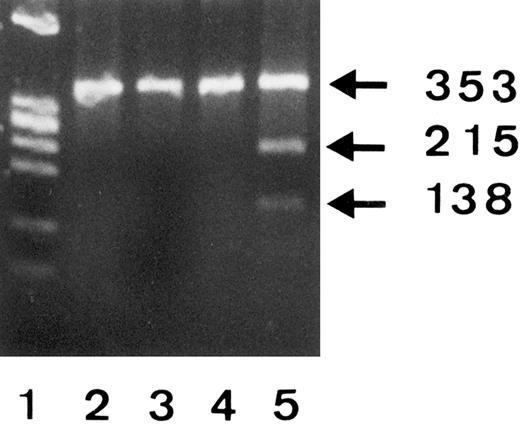
To confirm the C266A polymorphism, we amplified mRNA derived from three different SH(−) individuals and the SH(+) phenotyped donor no. 1 and analyzed the 353-bp products by RFLP using SfaN I endonuclease (Fig 4). Since SfaN I enzyme cleaves the 5′-GATGC-3′ but not 5′-GCTGC-3′ nucleotide sequences, restriction fragments (215 bp and 138 bp) were only obtained from the SH(+) donor no. 1 (lane 5), but not from the SH(−) individuals (lanes 2 through 4). The presence of an undigested 353-bp band in lane 5 was consistent with the observation that only one FcγRIIIB gene of donor no. 1 showed the SH mutation.
RFLP analysis of 353-bp PCR products from 3 SH(−) (lanes 2, 3, and 4) and one SH(+) (lane 5) phenotyped individuals using SfaN I endonuclease and 2% Separide gel stained with ethidium bromide. DNA size standards (PhiX174 RF DNA.HaeIII) are shown in lane 1.
RFLP analysis of 353-bp PCR products from 3 SH(−) (lanes 2, 3, and 4) and one SH(+) (lane 5) phenotyped individuals using SfaN I endonuclease and 2% Separide gel stained with ethidium bromide. DNA size standards (PhiX174 RF DNA.HaeIII) are shown in lane 1.
Inheritance of SH.To investigate whether the SH polymorphism was inherited from the parents of donor no. 1, her family members were phenotyped for SH using the MAIGA assay and genotyped by RFLP analysis using the SfaN I endonuclease. The neutrophils from her father and sister were typed SH(−), but the maternal cells were SH(+) (data not shown). These results showed that the SH antigen had been passed down from the mother to her daughter and that the presence of the mutation at 266 was associated with the SH phenotype.
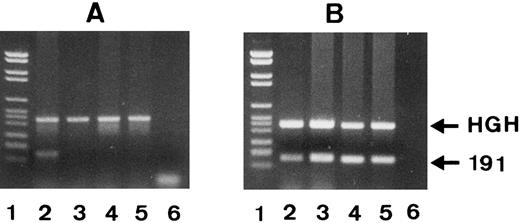
Genotyping by PCR-SSP.Since phenotyping requires a large number of freshly isolated neutrophils, we developed a technique based on PCR-SSP for genotyping of genomic DNA. We constructed the SH(+)-specific primer no. 7 with a T at the 3′ end and the SH(−)-specific primer no. 8 with a G at the 3′ end. As shown in Fig 5A, PCR-SSP with SH-specific primer no. 7 and genomic DNA from an SH(+) individual resulted in an SH-specific 191-bp PCR product (lane 2), whereas this primer failed to amplify genomic DNA from three SH(−) individuals (lanes 3 through 5). The 439-bp fragment of the HGH gene represents the internal PCR control. As shown in Fig 5B, the SH(−)-specific primer no. 8 amplified genomic DNA from all individuals, the SH(+) but heterozygous donor no. 1 (lane 2), and the three SH(−) individuals (lanes 3 through 5).
PCR-SSP analysis of 1 SH(+) (lane 2) and 3 SH(−) phenotyped individuals (lanes 3 to 5) using SH(+)-specific primer no. 7 (A) and SH(−)-specific primer no. 8 (B). The 191-bp bands represent the allele-specific PCR products and the HGH bands the internal controls. PCR products were analyzed on 1.5% agarose gel stained with ethidium bromide. DNA size standards (pBr 328 DNA.BglI + pBr 328 DNA.HinfI) are shown in lane 1. Only in donor 1 was a SH(+)-specific PCR product (A, lane 2) formed.
PCR-SSP analysis of 1 SH(+) (lane 2) and 3 SH(−) phenotyped individuals (lanes 3 to 5) using SH(+)-specific primer no. 7 (A) and SH(−)-specific primer no. 8 (B). The 191-bp bands represent the allele-specific PCR products and the HGH bands the internal controls. PCR products were analyzed on 1.5% agarose gel stained with ethidium bromide. DNA size standards (pBr 328 DNA.BglI + pBr 328 DNA.HinfI) are shown in lane 1. Only in donor 1 was a SH(+)-specific PCR product (A, lane 2) formed.
Four additional SH(+) individuals were found by genotyping of 107 unrelated blood donors. Neutrophils from these four donors were found to have the SH(+) phenotype using serum no. 1 in the MAIGA assay, suggesting an antigen frequency of approximately 4%. These results confirmed that the C266A polymorphism of the FcγRIIIB gene is associated with the expression of the SH antigen.
Relation between SH and NA2.Since the FcγRIIIb shows the NA polymorphism, we investigated whether the SH-specific mutation is associated with the NA1- or NA2-specific nucleotide substitutions. For this, we used the fact that the SH-associated mutation site is located within the DNA segment that is amplified by our NA-specific primers in the PCR-SSP technique, and subjected the NA-specific PCR products from five SH(+) donors to RFLP analysis using the SH-specific endonuclease SfaN I.
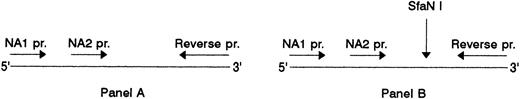
Association of SH with NA2. (A) NA1(−) (lanes 2 to 4) and NA2(−) (lanes 5 to 7) specific PCR products before and, (B), after digestion with SfaN I. The locations of the NA-specific sense primers (NA1 pr., NA2 pr.) and the reverse primer (Reverse pr.) used for PCR amplification of the NA-specific DNA fragments, as well as the cleavage site of the SfaN I enzyme, are illustrated. Restriction fragments were analyzed on 1.5% agarose gel using DNA size standards pBr 328 DNA.BglI + pBr 328 DNA.HinfI (lane 1). Three individuals were tested who were typed as follows: SH(+), NA1(+),NA2(+) (lanes 2 and 5); SH(+), NA1(−),NA2(+) (lanes 3 and 6); SH(−), NA1(+),NA2 (+) (lanes 4 and 7).
Association of SH with NA2. (A) NA1(−) (lanes 2 to 4) and NA2(−) (lanes 5 to 7) specific PCR products before and, (B), after digestion with SfaN I. The locations of the NA-specific sense primers (NA1 pr., NA2 pr.) and the reverse primer (Reverse pr.) used for PCR amplification of the NA-specific DNA fragments, as well as the cleavage site of the SfaN I enzyme, are illustrated. Restriction fragments were analyzed on 1.5% agarose gel using DNA size standards pBr 328 DNA.BglI + pBr 328 DNA.HinfI (lane 1). Three individuals were tested who were typed as follows: SH(+), NA1(+),NA2(+) (lanes 2 and 5); SH(+), NA1(−),NA2(+) (lanes 3 and 6); SH(−), NA1(+),NA2 (+) (lanes 4 and 7).
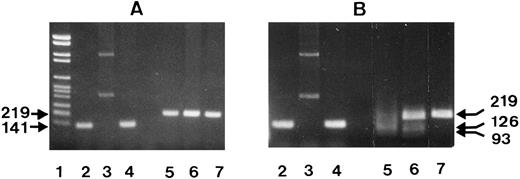
Figure 6 shows the NA1- and NA2-specific PCR products from three individuals before (Fig 6A) and after endonuclease digestion (Fig 6B). To simplify RFLP analysis, NA-specific PCR-SSP was performed in the absence of the HGH internal control. Two individuals were SH(+) with NA phenotypes NA1(+),NA2(+) and NA1(−),NA2(+), and one individual SH(−), NA1(+),NA2(+). As expected, the NA2-specific, 219-bp product was obtained from all three donors (Fig 6A, lanes 5 through 7) and the NA1-specific, 141-bp product only from the NA1(+) individuals (lanes 2 and 4), but not from the NA1(−) donor (lane 3).
After digestion with SfaN I, the NA2-specific PCR products of the two SH(+) individuals were cut into the restriction fragments of 126 and 93 bp (Fig 6B, lanes 5 and 6), whereas their NA1-specific PCR products were not cleaved (Fig 6B, lanes 2 and 4). In addition, no digestion products were obtained from the NA2(+), but SH(−) individual (lane 7). The additional undigested NA2-specific product detected in the SH(+), NA1(-),NA2(+), individual (lane 6) is the result of the presence of the SH-specific mutation only in one of both FcγRIIIB genes.
The same results were obtained with the PCR products from three additional SH(+), NA1(+), NA2(+) individuals (data not shown). Table 1 summarizes typing and RFLP analysis results, which demonstrated that the SH-specific nucleotide substitution is associated with NA2.
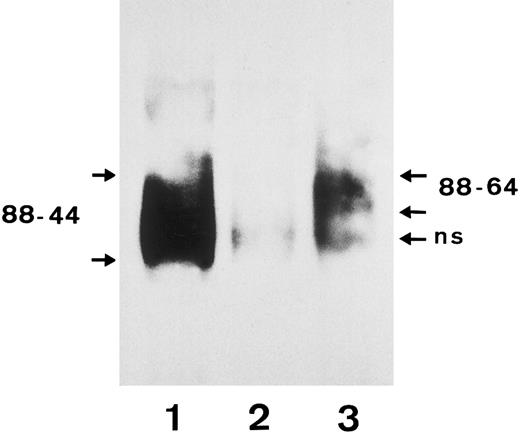
Immunochemical studies confirmed the results of the RFLP analysis. Anti-SH precipitated the FcγRIIIb with a molecular weight of 64 to 88 kD from SH(+), NA1(+),NA2(+) typed neutrophils, which corresponds to the NA2 isoform (Fig 7, lane 3), whereas a human polyclonal isoantibody against FcγRIIIb precipitated both the NA1 and the NA2 FcγRIIIb isoforms with a molecular weight of 44 to 88 kD (lane 1).
Immunoprecipitation analysis of FcγRIIIb from a SH(+), NA1(+),NA2(+) individual using biotin-labeled granulocytes. After granulocytes were biotin-labeled and lysed, the FcγRIIIb was precipitated with a serum containing an isoantibody to FcγRIIIb (lane 1), a negative control serum (lane 2), and the SH-specific serum (lane 3). Immunoprecipitates were analyzed by 10% SDS-PAGE under nonreducing conditions, transferred onto nitrocellulose, and visualized using streptavidin-horseradish peroxidase and chemiluminescence substrate.
Immunoprecipitation analysis of FcγRIIIb from a SH(+), NA1(+),NA2(+) individual using biotin-labeled granulocytes. After granulocytes were biotin-labeled and lysed, the FcγRIIIb was precipitated with a serum containing an isoantibody to FcγRIIIb (lane 1), a negative control serum (lane 2), and the SH-specific serum (lane 3). Immunoprecipitates were analyzed by 10% SDS-PAGE under nonreducing conditions, transferred onto nitrocellulose, and visualized using streptavidin-horseradish peroxidase and chemiluminescence substrate.
Three additional cases associated with SH alloimmunization.After characterization of the SH antigen by the use of serum no. 1, anti-SH was also detected in the maternal sera of three other cases of alloimmune neonatal neutropenia (case reports no. 2, 3, and 4). Two sera, no. 2 and no. 4, contained additional NA1-specific alloantibodies, which is why the three sera were tested against a panel of 61 NA1(−),NA2(+) typed neutrophils. Of these cells, 50 were typed SH(−) and 11 SH(+) using serum no. 1 in GIFT and GAT for phenotyping and PCR-SSP for genotyping. All three sera reacted in the GAT with the 11 SH(+), but not with the SH(−) neutrophils. Antibody binding to the FcγRIIIb in the MAIGA assay could be shown in two of the three sera, indicating that not all SH alloantibodies are detectable by MAIGA, a fact which is well known for the detection of NA-specific alloantibodies by this assay.17
SH phenotype frequency.Before SH alloantibody identification, the neutrophils from 309 randomly selected individuals had been tested with serum no. 3 in GAT and 14 of them had shown agglutination. We confirmed the SH(+) phenotype of the 14 neutrophils by SH genotyping of the donor DNA. The observed phenotype frequency of 5% is similar to the antigen frequency of 4% concluded from the genotyping findings in 107 individuals.
To check our finding that SH goes along with the NA2 phenotype, the 14 SH(+) individuals were typed for NA1 and NA2 and all were found to be NA2(+), 6 were NA1(−),NA2(+), and 8 were NA1(+),NA2(+).
DISCUSSION
The development of the MAIGA assay has vastly improved granulocyte serology. The assay has not only simplified the differentiation of multiple granulocyte-reactive antibodies in a single serum sample,17 but also the identification of the glycoprotein recognized by an antibody, as demonstrated by our detection of the SH antigen on the FcγRIIIb. Using this assay, the antigens LAN and SAR have found to be located on the FcγRIIIb, and the MART antigen on the subunit CD11b of the leukocyte adhesion molecule CD11b/CD18.20-22 The application of the MAIGA assay has also shown that the NC1 antigen is identical to NA2.23
In addition to the polymorphic sites, which are recognized by alloantibodies, the FcγRIIIb is also the target of autoantibodies17 and up to 23% of the autoantibodies show preferential binding to NA1(+) neutrophils.24 The high expression of the FcγRIIIb of about 200,000 copies per cell25 likely contributes to its high immunogenicity.
The elucidation of the molecular basis of SH allowed the development of a DNA-based technique using sequence (SH)-specific primers (PCR-SSP) for SH typing, which avoids cumbersome and time consuming isolation of fresh granulocytes required for serologic typing. We have already developed such a PCR-SSP method for NA typing, which has been successfully used in the Second International Granulocyte Serology Workshop. Others have described similar typing techniques for determination of platelet or HLA class II antigen genotypes.26 27
In contrast to the NA antigens, the SH antigen is associated with a single base mutation in the FcγRIIIB gene. The mutation leading to the SH polymorphism seems likely to be relatively recent. Since the nucleotide sequence of the SH allele is identical with the NA2 sequence, except for the nucleotide difference at the position 266, this mutation in the NA2 allele most likely occurred after the emergence of the NA allelism (Table 2). This hypothesis is in accordance with the low frequency of SH in the human gene pool since, in contrast to the white population, the NA1 gene is more frequent than NA2 in Chinese and Japanese populations (Table 2).19,28 29 Although on the molecular level the SH polymorphism is caused by a new allele in addition to NA1 and NA2 at the FcγRIIIb gene locus, for serologic reasons the Second International Granulocyte Serology Workshop group agreed to term the new antigen not in accordance with the N-terminology.
The C →A mutation results in an amino acid substitution of alanine by aspartic acid. It is likely that the replacement of a neutral by an acidic amino acid directly changes the tertiary structure of the FcγRIIIb forming the SH alloantigen. The SH-associated Ala78Asp substitution is situated in the membrane distal domain of the FcγRIIIb, which also contains three of the five nucleotide substitution sites associated with the NA polymorphism. Two of these nucleotide substitutions result in amino acid substitutions (codon 65 and 82), which lead to two additional N-linked glycosylation sites in the NA2 isoform.12,13 Since the inner membrane proximal domain carries the IgG binding site30 and individuals with FcγRIIIb deficiency do not suffer from major infections or immune diseases,15 it is possible that other, still unknown, mutations in the FcγRIIIB gene, especially in the outer membrane distal domain, are conserved, which could cause polymorphism and alloimmunization. We do not know whether SH polymorphism has an influence on the impaired phagocytic function reported for the NA2 phenotype.31
Alloantibodies to the SH antigen were detected in the maternal sera from four cases of alloimmune neonatal neutropenia, indicating that SH alloimmunization is not a rare event. Three sera contained additional granulocyte-reactive alloantibodies, anti-NA1 (2×) and anti-SL, but one case was only associated with SH alloimmunization, demonstrating that SH alloantibodies can cause alloimmune neonatal neutropenia. Although an infrequent disease, alloimmune neonatal neutropenia can cause severe infections such as meningitis or pneumonia, especially when neutropenia is not recognized adequately early after birth.32
ACKNOWLEDGMENT
We gratefully acknowledge the excellent technical support of Christine Hofmann. This work is part of an academic thesis (PhD) from Ernst-Ludwig Stein. We thank also the participants of the Second International Granulocyte Serology Workshop for their helpful comments concerning the nomenclature of new granulocyte antigens.
Supported by a grant from the Deutsche Forschungsgemeinschaft DFG Bu 770/3-2.
Address reprint requests to Juergen Bux, MD, Institute for Clinical Immunology and Transfusion Medicine, Justus Liebig University, Langhansstrasse 7, D-35385 Giessen, Germany.