Abstract
The interaction between interleukin-6 (IL-6) and IL-6 receptor (IL-6R) is the initial and most specific step in the IL-6 signaling pathway. Understanding its mechanism at the amino acid level is the basis for developing small IL-6–inhibiting molecules. We studied the human IL-6 (hIL-6)/hIL-6R binding interface by a combination of molecular modelling and site-directed mutagenesis. Our model suggests that the center of the interface between the two molecules consists of hydrophobic contacts predicted to account for most of the binding-free energy. These contacts can be regarded as a hydrophobic core shielded by hydrophilic residues that are also needed for recognition. Following this hypothesis, we altered in hIL-6 and hIL-6R residues predicted to reside in the contact region and to interact with each other. We studied the capacity of these mutants to form an IL-6/IL-6R complex and their ability to transduce the signal. This combined approach has led to the identification of certain residue-clusters in the binding interface and to a rational explanation of their specific interactions, suggesting therein a likely mechanism of complex formation. The results confirm the predictive model and strongly support our hypothesis. Comparison with other cytokines and their α-subunit receptors suggests that the structural location of certain binding sites are conserved.
INTERLEUKIN-6 (IL-6) is a multifunctional cytokine produced by a variety of cell types.1 It exerts its many activities through the interaction with specific receptors expressed on the surface of target cells. IL-6 binds first to a low-affinity α-subunit, a 80-kD glycoprotein called IL-6 receptor (IL-6R) or gp80. This IL-6/IL-6R complex recruits the signal-transducing β-subunit, a 130-kD glycoprotein called gp130. The association of gp130 with IL-6 and IL-6R leads to the formation of the high-affinity IL-6R complex, to the covalent linkage of two gp130 subunits, and to signal transduction. gp130 does not bind IL-6 or IL-6R on their own. It associates only with the preformed IL-6/IL-6R complex.2,3 Recent studies have suggested that the high-affinity IL-6R complex is a hexamer formed by two distinct IL-6, IL-6R, and gp130 molecules.4 5
Furthermore, the cytoplasmic and transmembrane domains of IL-6R are not needed for the transduction of the signal. IL-6R is often secreted in a soluble form (sIL-6R) that binds IL-6 and is biologically active as an agonist, rendering cells expressing gp130 but not IL-6R responsive to IL-6.6 7
IL-6 is predicted to fold as a four α-helix bundle protein belonging to the hematopoietin cytokine family. Like growth hormone (GH), it was classified to be one of the long chain members of the family.8,9 The two IL-6 receptor subunits, IL-6R and gp130, were classified in the hematopoietin cytokine receptor family. Their extracellular part is predicted to contain the family's typical module consisting of two fibronectin-III like domains.9-11 IL-6 is related to and shares activities with IL-11, cilliary neurotrophic factor (CNTF), leukemia-inhibitory factor (LIF), oncostatin M (OSM), and cardiotrophin-1 (CT-1),12 all of which were grouped into the IL-6–type subfamily due to the participation of gp130 as a common signal-transducing subunit in their respective high-affinity receptor complexes.12
The biologic activities of IL-6 include the stimulation of B- and T-cell growth and differentiation, production of acute-phase proteins by hepatocytes, multilineage hematopoiesis, osteoclast formation, maturation of megakaryocytes, and platelet production.1 An abnormal expression of IL-6 may be involved in the pathogenesis of a variety of diseases, among which are multiple myeloma, rheumatoid arthritis, postmenopausal osteoporosis, chronic autoimmune diseases, Castleman's disease, and acquired immunodeficiency syndrome (AIDS).1
Attempts to specifically limit the overproduction of IL-6 or its consequences can be made either at its site of production or at its target. The IL-6 overproduction can be specifically decreased by inhibition of its mRNA transcription or translation by antisense oligonucleotides or ribozyme action. These methods were shown to be effective on cells in culture but are not yet developed enough for use in vivo.13,14 Two approaches have been used to inhibit the action of IL-6. In the first approach, mutational analyses of IL-6 and sIL-6R have yielded several antagonists that bind to the corresponding wild-type molecules and form an IL-6/IL-6R complex but fail to transduce the signal. These mutants were shown to inhibit IL-6 activity in cell cultures but have not yet been tested in the organism, mostly because the amounts needed are too high.15-19 In a second approach, murine monoclonal antibodies (MoAbs) directed against human IL-6 (hIL-6) or hIL-6R that block either the binding between hIL-6 and hIL-6R or between the hIL-6/hIL-6R complex and gp130 were used. Clinical trials have shown that such MoAbs were successful in decreasing the IL-6–related symptoms in patients of certain diseases.20-23 Although the use of such MoAbs seems most promising at the moment, the amount of MoAbs needed is very high, their mode of administration is not very convenient, and human antimouse Ig antibody responses were encountered.23 Therefore, the search for other means to inhibit hIL-6 activity should continue.
Understanding the mechanism of interaction between hIL-6 and hIL-6R by identifying the amino acids involved may lead to the development of small and simple IL-6–inhibiting molecules. Although some x-ray crystallography and nuclear magnetic resonance (NMR) structural information is available for a few members of the cytokine family,9,24 only two x-ray structures of a cytokine/receptor complex were reported so far. These are the structures of hGH bound to an hGH receptor (hGHR) dimer or to human prolactin receptor (hPRLR).25,26 No such structural information is available for IL-6, and the cocrystallisation and x-ray analysis of the four or more proteins included in its high-affinity receptor complex are unlikely to be achieved soon. We have previously shown that a computer-assisted model of the IL-6/IL-6R complex could be built using the hGH/hGHR structure as a template.27,28 This model proved to be useful for the interpretation of our previous mutagenesis results and for the localization of some ligand binding sites on the receptor.28
We decided to continue here the structure/function study of hIL-6 and hIL-6R by molecular modelling and site-directed mutagenesis. Emphasis was on the binding interface between the two molecules. Previous results and our model suggested that the amino acids making up the hIL-6/hIL-6R interface could be divided mainly into three types: aromatic and hydrophobic amino acids that may contribute to the binding energy and hydrophilic residues that could be responsible for the recognition and the specificity of the interaction.27-30 Following this hypothesis and the predictions of our model, we analyzed in detail a series of hIL-6 and hIL-6R mutants; certain members of this series permitted us to identify binding sites that had not been discovered earlier. Our results shed light on how the IL-6/IL-6R complex is assembled in detail. They show that the predictions of the model were most probably correct and strongly support our hypothesis. Comparison with other cytokines and their corresponding α-subunit receptors suggests that the topography of the related binding epitopes is partially conserved.
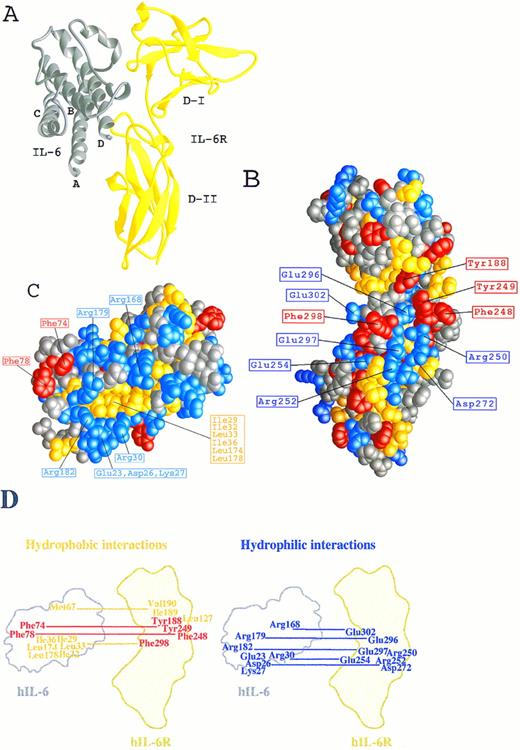
The three-dimensional model of the hIL-6/hIL-6R complex. (A) A ribbon representation of hIL-6 in grey bound to shIL-6R in yellow (side view). (B and C) Space-fill representations showing the complementary structural and functional epitopes for the binding of hIL-6 onto the predicted structure of hIL-6R (B) and for the binding of hIL-6R onto the predicted structure of hIL-6 (C). Side chains are colored according to their chemical properties: aromatic, red; hydrophobic, yellow; hydrophilic, blue; and polar-uncharged, grey. (D) A schematic illustration of the hydrophobic and hydrophilic interactions predicted in the hIL-6/hIL-6R binding interface. hIL-6R and hIL-6 are positioned as in (B) and (C), respectively.
The three-dimensional model of the hIL-6/hIL-6R complex. (A) A ribbon representation of hIL-6 in grey bound to shIL-6R in yellow (side view). (B and C) Space-fill representations showing the complementary structural and functional epitopes for the binding of hIL-6 onto the predicted structure of hIL-6R (B) and for the binding of hIL-6R onto the predicted structure of hIL-6 (C). Side chains are colored according to their chemical properties: aromatic, red; hydrophobic, yellow; hydrophilic, blue; and polar-uncharged, grey. (D) A schematic illustration of the hydrophobic and hydrophilic interactions predicted in the hIL-6/hIL-6R binding interface. hIL-6R and hIL-6 are positioned as in (B) and (C), respectively.
MATERIALS AND METHODS
Molecular Modelling of the hIL-6/hIL-6R Complex
The methods used for the homology-based modelling of hIL-6 and of hIL-6R using the x-ray structures of granulocyte colony-stimulating factor (G-CSF) and hGHR, respectively, as templates were described previously in Ehlers et al,17 Grötzinger et al,27 and Kalai et al.28 For graphic representation of the structures, the programs GRASP31 and RIBBONS 2.032 running on an Indigo2 SGI were used (Silicon Graphics Inc, Mountain View, CA).
Generation of shIL-6R and hIL-6 Mutant cDNAs
Site-directed mutagenesis was performed using an oligonucleotide-directed in vitro mutagenesis system (Amersham, Amersham, UK) and synthetic oligonucleotides as primers, as described before.28-30,33 34 The single-stranded DNA (ssDNA) of M13mp19–IL-6R was used as template for the hIL-6R mutations. The ssDNA of pBS–IL-6− was used as template for mutation hIL-6, with the exception of mutant F74E,F78E, for which the ssDNA of mutants F74E and F78E was used. The sequence of the primers used for mutagenesis of hIL-6R and hIL-6 are available on request.
The resulting hIL-6R mutant cDNAs were amplified by polymerase chain reaction (PCR) according to standard protocols35 using primers M13(−20) universal and ASN226-5′TCGTGGATGACACAGTGATGCTGGAGGTCC3′ and primers SN227-5′ATGTTGTTTGTGAGTGGGGTCC3′ and G165-5′TGTCCAAGGCGTGCCCATGGC3′ for mutations of residues from domain I and II, respectively. Amplification of mutated DNAs was performed either for 30 cycles using Taq DNA polymerase (Promega, Madison, WI) or for 25 cycles using Pfu DNA polymerase (Stratagene, La Jolla, CA). Each cycle consisted of incubations for 1 minute at 94°C, 1.5 minutes at 55°C, and 2 minutes at 72°C. The amplified products were digested with BssHI and EcoRV for mutations of domain I residues, Phe248, and mutation Y249V and with Bal I and Nco I for mutations Y249R, E296R, E297R, and E296R,E297R. The resulting fragments were inserted in place of the corresponding fragment of the wild-type (WT) pSP64T-sgp80R. The Bal I and Nco I fragment of mutant Y188R was replaced with the corresponding fragment of mutant Y249R to obtain mutant Y188R,Y249R. The sequences of all shIL-6R and hIL-6 mutants were confirmed using a Sequenase sequencing kit (US Biochemicals, Cleveland, OH).
Production of WT and Mutant shIL-6 R and hIL-6 Proteins
Determination of hIL-6 and shIL-6R Protein Concentrations
hIL-6 and shIL-6R protein concentrations were determined using the following enzyme-linked type antibody sandwich immunometric assays.
For hIL-6, we used microtiter plates coated with 100 μL/well of 5 μg/mL of anti–hIL-6 MoAb AH-65 (Immunotech SA, Marseilles, France) to capture and a biotinylated anti–hIL-6 MoAb B-E4 (B-E4-Bio) to detect bound hIL-6 molecules. B-E4 (kindly provided by Dr J. Wijdenes, Diaclone, Besançon, France) was biotinylated using a protein biotinylation kit according to manufacturer's instructions (Boehringer Mannheim, Mannheim, Germany). Bound biotinylated B-E4 was detected with an alkaline phosphatase-conjugated Extravidin (Sigma, St Louis, MO).
For shIL-6R, we used microtiter plates coated with 100 μL/well of 5 μg/mL of anti–hIL-6R MoAbs M-164, M-195 (kindly provided by Dr J. Brochier, INSERM U291, Montpellier, France), and 22.1 (kindly provided by Dr D. Novick, The Weizmann Institute of Science, Rehovot, Israel) to capture and an alkaline phosphatase-conjugated M-91 anti–hIL-6R MoAb (M-91-AP; Immunotech) to detect bound shIL-6R molecules.
In Vitro hIL-6 and hIL-6R Binding Assay
The potential of mutant and WT hIL-6 and shIL-6R to form an hIL-6/hIL-6R complex in solution with the corresponding WT partners was tested by an enzyme-ligand immunoassay using microtiter plates coated with anti–hIL-6 MoAb AH-6537 (Immunotech) to capture and with anti–hIL-6R MoAb M-91-AP (Immunotech) to detect formed hIL-6/shIL-6R complexes, as described.28 38
Murine 7TD1 Hybridoma Cell Growth Assay
Human XG-1 Myeloma Cell Growth Assay
The IL-6–induced proliferation of XG-1 cells was measured essentially as described.40 Briefly, cells were washed three times and incubated overnight at 37°C without IL-6 in growth medium (RPMI 1640 supplemented with 10% fetal calf serum [FCS] and 50 μmol/L 2-mercaptoethanol [2-ME; SERVA, Heidelberg, Germany]) and then washed again. A total of 5 × 103 cells/well in 200 μL of growth medium in 96-well flat-bottom microtiter culture plates was subsequently incubated in triplicate with a serial dilution of WT or mutant hIL-6 for 3 days at 37°C in 5% CO2 , humidified. Cell proliferation was measured after incubation using the XTT cell proliferation kit II (Boehringer Mannheim) according to the manufacturer's instructions.
Human A375 Melanoma Cell Growth Arrest Assay
The growth arrest of the IL-6–sensitive A375 cells was performed as previously described.30 Briefly, cells were seeded at a density of 2 × 103 cells/well in 200 μL of Iscove's medium supplemented with 5% FCS in 96-well flat-bottom microtiter culture plates. They were incubated in triplicate with a serial dilution of WT or mutant hIL-6 at 37°C in 5% CO2 , humidified. Cell proliferation was measured after 5 days of incubation. Cells were washed twice with cold phosphate-buffered saline (PBS) and were then fixed for 15 minutes at room temperature (RT) with methanol.
After the removal of methanol, the cells were stained with 75 μL/well of 1% methylene blue in PBS for 1 hour at RT. Plates were washed three times with tap water and then dried upside down. The stained cells were resuspended for 30 minutes at 42°C in 200 μL/well 0.1 N HCl. Absorbance was measured at 620 nm.
Dot Blot Analysis of Native and Denatured WT hIL-6 and shIL-6R
Dot blots were performed using nitrocellulose membranes Trans-Blot (Bio-Rad, Hercules, CA) and analyzed using ProtoBlot Western blot AP systems (Promega) following the manufacturer's instructions. Membranes were blocked by tris-buffered saline (TBS) supplemented with 5% dried milk. The washing solution was TBS. The working concentration of primary MoAbs was 10 μg/mL. Rabbit anti–shIL-6R antiserum (PC-Rb) was diluted 1/1,000. Primary and secondary Abs were diluted in TBS supplemented with 1% dried milk.
Immunoprecipitation of S35-Labeled shIL-6R
The production and immunoprecipitation of S35-labeled shIL-6R were performed as described before.28 The amount of anti–shIL-6R antibody-specific radioactivity was determined after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (8% to 16%) and Tris/glycine (Novex, San Diego, CA). Gel portions containing the immunoprecipitated S35-labeled shIL-6R protein were identified by autoradiography and were cut and their radioactivity was measured with a liquid-scintillation β-counter.
Analysis of the Conformational Integrity of hIL-6 and shIL-6R Variants
The conformational integrity of the hIL-6 variants was assessed using the enzyme-linked–type antibody sandwich immunometric assays we described above by requantification of equal amounts of each variant at a subsaturation concentration using a larger group of MoAbs, including at least some that are conformation-dependent.
For hIL-6, we used MoAbs AH-64, AH-65 (Immunotech), 8, 16 (kindly provided by Dr J.P.J. Brakenhoff, Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, Amsterdam, The Netherlands),29,30 and B-E8 (kindly provided by Dr J. Wijdenes)41 as phase and B-E4-BIO and alkaline phosphatase-conjugated Extravidin (Sigma) as tracers.
For shIL-6R, we used MoAbs M-37, M-113, M-164, M-182, and M-195 (kindly provided by Dr J. Brochier)28 and MoAbs 17.6, 22.1, 32.3, 34.4, 38.8, 48.8, and 50.6 (kindly provided by Dr D. Novick)42 as phase and M-91-AP as tracer.
The values obtained for each variant by each MoAb were corrected after their comparison to the mean of the values obtained by all of the MoAbs that recognized the given variant well.
RESULTS
Architecture of the hIL-6/hIL-6R Binding Interface
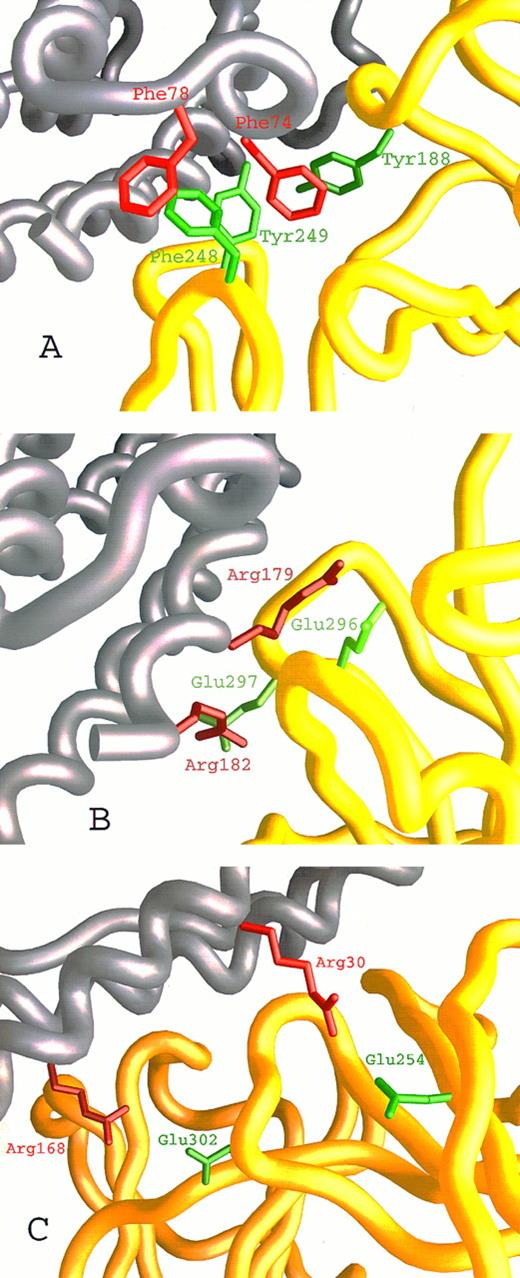
We have recently described27,28 the computer-assisted modelling of the structure of the hIL-6/hIL-6R complex using the x-ray structures of hGCSF43 and of hGH bound to the hGHR25 as templates. We used this model here (Fig 1A) to identify amino acids participating in the contact region between the two proteins and to explore their contribution to the interaction. Inspection of the hIL-6/hIL-6R interaction regions on both molecules has shown that they are made up of hydrophobic areas surrounded by several clusters of hydrophilic residues contributed by the two proteins (Fig 1B through D). The first hydrophobic area consists of five aromatic residues interacting with each other. These include Tyr188 from the EF loops of domain I and Phe248 and Tyr249 from the B′C′ loop of domain II of hIL-6R (Fig 1B and D and Fig 2A) and Phe74 and Phe78 from the AB loop of hIL-6 (Fig 1C and D and Fig 2A). Some of the hydrophobic amino acids included in the background of these aromatic residues may also contribute to the binding. These are Leu127, Val190 of hIL-6R, and Met67 of hIL-6 (Fig 1D). In the second hydrophobic area, Phe298 from the F′G′ loop of domain II of hIL-6R (Fig 1B and D) is embedded in a pocket consisting of residues Ile29, Ile32, Leu33, and Ile36 from the A-helix and Leu174 and Leu178 from the D-helix of hIL-6 (Fig 1C and D). The residues located in the hydrophilic clusters are predicted to form salt bridges between the two proteins. Residues suggested to participate in this type of interaction are Glu23, Asp26, Lys27, and Arg30 from the A-helix and Arg168, Arg179, and Arg182 from the D-helix of hIL-6 (Fig 1C and D and Fig 2B and C) and Arg250, Arg252, Glu254, Asp272, Glu296, Glu297, and Glu302 of hIL-6R (Fig 1B and D and Fig 2B and C).
Close-up on the hIL-6/hIL-6R binding interface according to our model. hIL-6 is colored grey and hIL-6R is colored yellow. Side chains of residues contributed by hIL-6 are red, whereas those contributed by hIL-6R are green. (A) The hydrophobic and aromatic cluster consisting of Tyr188, Phe248, and Tyr249 from the EF and B′C′ loops of hIL-6R and Phe74 and Phe78 from the AB loop of hIL-6. (B and C) Hydrophilic interactions. (B) Glu296 and Glu297 from the F′G′ loop of hIL-6R interact with Arg179 and Arg182 from the end of the D-helix of hIL-6. (C) Glu254 from the beginning of the C′-β-strand and Glu302 from the F′G′ loop of hIL-6R interact with Arg30 from the A-helix and Arg168 from the D-helix of hIL-6, respectively.
Close-up on the hIL-6/hIL-6R binding interface according to our model. hIL-6 is colored grey and hIL-6R is colored yellow. Side chains of residues contributed by hIL-6 are red, whereas those contributed by hIL-6R are green. (A) The hydrophobic and aromatic cluster consisting of Tyr188, Phe248, and Tyr249 from the EF and B′C′ loops of hIL-6R and Phe74 and Phe78 from the AB loop of hIL-6. (B and C) Hydrophilic interactions. (B) Glu296 and Glu297 from the F′G′ loop of hIL-6R interact with Arg179 and Arg182 from the end of the D-helix of hIL-6. (C) Glu254 from the beginning of the C′-β-strand and Glu302 from the F′G′ loop of hIL-6R interact with Arg30 from the A-helix and Arg168 from the D-helix of hIL-6, respectively.
Importance of Type of Residue, Size, and Charge for Interactions at the hIL-6/hIL-6R Binding Interface
According to the structural model, the interactions described above should be crucial for maintaining the binding capacities of hIL-6 and its receptor.
Following this assumption, we selected amino acids from each of the two proteins and modified their type or size by site-directed mutagenesis. Hydrophobic residues were replaced by hydrophilic amino acids and vice versa. The charge of certain hydrophilic amino acids was reversed from positive to negative and vice versa. Large residues were replaced by smaller ones and small residues were replaced by larger ones.
Mutant proteins were produced and the hIL-6R binding and signal-transduction capacities of the resulting hIL-6 mutants and the hIL-6 binding capacity of the resulting shIL-6R mutants were tested.
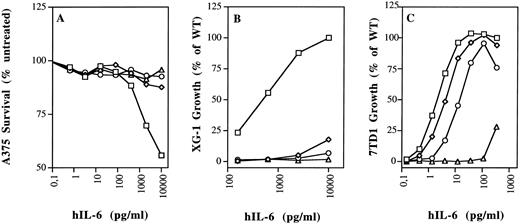
Aromatic and hydrophobic interactions between hIL-6 and hIL-6R.We showed previously that Tyr188, Phe248, and Tyr249 of hIL-6R are necessary for hIL-6 binding and signal transduction.28 It was recently shown that two synthetic peptides with amino acid sequences corresponding to residues 237-250 and 240-253 of hIL-6R, respectively, can bind hIL-6.44 As seen in Fig 2A, Tyr188, Phe248, and Tyr249 of IL-6R are predicted to interact with two phenylalanine residues, 74 and 78, located at the end of the AB loop of hIL-6. Previous studies suggested that an hIL-6R binding site could be found between residues 77 and 9517 or between residues 63 and 113 of hIL-6.45 We produced mutants F74E, F78E, and the double mutant F74E,F78E, thus replacing the aromatic and hydrophobic phenylalanines by the hydrophilic, charged glutamic acids. As seen in Figs 3 and 4, these replacements have nearly abolished the binding to shIL-6R. Unlike wild-type hIL-6, corresponding mutants did not significantly decrease the growth of the human A375 melanoma cells (Fig 4A), and they did not stimulate the growth of the human XG-1 myeloma cells (Figs 3 and 4B). The effect of mutagenesis was more pronounced in the case of the double mutant F74E,F78E that did not bind to hIL-6R and failed to show any XG-1 cell growth stimulation, even at a concentration of 25 ng/mL (Fig 3). These results support the prediction that both residues participate in the binding to hIL-6R.
Functional characterization of the hIL-6 mutant proteins. Amino acid substitution is indicated by a single letter code. (−) uninjected oocyte-conditioned medium. Binding to shIL-6R is presented as percentage of the value obtained with WT hIL-6. Biologic activity was measured using human XG-1 myeloma and murine 7TD1 hybridoma cell growth stimulation assays. D50 represents the effective concentration giving half of the maximal response obtained with the WT hIL-6. Values represent means of at least two independent experiments performed in duplicate. Low activity (LA) represents maximal activities below the D50 of the WT. Values in parenthesis were calculated by extrapolation when some activity was still detectable. NA, no activity.
Functional characterization of the hIL-6 mutant proteins. Amino acid substitution is indicated by a single letter code. (−) uninjected oocyte-conditioned medium. Binding to shIL-6R is presented as percentage of the value obtained with WT hIL-6. Biologic activity was measured using human XG-1 myeloma and murine 7TD1 hybridoma cell growth stimulation assays. D50 represents the effective concentration giving half of the maximal response obtained with the WT hIL-6. Values represent means of at least two independent experiments performed in duplicate. Low activity (LA) represents maximal activities below the D50 of the WT. Values in parenthesis were calculated by extrapolation when some activity was still detectable. NA, no activity.
Biologic activity of hIL-6 mutants F74E, F78E, and R179E on human and murine cells. (A) Growth arrest of human A375 melanoma cells. (B) Growth stimulation of human XG-1 myeloma cells. (C) Growth stimulation of murine 7TD1 hybridoma cells. Values are the mean of at least two independent experiments performed in triplicate, with standard deviation less than 10%. (□) WT; (⋄) F74E; (○) F78E; (▵) R179E.
Biologic activity of hIL-6 mutants F74E, F78E, and R179E on human and murine cells. (A) Growth arrest of human A375 melanoma cells. (B) Growth stimulation of human XG-1 myeloma cells. (C) Growth stimulation of murine 7TD1 hybridoma cells. Values are the mean of at least two independent experiments performed in triplicate, with standard deviation less than 10%. (□) WT; (⋄) F74E; (○) F78E; (▵) R179E.
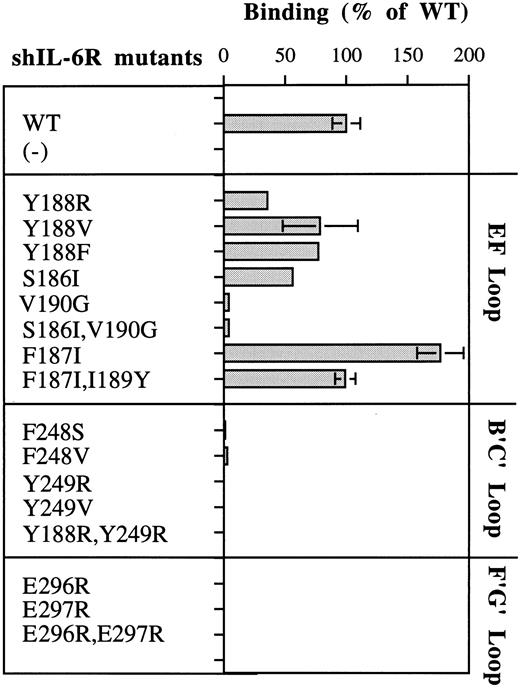
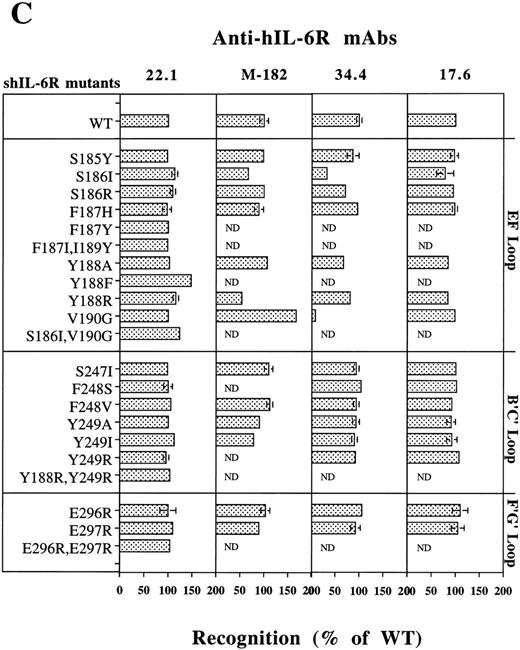
As seen in Fig 2A, Phe248 and Tyr249 are at the center of the predicted binding site, whereas Tyr188 is more peripheral and thus should be a minor participant in the binding process. These predictions are fully supported by the results shown in Fig 5. Replacement of Phe248 by the polar and uncharged serine (mutant F248S) and of Tyr249 by the hydrophilic arginine (mutant Y249R) decreased the hIL-6 binding capacity of the mutants to an almost undetectable level (Fig 5). The observation that similar results were obtained when these residues were replaced by the hydrophobic valine (F248V and Y249V) suggests that the aromatic nature of these residues is more important than their hydrophobicity for interaction with hIL-6 (Fig 5). Replacement of Tyr188 by the hydrophilic arginine (Y188R) resulted in a stronger effect on the IL-6 binding potential of the corresponding mutant than that of its replacement by the hydrophobic valine (Y188V) or the aromatic and hydrophobic phenylalanine (Y188F; Fig 5). This finding suggests that the interaction at this site is hydrophobic, as predicted. The observation that mutant Y188R still kept about 35% of the WT IL-6 binding capacity supports the suggestion that Tyr188 is a less-important participant in this binding site. As expected, the double mutant Y188R,Y249R did not bind hIL-6 at all. These results show that these three residues are crucial for IL-6 binding and stress the importance of their aromatic nature.
Analysis of the hIL-6 binding capacity of shIL-6R mutants. Results are presented as percentages of the value obtained for WT shIL-6R. Values represent the mean of at least two independent experiments performed in duplicate. (−) uninjected oocyte-conditioned medium.
Analysis of the hIL-6 binding capacity of shIL-6R mutants. Results are presented as percentages of the value obtained for WT shIL-6R. Values represent the mean of at least two independent experiments performed in duplicate. (−) uninjected oocyte-conditioned medium.
Furthermore, our model suggests that other hydrophobic amino acids may also contribute to this interaction zone. Leu127 of hIL-6R may increase the hydrophobic background of the interaction between Tyr188, Tyr249 (hIL-6R), and Phe74 (hIL-6), whereas Ile189 and Val190 of hIL-6R may interact with Met67 of hIL-6. The amount of experimental support to this prediction is still limited. No mutants of Met67 of hIL-6 were studied so far. Removal of the side chain of Val190 (mutants V190G and S186l,Val190G) decreased the binding to hIL-6 significantly (Fig 5). An earlier study showed that a Val190G hIL-6R mutant did not bind gp130 and was biologically inactive.46 These results may indicate that Val190 is directly involved in the binding of hIL-6; however, the removal of the side chain might have disrupted the local structure of the EF-loop of hIL-6R, as we suggested previously.28 The hIL-6 binding capacity of mutant F187I was twice as high as that of WT shIL-6R. The addition of the substitution of Ile189 to the larger tyrosine (mutant F187I,I189Y) brought it back to the WT level (Fig 5).
As described above, our model predicted a second hydrophobic interaction zone between hIL-6 and hIL-6R. This hydrophobic cluster includes residues Phe298 of hIL-6R (Fig 1B and D) and residues Ile29, Ile32, Leu33, Ile36, Leu174, and Leu178 of hIL-6 (Fig 1C and D). Several results indicate the existence of this second hydrophobic interaction zone. In an earlier hIL-6 mutagenesis study, we reported that replacing the hydrophobic Leu174 by the hydrophilic arginine resulted in the complete loss of activity. Replacement of Leu178 by glutamine reduced the activity to less than 50%.30 Furthermore, Yawata et al46 reported that the substitution of Glu297 by alanine coupled with the substitution of Phe298 by isoleucine resulted in the complete loss of hIL-6 binding.
Hydrophilic residues and salt bridge formation between hIL-6 and hIL-6R.Our model predicted that salt bridges form between the positively charged Arg179 and Arg182 of hIL-6 and the negatively charged Glu296 and Glu297 of hIL-6R (Figs 1D and 2B). Thus, in addition to the possible hydrophobic interactions involving the long side chains (methylene groups) of these arginines, their positively charged head groups are fixed in salt bridges keeping the side chains in such a position that they can shield the hydrophobic core of the interaction from the access of water. Replacing Arg179 by the aromatic hydrophobic tryptophan (R179W) resulted in the loss of binding to hIL-6R and biologic activity (Fig 3). The effect obtained when the charge was reversed from positive to negative by replacing Arg179 and Arg182 by glutamic acids (R179E and R179E,R182E) was somewhat stronger (Figs 3 and 4). Changing Arg to Glu resulted in the shortening of the side chain and thus in unshielding of the hydrophobic core. It may have also resulted in the repulsion of the two negative charges contributed now by both IL-6 and its receptor.
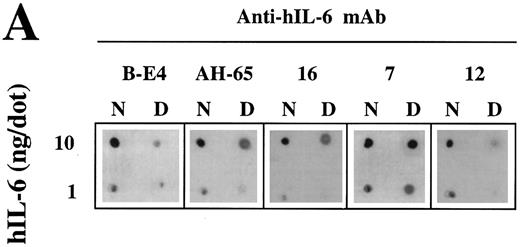
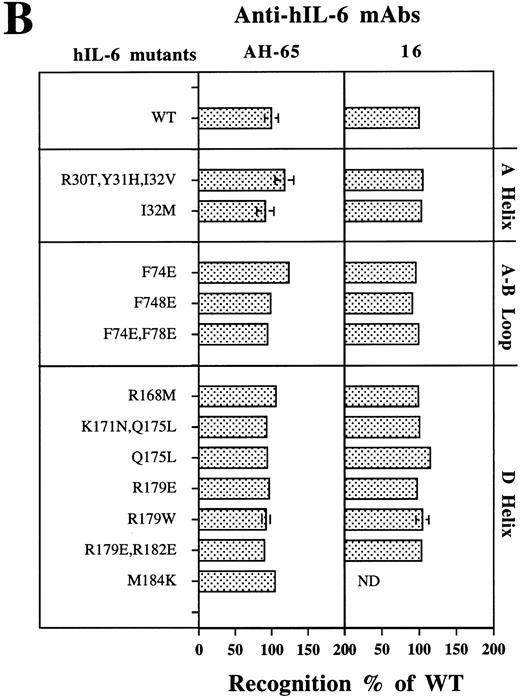
Analysis of the structural integrity of the hIL-6 mutants using conformation-dependent anti–hIL-6 MoAbs. (A) Differential recognition of dot blots of native (N) and denatured (D) WT hIL-6 by anti–hIL-6 MoAbs. Dot blot analysis was performed at two concentrations of untreated (native) or boiled hIL-6. Boiling was performed in the presence of 0.1% SDS and 0.167% (vol/vol) β-mercaptoethanol for 7 minutes (denatured). Results of one of two independent experiments are shown. Both experiments gave very similar results. (B) Recognition of hIL-6 mutants by three different conformation-dependent anti–hIL-6 MoAbs as determined by sandwich ELISA. The concentrations of equal amounts of each hIL-6 variant were determined using microtiter plates coated with either anti–hIL-6 MoAbs AH-65 or MoAb 16 as phase and biotinylated anti–hIL-6 MoAb B-E4 as tracer. Results are presented as the percentage of the values obtained for WT hIL-6. Values are the mean ± SD of at least two independent experiments performed in duplicate. ND, not determined.
Analysis of the structural integrity of the hIL-6 mutants using conformation-dependent anti–hIL-6 MoAbs. (A) Differential recognition of dot blots of native (N) and denatured (D) WT hIL-6 by anti–hIL-6 MoAbs. Dot blot analysis was performed at two concentrations of untreated (native) or boiled hIL-6. Boiling was performed in the presence of 0.1% SDS and 0.167% (vol/vol) β-mercaptoethanol for 7 minutes (denatured). Results of one of two independent experiments are shown. Both experiments gave very similar results. (B) Recognition of hIL-6 mutants by three different conformation-dependent anti–hIL-6 MoAbs as determined by sandwich ELISA. The concentrations of equal amounts of each hIL-6 variant were determined using microtiter plates coated with either anti–hIL-6 MoAbs AH-65 or MoAb 16 as phase and biotinylated anti–hIL-6 MoAb B-E4 as tracer. Results are presented as the percentage of the values obtained for WT hIL-6. Values are the mean ± SD of at least two independent experiments performed in duplicate. ND, not determined.
Replacements of glutamic acids 296 and 297 of hIL-6R by arginine (E296R, E297R, and E296R,E297R) resulted in complete loss of binding to hIL-6 (Fig 5). These results show that these four amino acids are crucial for the recognition and binding of IL-6 and IL-6R. The effects observed for the charge reversal experiments support the suggestion that these amino acids are involved in the formation of salt bridges between the two molecules.
It was recently shown that minibody β-polypeptides carrying an amino acid sequence similar to that found between residues Arg293 and Glu297 of hIL-6R in one of their β-loops can bind IL-6 and competitively inhibit the formation of an IL-6/IL-6R complex.47 Evidence for the involvement of Arg250 and Arg252 of hIL-6R in hIL-6 binding was obtained when this inhibitor was improved by the addition of an amino acid sequence similar to that found between residues Arg250 and Phe253 of hIL-6R in another β-loop.48 In addition, synthetic peptides with amino acid sequences corresponding to Tyr249 to Arg258 in hIL-6R were shown to bind hIL-644 or to decrease its activity.49
Another pair of amino acids predicted to form a salt bridge between hIL-6 and hIL-6R is shown in Fig 2C. Arg30 of hIL-6 is predicted to interact with Glu254 of hIL-6R. The triple mutant R30T,Y31H,I32V, in which the hydrophilic Arg30 is replaced by the polar uncharged threonine, has lost most of its hIL-6 binding capacity (Fig 3). In this mutant, the region between amino acids 30 and 32 in hIL-6 was exchanged for the analogous region of mIL-6. When compared with the WT, this mutant was only three times less active in the murine 7TD1 cell growth assay. In contrast, its D50 value obtained for the human XG-1 cells was at least 10 times larger than that of the WT. The decrease in binding to hIL-6R can be attributed mainly to the substitution of Arg30 because (1) previous studies showed that Tyr31 is involved in gp130 binding and not in IL-6R binding18,50; (2) mutant I32M showed a slight increase in hIL-6R binding and was twice as active on the human XG-1 cells as the WT control, without showing any significant change in the activity on the murine cells; (3) deletion of residues 29 and 30 in hIL-6 resulted in a significant decrease in activity34,51; and (4) another extensive mutagenesis study of hIL-6R suggested that Glu254 is involved in IL-6 binding.46
The residue predicted by our hIL-6/hIL-6R model to interact directly with Arg168 of hIL-6 is Glu302 of hIL-6R (Fig 2D). Substitution of Arg168 by the hydrophobic methionine (mutant R168M) resulted in an almost complete loss of binding to hIL-6R and growth stimulation of human XG-1 cells. In an earlier report, we showed that this mutant bound very weakly to human A375 melanoma cells and had only a mild effect on their growth.30 Mutation of Glu302 of hIL-6R was reported earlier to result in complete loss of binding to hIL-6.46
Gln175 and Met184 from hIL-6 are examples of amino acids located at the hIL-6/hIL-6R binding interface that were not predicted by our model to be directly involved in the binding process. The substitutions of these residues by an hydrophobic or hydrophilic amino acid (mutants Q175L and M184K, respectively) had a relatively small effect on their hIL-6R binding and signal-transducing capacities (Fig 3).
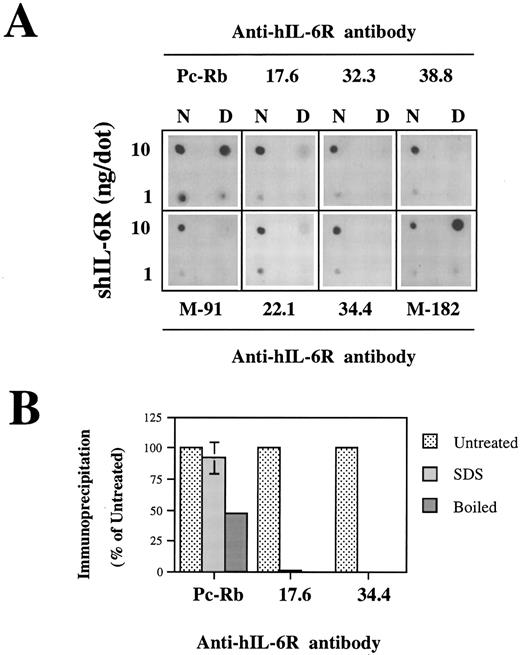
Analysis of the structural integrity of the shIL-6R mutants using conformation-dependent anti–hIL-6R MoAbs. (A and B) Differential recognition of native (N) and denatured (D) WT shIL-6R by anti–hIL-6R murine MoAbs 17.6, 22.1, 32.3, 34.4, 38.8, M-91, and M-182 and Pc-Rb, a rabbit anti–hIL-6R polyclonal antiserum. (A) Dot blot analysis of two concentrations of untreated (native) or denatured shIL-6R as described in Fig 6A. (B) Immunoprecipitation of equal amounts of S35-labeled shIL-6R by anti–shIL-6R Abs in the absence (untreated) or presence (SDS) of 0.1% SDS or after 10 minutes of boiling (boiled). The amounts of immunoprecipitated shIL-6R were measured and are presented as the percentage of the values obtained with untreated shIL-6R. The effect of boiling was tested once, whereas that of SDS was tested twice.
Analysis of the structural integrity of the shIL-6R mutants using conformation-dependent anti–hIL-6R MoAbs. (C) Recognition of shIL-6R mutants by five different conformation-dependent anti–hIL-6R MoAbs as determined by sandwich ELISA. The concentrations of equal amounts of each shIL-6R variant were determined using microtiter plates coated with anti–hIL-6R MoAbs 22.1, M-182, 34.4, or 17.6 as phase and alkaline-phosphatase conjugated anti–hIL-6R MoAb M-91 as tracer. Results are presented as the percentage of the values obtained for WT shIL-6R. Values are the mean ± SD of at least two independent experiments performed in duplicate. ND, not determined.
Analysis of the structural integrity of the shIL-6R mutants using conformation-dependent anti–hIL-6R MoAbs. (A and B) Differential recognition of native (N) and denatured (D) WT shIL-6R by anti–hIL-6R murine MoAbs 17.6, 22.1, 32.3, 34.4, 38.8, M-91, and M-182 and Pc-Rb, a rabbit anti–hIL-6R polyclonal antiserum. (A) Dot blot analysis of two concentrations of untreated (native) or denatured shIL-6R as described in Fig 6A. (B) Immunoprecipitation of equal amounts of S35-labeled shIL-6R by anti–shIL-6R Abs in the absence (untreated) or presence (SDS) of 0.1% SDS or after 10 minutes of boiling (boiled). The amounts of immunoprecipitated shIL-6R were measured and are presented as the percentage of the values obtained with untreated shIL-6R. The effect of boiling was tested once, whereas that of SDS was tested twice.
Analysis of the structural integrity of the shIL-6R mutants using conformation-dependent anti–hIL-6R MoAbs. (C) Recognition of shIL-6R mutants by five different conformation-dependent anti–hIL-6R MoAbs as determined by sandwich ELISA. The concentrations of equal amounts of each shIL-6R variant were determined using microtiter plates coated with anti–hIL-6R MoAbs 22.1, M-182, 34.4, or 17.6 as phase and alkaline-phosphatase conjugated anti–hIL-6R MoAb M-91 as tracer. Results are presented as the percentage of the values obtained for WT shIL-6R. Values are the mean ± SD of at least two independent experiments performed in duplicate. ND, not determined.
Differences Between the hIL-6/hIL-6R and hIL-6/mIL-6R Binding Interfaces
The murine IL-6R (mIL-6R) binds and transduces the signal of both human and mIL-6, whereas the hIL-6R is species-specific.36,52 The homology between hIL-6 and mIL-6 and between hIL-6R and mIL-6R is limited. An explanation of the species specificity of the IL-6/IL-6R interaction is described by us in another report.27 We analyzed here the effects of substitutions of several amino acids in hIL-6 predicted to be important for the interaction with hIL-6R, on the capacity of the mutants to stimulate the growth of 7TD1 murine hybridoma cells.
Differences were observed mainly for mutants F74E and F78E. The homologues of phenylalanines 74 and 78 in hIL-6 are the polar uncharged, aromatic tyrosines 73 and 77 in mIL-6. In addition, Tyr188 and Phe248 in the hIL-6R correspond to His186 and Tyr245 in the mIL-6R. This finding suggests that this contact region in the mIL-6/mIL-6R binding interface is dominated by aromatic interactions.
It is possible that the negatively charged glutamic acid that replaced Phe74 in mutant F74E is able to interact with the positively charged His186 of mIL-6R. The replacement of Phe78 by glutamic acid in mutant F78E and in the double mutant F74E,F78E should be more difficult to accommodate.
Indeed, mutant F74E was nearly as active as the WT in the 7TD1 growth stimulation assay, whereas mutant F78E was about six times less active. The double mutant F74E,F78E was about 46 times less active (Figs 3 and 4).
Substitution of Arg168 seems to affect the interaction with hIL-6R more than the interaction with mIL-6R. Mutant R168M could hardly stimulate the growth of human XG-1 cells and was about seven times less active in the murine 7TD1 cell growth assay as compared with the WT control (Fig 3). Probable equivalents for Arg168 of hIL-6 and Glu302 of hIL-6R in the murine proteines are Lys168 and Asp296, respectively. Arginines 179 and 182 of hIL-6 and glutamic acids 296 and 297 of hIL-6R are conserved residues in the corresponding murine molecules (Arg179, Arg182, Glu293, and Glu294, respectively). These amino acids are crucial for the function in the human and the murine systems (Figs 3 and 4). These results are supported by previous reports.29,45 47
Analysis of the Conformational Integrity of the hIL-6 and shIL-6R Mutants
To determine if the decrease in activity observed for some of the hIL-6 and shIL-6R mutants was due to a modification in the region of contact between the two molecules or due to a more drastic change in their tertiary structure, conformational analysis was performed for all of these proteins. Among the tools used as sensitive probes for structural alternations in mutant proteins are conformation-dependent MoAbs.28-30
Recognition of the hIL-6 mutants by conformation-dependent anti–hIL-6 MoAbs.A previous study has shown by immunoprecipitation of S35-labeled hIL-6 in the presence or absence of 0.1% SDS that anti–hIL-6 MoAb 12 recognized preferentialy native hIL-6, whereas anti–hIL-6 MoAb 7 reacted better with the denatured form of the antigen. To show that anti–hIL-6 MoAbs B-E4, AH-65, and 16 are conformation-dependent, we compared their potential to recognize the native and denatured hIL-6 by dot blot analysis with that of MoAbs 7 and 12 (Fig 6A). The only MoAb to react well with the denatured hIL-6 was MoAb 7; the other four reacted better with the native protein.
Analysis of the recognition of all of the hIL-6 mutants studied here by sandwich enzyme-linked immunosorbent assay (ELISA) using either AH-65 or MoAb 16 as phase and biotinylated B-E4 as tracer has shown that all of these hIL-6 variants were recognized as well as the WT hIL-6 by the three conformation-dependent anti–hIL-6 MoAbs (Fig 6B). This finding suggests that the these amino acid substitutions did not drastically affect the general tritiary structure of the proteins.
Recognition of the shIL-6R mutants by conformation-dependent anti–hIL-6R MoAbs.To evaluate the conformational integrity of our shIL-6R mutant, we followed a similar approach. The conformation dependence of eight anti–hIL-6R Abs was assessed by their potential to recognize the native and denatured hIL-6 by dot blot analysis (Fig 7A). The results obtained have shown that MoAbs 17.6, 22.1, 32.3, 34.4, 38.8, and M-91 are conformation-dependent and recognize preferentially the native form of shIL-6R. They also showed that anti–hIL-6R polyclonal rabbit antiserum (Pc-Rb) and MoAb M-182 also recognize well the denatured form of the antigen. The conformation dependence of MoAbs 17.6 and 34.4 was verified again by the immunoprecipitation of S35-labeled shIL-6R in the presence or absence of 0.1% SDS or after the boiling of the antigen for 10 minutes (Fig 7B). Both MoAbs failed to recognize the denatured shIL-6R, whereas Pc-Rb recognized it to a certain degree and even well, depending on the treatment (Fig 7B).
Analysis of the recognition of our shIL-6R mutants by a sandwich ELISA using MoAbs 17.6, 22.1,M-182, 34.4 (Fig 7C), 32.3, and 38.8 (not shown) as phase and the alkaline phosphatase-conjugated MoAb M-91 as tracer has shown that the conformational and structural integrity of most of these mutants was conserved.
The only exceptions were mutants of Ser186 and Val190, which were recognized less by MoAb 34.4 (Fig 7C) and MoAbs 32.3 and 38.8, which gave very similar results (not shown). Mutant V190G was recognized better than the WT shIL-6R by MoAb M-182 (Fig 7C). This MoAb recognized the denatured shIL-6R better than the native antigen in the dot blot analysis (Fig 7A). These results support our previous suggestion that these specific mutations might affect the structure of the protein at least locally.28
DISCUSSION
In this study, we analyzed in detail the predictions of a three-dimensional model of the hIL-6/hIL-6R complex. Analysis was performed by site-directed mutagenesis of hIL-6 and hIL-6R and focused on the proposed binding interface between these molecules. Substitutions of amino acids in hIL-6 and hIL-6R, which were predicted by our model to interact with each other, showed the expected results. The structural integrity of these mutants was confirmed by their recognition by at least three different conformation-dependent MoAbs. Thus, we were able to verify the involvement of amino acid residues suspected of being directly involved in the binding between hIL-6 and hIL-6R and to identify additional ones. Our results allow the identification of the different zones of interaction between the two proteins. They show that recognition epitopes for hIL-6R are located mainly in the beginning of the A-helix and in the ends of the AB loop and D-helix of hIL-6 (Fig 8A). Recognition epitopes for hIL-6 are located mainly in loops EF of domain-I and B′C′ and F′G′ of domain II of hIL-6R (Fig 8B). By changing the type, size, or charge of the residue in question, we showed that interaction zones in the hIL-6/hIL-6R binding interface are made up of relatively large aromatic, hydrophobic, or hydrophilic amino acids that are exposed on the surface of the interacting proteins. This approach has led to the localization of different residue clusters in the binding interface and to the identification of the type of interaction in which they are involved.
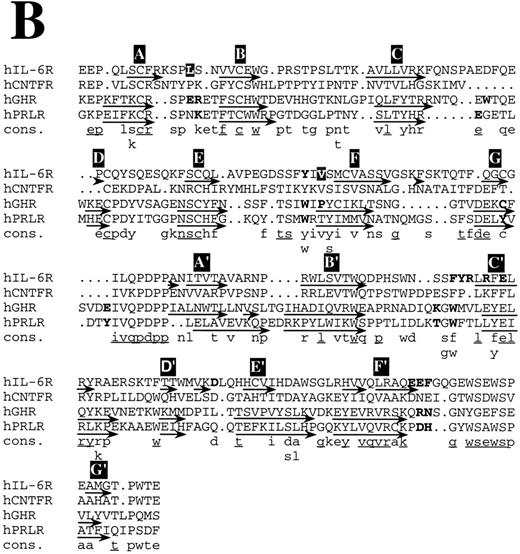
Structure-based amino acid sequence alignments. (A) Alignment of hIL-6 with hCNTF and hGH. Boxes indicate the location of α-helices according to the corresponding x-ray structures and our model.24,25 α-Helices are labeled in white letters over a black background, A through D. Residues in the first receptor binding epitope of each cytokine according to earlier studies,25,26 72 and our results are in bold. Model predictions for which experimental evidence in unavailable or poor are in white letters over a grey background. A consensus (Cons) of at least two is in lower case and of at least three is in lower case and underlined.
Structure-based amino acid sequence alignments. (B) Alignment of hIL-6R with hCNTFR, hGHR, and hPRLR. Arrows indicate the location of β-strands according to the corresponding x-ray structures and our model.25,26 β-Strands are labeled in white letters over a black background, A through G and A′ through G′ for receptor domains I and II, respectively. Residues in the ligand binding epitope of each receptor according to earlier studies,25,26 and our results are in bold. Model predictions for which experimental evidence is unavailable or poor are in white letters over a grey background. A consensus (Cons) of at least two is in lower case and of at least three is in lower case and underlined.
Structure-based amino acid sequence alignments. (A) Alignment of hIL-6 with hCNTF and hGH. Boxes indicate the location of α-helices according to the corresponding x-ray structures and our model.24,25 α-Helices are labeled in white letters over a black background, A through D. Residues in the first receptor binding epitope of each cytokine according to earlier studies,25,26 72 and our results are in bold. Model predictions for which experimental evidence in unavailable or poor are in white letters over a grey background. A consensus (Cons) of at least two is in lower case and of at least three is in lower case and underlined.
Structure-based amino acid sequence alignments. (B) Alignment of hIL-6R with hCNTFR, hGHR, and hPRLR. Arrows indicate the location of β-strands according to the corresponding x-ray structures and our model.25,26 β-Strands are labeled in white letters over a black background, A through G and A′ through G′ for receptor domains I and II, respectively. Residues in the ligand binding epitope of each receptor according to earlier studies,25,26 and our results are in bold. Model predictions for which experimental evidence is unavailable or poor are in white letters over a grey background. A consensus (Cons) of at least two is in lower case and of at least three is in lower case and underlined.
Structure-based amino acid sequence alignments of hIL-6 with hCNTF24 and hGH25 and of their corresponding first binding receptors,25,26 respectively, show that the sequence homology between the corresponding proteins is low (Fig 8). In general, the similarity between the receptors (Fig 8B) is higher than among the cytokines (Fig 8A). Insertions and deletions occurred probably in the loop regions, because their alignment is more difficult. Nevertheless, the general location of certain major binding sites is conserved in the protein structures and is similar to those used in hIL-6 and hIL-6R, respectively (Fig 8). The sequence differences observed in these sites reflect the specificity. Equivalents for some of the interaction regions in the hGH/hGHR were not found in the hIL-6/hIL-6R complex and probably will not be located in the CNTF/CNTFR complex (Fig 8). This is not unexpected, because GH is believed to interact with two identical receptor subunits,25 whereas IL-6 and CNTF are thought to interact with at least three different and distinct ones.12
A recent report suggested that the binding interface between hGH and its receptor resembles a cross-section through a globular protein. The center of this new protein, formed upon the binding between hGH and hGHR (hGHbp-I), is a hydrophobic region that accounts for most of the binding-free energy. This hydrophobic core that is dominated by two aromatic tryptophan residues, located in the EF and B′C′ loops of the receptor, is surrounded by hydrophilic and partially hydrated residues that seem to contribute less to the binding affinity.53 The architecture of the IL-6/IL-6R contact region is similar to the one described above. Aromatic residues from the EF and B′C′ loops of the receptor are at the center of the binding interface. Predominance of aromatic amino acids can be observed in the EF and B′C′ loops of most of the members of the cytokine receptor family.10 The involvement of some of these residues in the binding of the ligand by receptors other than hGHR, hPRLR, and hIL-6R (Fig 8B) was shown by mutagenesis.54-56 It seems highly probable that aromatic residues such as Phe, Trp, and Tyr are favored in the functional core, because their side chains provide conformational rigidity and functional plasticity needed for the fitting into critical regions in the binding interface. They probably contribute more to the binding energy and less to the specificity. The nature of the interactions in which they are involved is dependent on their environment. Affinity is dependent on the actual number of contacts formed between the two binding proteins and on their nature. Large hydrophobic interaction zones may stabilize the complex, whereas specificity is achieved by formation of salt bridges around these central binding regions.
Interestingly, IL-6R is often secreted in a soluble form (sIL-6R) that binds IL-6 and is biologically active as an agonist, rendering cells expressing gp130, but not IL-6R, responsive to IL-6. This form of the IL-6R shows structural and functional similarities to the P40 subunit of IL-12. In the functional IL-12 cytokine the P40 subunit is covalently linked to a second polypeptide, P35, which is structurally related to IL-6.57 The IL-12 signal is transduced via a membrane bound receptor resembling gp130.58 Taken together, the findings discussed above suggest that the IL-6/sIL-6R complex may be considered as a new protein that represents a functionally activated form of the cytokine with a larger target cell population. This may be applicable also to the complexes of CNTF and IL-11 and their corresponding α-subunit receptors.59-61
In view of the current lack of x-ray or NMR structural information on IL-6 and its receptor, the three-dimensional model of their complex, validated by mutagenesis, could be useful. Indeed, the existence of disease-related variants of hGH and hGHR was discovered recently. The availability of structural information on those molecules was of great value in understanding the implications and the mechanism of malfunction.62-64 Variants carrying mutations in the extracellular domain of hGM-CSFR and c-Mpl were also reported.65 66 Although no such variants of IL-6 nor of IL-6R have been reported so far, it is possible that some will be discovered in the near future.
The model can also be used for the rational design of antagonistic variants of IL-6 and sIL-6R.18,67 The topology of the binding site corresponding to Tyr188, Phe248, and Tyr249 of IL-6R and Phe74 and Phe78 of hIL-6 can be used for the screening of data banks in search of small synthetically produced molecules that might mimic the structure of these sites and act as hIL-6 inhibitors. Alternatively, such molecules could be designed and synthesized using our model as a template.68
Furthermore, we reported recently on the mapping of the epitopes of several neutralizing MoAbs directed against hIL-6R using mutants of the antigen and its molecular model.28 Following a similar approach, we analyzed a series of MoAbs directed against hIL-6R and hIL-6 (to be published elsewhere). Some of these (B-E8 and PM-1) are potent inhibitors of hIL-6 activity, which were already tested in vivo.22,23,69,70 In this perspective, our results contribute to the understanding of the mechanism of interaction of existing hIL-6 antagonists and may be of help in the decision regarding which antibodies should used and which antibody combinations will be most effective.71
ACKNOWLEDGMENT
We thank L. De Wit and E. Bautens for their technical assistance. We thank Dr D. Novick, Dr J. Brochier, Dr J. Wijdenes, and Dr J.P.J. Brakenhoff for their anti–hIL-6R and anti–hIL-6 MoAbs, which were used in this study.
Supported by grants from the CGER Fonds de Recherche Cancérologique and the Fund for Medical Scientific Research (Belgium). M.K. is a recipient of a fellowship from the “Université Libre de Bruxelles.”
Address reprint requests to Jean Content, MD, Institut Pasteur de Bruxelles, Département de Virologie, 642 Rue Engeland, B-1180, Bruxelles, Belgium.