Abstract
A unique cDNA for hexokinase (HK) was identified from poly(A)+ RNA of human reticulocytes by anchored polymerase chain reaction. This appeared to represent the cDNA for the red blood cell (RBC)–specific HK isozyme (HKR ) described in our previous study (Murakami et al: Blood 75:770, 1990). Its nucleotide sequence was identical to HKI cDNA except for the 5′ extreme end. It lacked the first 62 nucleotides of the HKI coding region: instead, it contained a unique sequence of 60 nucleotides at the beginning of the coding sequence as well as another unique sequence upstream of the putative translation initiation site. It lacked the porin-binding domain which facilitates binding to the mitochondria, thus explaining the exclusive cytoplasmic localization of HKR . It was the major cDNA derived from reticulocytes, consistent with the observation that HKR activity is predominant in reticulocytes. Northern blot analysis showed that it was expressed in the reticulocytes and in the K562 erythroleukemic cell line, but not in a lymphocytic cell line. In the extract of K562 cells, HKR activity co-eluted with the HKR of human RBCs on a MonoQ column (Pharmacia, Piscataway, NJ) chromatography, using a salt gradient elution. The separate genetic control of the RBC-specific HK isozyme explains the clinical reports of two types of HK deficiency, one in which the HK activity was reduced exclusively in the RBC (HKR defect) and another with general decrease of HK activity in several tissues (HKI defect).
THE RETICULOCYTE loses most of its intracellular organelles during maturation into a red blood cell (RBC). The RBC then remains dependent on the preformed enzymatic machinery throughout its life span. Thus, the metabolism in reticulocytes is distinct from that in RBCs.
Hexokinase (HK: E.C.2.7.1.1.) plays a key role in the metabolic regulation of the RBC because the energy production in RBCs is derived entirely through glycolysis and HK is a rate-limiting enzyme. Heterogenous forms of HKI have been described in RBCs.1-6 In human RBCs, it has been suggested that the multiple subtypes reflect posttranslational modification.7 In rabbit RBCs, an HK subtype that is well separated from HKI localizes exclusively in the cytoplasm.3,8 It is predominant in the reticulocyte and decays rapidly during maturation process. It was suggested that HKI , but not this subtype, is stabilized by binding to the mitochondria.8
In RBCs separated by age using buoyant density ultracentrifugatioin, several enzymes decline linearly with age, while HK decays in a biphasic manner, with an initial rapid decay followed with a slow decline.9,10 In our previous study,10 HK from RBCs was clearly separated into two isozymes using the MonoQ column (FPLC; Pharmacia, Piscataway, NJ). One eluted similarly to HKI , and the other (called HKR ) that eluted separately was unique to the RBC and predominant in the reticulocytes. Each decayed independently in RBCs separated by age, HKI with a long half-life, and HKR with a short half-life. From this evidence, together with the clinical observations reported,11 12 we suggested that the HKR isozyme is a gene product unique to the reticulocytes, rather than a posttranslational modification.
Mammalian HK consists of four isozymes that vary in kinetic properties and tissue distribution.13 HKI , HKII , and HKIII are single polypeptide chains of approximately 100 kD and are inhibited by glucose-6-phosphate. HKIV or glucokinase (GK) is about 50 kD and is insensitive to inhibition by glucose-6-phosphate, as is yeast HK. Recent clonings14-17 of HKII and GK of rats and humans have provided evidences for the gene duplication — fusion hypothesis previously suggested for the evolution of mammalian HKs. In this model, HKs evolved through duplication and fusion of an ancestral hexokinase that resembles yeast HK and GK. After the duplication event, the amino terminal half evolved into a regulatory domain, whereas the carboxy terminal half retained the catalytic function. The new HK gene further underwent duplication to produce three HKs: HKI , HKII , and HKIII .
The structures of the HKII , HKII , and GK genes have shown a remarkably similar organization of exons and introns.14-17 The gene for HKII consists of 18 exons, with the 10th exon attached to the 2nd exon through 10th exon of GK. The liver and pancreatic glucokinases are produced by alternate splicing of the 1st exon and utilization of alternate promoters, and the mRNA for liver GK, but not for pancreatic GK, is induced by insulin.18,19 HKI has a porin-binding domain (PBD) at its N-terminus, whose 15-amino acid sequence is absolutely conserved and which mediates its binding to the mitochondria.20 Spermatogenic cell-specific HKIs have been reported to contain unique sequences at the 5′ terminus lacking the PBD.21
HKR kinetically belongs to HKI .10 It localizes in the cytoplasmic fraction and does not bind to the mitochondrial membrane. This information, together with the genetic structures of HK and GK, suggested that HKR results from alternative splicing of the 1st exon incorporating a specific 5′ coding sequence and an unique promoter. From the reticulocyte mRNA, cDNA was constructed by anchored polymerase chain reaction (PCR)22 using a specific primer derived from the sequence of the human kidney HKI . This study reports the isolation of cDNA containing a unique sequence immediately 5′ to the common domain of HKI .
MATERIALS AND METHODS
Materials.Human HKI cDNA (phHK15-2/host-HB101/vector-pBR327)25 was kindly provided by Dr Greame I. Bell (University of Chicago, Chicago, IL). The primers used were obtained from Life Technologies Inc (Gaithersburg, MD). All cell lines were cultured in the RPMI-1640 medium in the presence of 5% CO2 at 37°C. General molecular biological techniques were performed according to “Molecular Cloning.”23
RNA isolation.Reticulocyte-rich blood (12% to 15% reticulocytes) was obtained from patients with sickle cell anemia. Platelets were removed by defibrination on glass beads and white blood cells by filtration through two layers of Whatman 2 filter paper (Hillsboro, OR). The resulting suspension contained exclusively RBCs. A reticulocyte-rich fraction (≥90% reticulocytes) was next isolated by slicing the top layer, after ultracentrifugation of the RBC suspension on 23.6% arabinogalactane.24 Finally, the RNA was isolated with Trizol reagent (Life Technologies). For the cDNA cloning, poly(A)+ RNA was isolated from the total RNA by oligo-(dT)-cellulose affinity chromatography. Total RNA from other cell lines was also prepared with the Trizol reagent (Life Technologies).
Northern blot analysis.The total RNA was fractionated in a 1.4% agarose gel containing formaldehyde and transferred to nitrocellulose membrane. HKR mRNA was probed with a 147-nucleotide (nt) fragment derived from HKR cDNA (−90 to +57 nt), and HKI mRNA was probed with an HindIII/Stu I fragment derived from nt 1043-2596 of the HKI cDNA. Both probes were 32P random labeled. After hybridization at 42°C overnight, the membranes were washed at 42°C.
Cloning of HKR cDNA with anchored PCR.The poly(A)+ RNA was obtained from 400 μg of total RNA from the reticulocytes. The first-strand cDNA for reticulocyte HK was prepared from the poly(A)+ RNA using the HKI -specific primer derived from nt 634-618 of the HKI cDNA25 (oligonucleotide 1; 5′CCACTGTGTCATTCACC3′) and isolated by Sepharose 4B spin column (Pharmacia). The purified cDNA was tailed with dATP and terminal deoxynucleotidyl transferase and amplified by PCR (Perkin-Elmer Cetus, Norwalk, CT) with a 3′ primer (oligonucleotide 1) and two 5′ primers [oligonucleotide 2: (dT)-adaptor; CTGCAGGTCTCTAGATTTTTTTTTTTTTTT and oligonucleotide 3: adaptor; CTGCAGGTCTCTAGA] according to the RACE protocol described by Frohman et al.22 The amplification was performed for 40 cycles of 40 seconds at 94°C, 40 seconds at 55°C, and 2 minutes at 72°C. The amplified cDNA was combined and purified with the GELase method (Epicentre Technologies, Madison, WI) for sequencing.
DNA sequencing.The DNA sequence was determined by the dideoxy method at the Cancer Center Core Facility at the Columbia University using oligonucleotide 4 (nt 177-194 of the HKI cDNA), oligonucleotide 5 (nt 229-245 of the HKI cDNA), and oligonucleotide 6 (nt −137-123 of the HKR cDNA). The sequence alignment was analyzed by the program FASTA (Genetics Computer Group, Madison, WI).
MonoQ column chromatography of HK.The lysate of RBCs obtained from a healthy volunteer was applied on DE52 equilibrated with 20 mmol/L Tris-Cl, pH 7.5, 0.5 mmol/L glucose, 0.5 mmol/L dithiothreitol, and 10 mmol/L KCl (buffer A). The protein absorbed was eluted with 0.5 mol/L KCl in the buffer A and precipitated with 60% saturated ammonium sulfate. K562 cells (∼1.5 × 107 cells) were lysed in buffer A and the homogenate was centrifuged at 14,000 rpm for 30 minutes. Both protein preparations were passed through a 200-μm membrane before applying on the MonoQ column. The MonoQ column was run with a gradient of 50 to 350 mmol/L KCl in 50 mmol/L Tris-Cl pH 7.5, 0.5 mmol/L glucose, and 0.5 mmol/L dithiothreitol at 0.5 mL/min. HK activity was measured for each 0.25-mL fraction as described previously. KCl concentration in each fraction was measured by conductivity meter (Radiometer America, Cleveland, OH).
RESULTS
Identification of RBC-specific HKR cDNA.Total RNA was extracted from the reticulocyte-rich fraction. Northern analysis of total RNA showed a strong expression of the HK mRNA (data not shown), using an Stu I/HindIII fragment (nt 1043-2596) of human HKI as a probe. In the reticulocyte-rich fraction, HKR represents more than 90% of the total HK activity. Furthermore, we had shown that HKI is more stable with a longer half-life. We assumed then that HK mRNA expressed in the reticulocyte should consist mostly of mRNA for HKR . Therefore, RNA was extracted from the reticulocyte-enriched fraction and poly(A)+ RNA was obtained by oligo(dT) affinity column for preparation of cDNA.
Since HKR is kinetically a subtype of HKI we anticipated that it may contain a unique sequence at 5′ end of HKI . Therefore, we used an anchored PCR protocol as described by Frohman et al22 to isolate HKR cDNA. The specific primer used for preparation of the first-strand cDNA was derived from nt 634-618 of human HKI . Following tailing with poly(dT) on the first-strand cDNA, PCR amplification produced a fragment of ∼800 bp. The fragment was purified by GELase (Epicentre Technologies) and sequenced in both directions. Figure 1A shows the sequence at 5′ end and its relationship to that of HKI .25 The 3′ side sequence is identical to that of HKI while the 5′ side sequence is quite different. The first 62 nt at the 5′ end of HKI coding region, including the PBD, are missing. The ATG starting at the 120th nt could be a translation initiation site, although not favored from the Kozak consensus,26 inserting 60 unique nts. This ATG is preceded by an in-frame stop codon at the nt −27. The upstream sequence to the translation initiation site is unique to this cDNA. Thus, this cDNA contains a sequence identical to that of HKI in the most coding region, lacks PBD, and contains a unique sequence at the 5′ end of coding region and another unique sequence at the 5′ noncoding region. Although the entire sequence of cDNA was not determined, the sequence at the 3′ end is likely to be identical to that of HKI because PCR amplification of the 3′ side of cDNA showed the identity to HKI .
Nucleotide and predicted amino acid sequences of HKR . (A) The nucleotide sequence of the 5′ end of cDNA is compared with that of human HKI , ×61091.Gb_Pr.25 The predicted translation initiation site is indicated with bold letters and the in-frame stop codon is underlined. The two sequences are aligned with the FASTA program. (B) The predicted amino acid sequence of the N-terminus which differs from that of HKI is shown. The subsequent sequence is identical to HKI .
Nucleotide and predicted amino acid sequences of HKR . (A) The nucleotide sequence of the 5′ end of cDNA is compared with that of human HKI , ×61091.Gb_Pr.25 The predicted translation initiation site is indicated with bold letters and the in-frame stop codon is underlined. The two sequences are aligned with the FASTA program. (B) The predicted amino acid sequence of the N-terminus which differs from that of HKI is shown. The subsequent sequence is identical to HKI .
Figure 1B shows the unique amino acid sequence predicted from the putative translation initiation site, in comparison with that of HKI .25 The leading sequence comprises 20 amino acids, one amino acid less than in HKI .
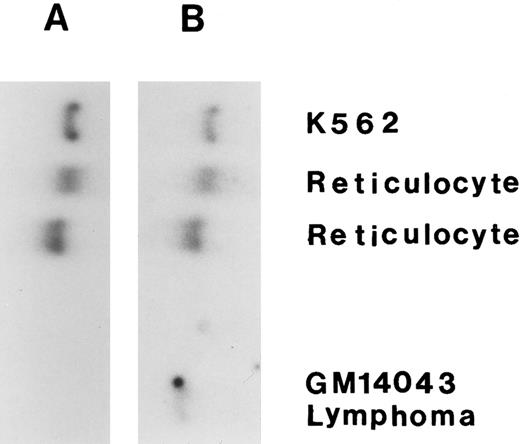
The expression of HKR mRNA versus HKI mRNA.To confirm the specific expression of HKR in the RBCs, Northern analysis was performed in comparison to HKI expression. The expression of HKR and HKI was tested with specific probes: one derived from nt −90 to +57 of the HKR cDNA (HKR -specific) and another from the HindIII/Stu I fragment of HKI cDNA (HKI -specific). As shown in Fig 2, HKI mRNA was detected in reticulocytes and in the K562 erythroleukemic human cell line, as well as in a lymphocytic cell line. On the other hand, HKR mRNA was detected only in reticulocytes and in the K562 erythroleukemic human cell line. Thus, the expression of HKR is restricted to erythropoietic cells, while the expression of HKI is general.
Expression of HKR gene. The Northern blot analysis was done for total RNA (∼20 μg each) from reticulocyte, K562 cells, and lymphocytic cell line. (A) Hybridization with HKR -specific probe; (B) Hybridization with HKI -specific probe.
Expression of HKR gene. The Northern blot analysis was done for total RNA (∼20 μg each) from reticulocyte, K562 cells, and lymphocytic cell line. (A) Hybridization with HKR -specific probe; (B) Hybridization with HKI -specific probe.
Identification of HKR activity in extracts of K562 cells.Northern analysis suggested the expression of the HKR gene in K562 erythro-leukemic cells. To confirm this, we analyzed the K562 HK isozymes by MonoQ anionic exchange column chromatography. In our previous study10 with the fine resolution provided by this ionic exchange column, the HKR of RBCs was distinctively identified from HKI and eluted between HKI and HKII . As analyzed with the same system, white blood cells contained HKI and HKII whereas platelets contained only HKI .10 The extracts of human RBC and of K562 cells were applied on MonoQ column and the protein was eluted with a linear gradient of 50 to 350 mmol/L KCl. The HK activity profile of the K562 cells was compared with that of RBCs. The K562 cells contained heterogeneous forms of HKI , apparently consisting of three peaks. The middle peak eluted at the same ionic strength as the HKI of RBC, followed by the major peak which comigrated with RBC HKR . In K562 there appears to be another subtype of HKI which preceded HKI .
DISCUSSION
From a reticulocyte-enriched preparation we have identified a unique cDNA which shares most of the 3′ side sequence with HKI and contains unique sequences at the 5′ end as well as at upstream of the translation initiation site. We conclude that this represents the cDNA for HKR , which we previously suggested to be an RBC-specific HK isozyme.10 This conclusion is based on the following: (1) the HKR cDNA was the major HK cDNA identified from the reticulocytes; (2) the mRNA preparation used was derived from WBCs and platelet-free reticulocyte-rich suspension; (3) the HKR mRNA expression was restricted to reticulocytes and the K562, an erythro-leukemic human cell line; (4) HKR activity was identified in the extract of K562 cells; and (5) HKR does not bind to the mitochondria and the cDNA lacks the first 62 nt of HKI coding sequence including PBD, which mediates mitochondrial binding.26
We suggest, although not favored by the Kozak consensus,25 that the ATG starting at the 120th nucleotide is the translation start site, preceded by the in-frame stop codon at the nucleotide −27. HKR contains a unique sequence of 60 nt at the 5′ end of the putative coding region, and thus a molecular size similar to HKI . In our previous report we thought that HKR must be larger than HKI since HKI was retarded by one fraction on column chromatography using BioGelA 0.5m. HKI is most likely retarded on the BioGelA (Bio-Rad Laboratory, Hercules, CA) 0.5m filtration column by the interaction with the matrix because the amino terminal peptide of HKI is more hydrophobic than that of HKR due to the PBD included in HKI , but not in HKR .
Similar unique mRNAs for HKI with identity to most of the coding region of HKI except the 5′ terminus have been observed in mouse spermatogenic cells. They also lack PBD and contain instead a unique sequence at the 5′ end as well as upstream of the putative translation start site, and are differentially expressed during the spermatogenesis.21 An evolutionary model for HK, proposed by Griffin et al,20 suggested that recruitment of an exon may have been responsible for the addition of PBD to the N-terminus of eukaryotic HKI . The cDNAs for HKR and for the spermatogenic HKIs appear to support this current model for the evolution of hexokinase genes.
The cDNAs for glucokinase (GK) from the liver and pancreatic β cells were reported to be identical in the 3′ untranslated region and most of the 3′ side coding region, but to differ at the 5′ untranslated sequence and at the first 45 nts in the coding region.18,19 The divergence of mRNA sequences has been traced to two distinct tissue-specific promoters, with alternative splicing of different 5′ end exons in liver and pancreatic β cells. Furthermore, multiple GK transcripts have been reported to result from a similar mechanism in rat27 and human liver.28 We assume that a similar mechanism operates for production of HKR and HKI mRNAs. From a human genomic library, we have identified several clones containing the HKR -specific sequence. The analysis of gene structure should demonstrate the mechanism for generation of HKR mRNA as well as the factors involved in the induction of HKR mRNA expression.
A number of erythroid-specific enzymes have been reported to result from a similar mechanism. Erythroid- and liver-specific pyruvate kinases are produced from different transcription units operating with two cell-restricted promoters that are mutually exclusive.29 Two forms of NADH-cytochrome b5 reductase are produced from one gene, a myristylated membrane-bound enzyme expressed in all tissues and a soluble, erythrocyte-specific isoform.30 The two forms are identical except at the amino terminus which mediates binding to the bilayer in the membrane and contains the myristylation consensus sequence. They are produced by the use of alternative promoter/alternate exons.
In a case of nonspherocytic hemolytic anemias associated with HK deficiency,12 the HK activity was generally reduced in RBCs as well as white blood cells and fibroblasts. Another case of hemolytic anemia was reported where the HK deficiency was limited to the RBCs.11 These differences can be clearly explained if HKI and HKR are under separate genetic control. HKI is a ubiquitous isozyme present in most tissues whereas HKR expression is restricted to the RBCs. Since HKR activity is highly induced and predominant in the reticulocyte, its deficiency could contribute to the quality of the reticulocytes and, in turn, to the anemia.
ACKNOWLEDGMENT
We thank Timothy Cheng and Jennifer Huang for their professional support. We are grateful to Dr Greame I. Bell at the University of Chicago for his generous donation of human HKI cDNA (phHK15-2/host-HB101/vector-pBR327).
Supported by Grant No. RO1 HL48996-15 from the National Institutes of Health, Heart, Lung and Blood Institute, Bethesda, MD.
Address reprint requests to Sergio Piomelli, MD, Division of Pediatric Hematology, College of Physicians & Surgeons of Columbia University, 630 W 168th St, New York, NY 10032.