Abstract
Interleukin-12 (IL-12), a cytokine with in vitro and in vivo immunomodulatory effects, is produced mostly by activated monocytes and macrophages. To study the effect of human immunodeficiency virus (HIV) infection on IL-12 production, we investigated the expression of IL-12 at mRNA and protein levels by human monocytes preincubated with HIV-gp120. In these conditions, we show that monocytes have a decreased ability to express IL-12 mRNA subunits and to produce IL-12 p40 and bioactive p70 proteins in response to Staphylococcus aureus strain cowan I (SAC). We showed that in human monocyte cultures, HIV-gp120 induces a significant IL-10 synthesis, which in turn inhibits IL-12 subunits mRNA accumulation and protein secretion after SAC-activation. Similar data were obtained with human macrophages. These results suggest that, during HIV infection, gp120 induces in uninfected monocytes and macrophages IL-10/IL-12 disregulation, which can alter immune response.
INTERLEUKIN-12 (IL-12) is a heterodimeric cytokine of 70 kD (p70) formed by two covalently linked glycosylated chains of 40 kD (p40) and 35 kD (p35) encoded by two separated genes.1-4 The coexpression of the two chains in one cell is required for the production of the bioactive IL-12 heterodimer.3,4 The p40 gene seems to be transcribed only in IL-12–producing cells5,6 in contrast with p35, which appears to be expressed more ubiquitously.5,6 IL-12 production was first noted in Epstein-Barr virus–transformed B-cell lines, either constitutively or in response to phorbol diesters.1,2 However, the major sources of IL-12 are monocytes and macrophages5,6 and, to some degree, other accessory cells.7,8 IL-12 is produced by antigen-presenting cells (APCs) as the result of nonspecific interaction with infectious agents5 or specific interaction with antigen-activated T cells through CD40 ligation.9,10 IL-12 has multiple effects on both natural killer (NK) and T cells, including stimulation of interferon-γ (INF-γ) production, proliferation, and cytolytic activity.11-15 In addition, APC-produced IL-12 is critically involved in the differentiation of naive T cells to Th1 phenotype via a mechanism dependent in part on INFγ.15-18 Th1 response is associated with an enhancement of cellular immunity directed, in particular, toward intracellular pathogens.19 20
There is a defect of IL-12 production during human immunodeficiency virus (HIV) infection.21 The addition of exogenous IL-12 on peripheral blood mononuclear cells (PBMCs) from individuals infected with HIV can enhance, in vitro, NK cytotoxic activity22 and restore IL-2 production and HIV-specific cell cytotoxicity.23 PBMCs from HIV-infected patients at different stages of the disease have a significant decreased ability to produce IL-12 p40 and IL-12 heterodimer in response to Staphylococcus aureus strain cowan I (SAC).21,24 The exact molecular mechanism involved in this defect remains unclear. Because in vivo and in vitro, a minor proportion of monocytes is infected,21 this deficit does not seem directly related to the cellular infection but rather secondary to the action of one or several viral or cellular factors produced during HIV infection. In this report we have investigated the effect of HIV-gp120 on IL-12 production by monocytes. Our results show that HIV-gp120 indirectly inhibits monocyte/macrophage IL-12 production by inducing synthesis of IL-10, which, in turn, inhibits after SAC-activation IL-12 expression at the RNA level.
MATERIALS AND METHODS
Preparation of human monocytes and macrophages.Monocytes were isolated from buffy-coat prepared from normal volunteer donors. PBMCs were prepared with the use of Ficoll-Hypaque density gradient centrifugation (Lymphoprep; Nygaard and Co, Oslo, Norway) and resuspended in RPMI 1640, medium supplemented with 5% heat-inactivated fetal calf serum (FCS), (Seromed; Biochrom KG, Berlin, Germany). Cells were incubated in Petri dishes (Nunc, Rockville, Denmark) at 37°C for 1 hour. Nonadherent cells were removed by several washes with phosphate-buffered saline. The adherent cells were incubated in RPMI 1640 with 10% heat-inactivated FCS (Seromed) overnight and then harvested by scraping the plates with a rubber policeman. Monocytes (>85% pure) were resuspended at 106 cells/mL in Iscove's medium (Seromed) supplemented with 10% heat-inactivated FCS. In some experiments, purified monocytes (>95% pure) were prepared from PBMC by conterflow centrifugal elutriation (Beckman elutriator equipped with a JE-5.0 rotor; Beckman Instruments, Munich, Germany). Monocyte-enriched adherent cells or elutriated monocytes were characterized by flow cytometry analysis (fluorescence-activated cell sorter; Becton Dickinson & Co, Mountain View, CA) after labeling with fluorescein isothiocyanate-conjugated anti-CD14 (Becton Dickinson). Macrophages were obtained by culture of monocyte-enriched adherent cells at 106 cells/mL for 7 days in Iscove's medium with 10% heat-inactivated FCS and 200 U/mL of recombinant human macrophage colony-stimulating factors (M-CSF; Sigma-Aldrich, St Louis, MO).
Reagents.SAC was used in culture at 1/10.000 (vol/vol; Pansorbin; Calbiochem-Berhing Corp, La Jolla, CA). HIV-1 recombinant gp120 (IIIB) produced in baculovirus expression system was used at 3 μg/mL. This gp120 (purchased from Neosystem, Strasbourg, France) migrates on standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels as 115 kD and more than 95% pure as estimated by analysis of Coomassie blue stained SDS-PAGE under reduced and nonreduced conditions. Human recombinant IL-10 was purchased from Genzyme (Cambridge, MA) and monoclonal neutralizing antihuman IL-10 from R&D Systems (Oxon, UK).
Primers and probes.Primers and probes were designed using Macvector software (Kodak Scientific Imaging Systems, New Haven, CT) and synthesized by Genset (Paris, France). Probes were digoxigenin labeled using a Digoxigenin oligonucleotide 3′labeling kit (Boerhinger Mannheim, Mannheim, Germany) according to the manufacturer's instructions.
The following primers and probes were used for quantitative polymerase chain reaction (PCR): P40 5′ biotinylated primer, GAGAAATGGTGGTCCTCACCTGTG; P40 3′ primer, GAGTGTAGCAGCTCCGCACGTC; P40 probe, TGGTCCAAGGTCCAGGTG; P35 5′ biotinylated primer, AGCCTCCTCCTTGTCGCTACC; P35 3′ primer, GCCTCCACTGTGCTGGTTTTATC; P35 probe, GGGAGTGGTGAAGGCATG; IL-10 5′ biotinylated primer, GCAACCTGCCTAACATGCTTCG; IL-10 3′ primer, GAAGATGTCAAACTCACTCATGGC; IL-10 probe, GCTGAAGGCATCTCGGAG; glyceraldehyde phosphodeshydrogenase (GAPDH) 5′ biotinylated primer, GGTGAAGTCGGAGTCAACGGA; GAPDH 3′ primer, GAGGGATCTCGCTCCTGGAAGA; and GAPDH probe, AAAGCAGCCCTGGTGACC.
Quantitative PCR.mRNA levels were quantitated by using a kinetic ELISA PCR.25-27
mRNA extraction and cDNA synthesis.Poly-A+ mRNA samples were prepared from 106 cells by using Oligotex Direct mRNA kit (Quiagen, Hilden, Germany). RNA was harvested in 20 μL of diethyl pyrocarbonate–treated water and stored at −80°C until use. The cDNA was obtained using random hexamers and avian myeloblastosis virus reverse transcriptase as previously described.25 Controls for contamination containing no RNA and RNA but no reverse transcriptase were always used and were negative by PCR analysis.
PCR.PCR products were amplified in 50 μL of amplification buffer (Bioprobe, Montreuil, France) containing 1 μL of cDNA, 0.25 μmol/L each primer, and 1.25 U of Taq polymerase, preincubated with 1.75 pmol per reaction of Taq polymerase antibody (Taq Start Antibody; Clontech, Paolo Alto, CA) at room temperature for 15 minutes to reduce nonspecific amplifications. Samples were overlayed with mineral oil and amplified in 96-well plates (Costar, Cambridge, MA). The amplification was performed in a PTC-100 thermal cycler (MJ Research, Watertown, MA). The thermal profile was initially 93°C for 3 minutes, followed by 42 cycles at 94°C for 30 seconds, 60°C for 1 minute, 72°C for 1 minute, and a final extension at 72°C for 5 minutes.
Quantitation of PCR products.The quantitation of the PCR products was performed as follows. Five-microliter aliquots were removed every three cycles of the PCR, starting at the indicated cycle and transferred onto avidin-coated microplates. The microplates for colorimetric detection (Maxisorp; Nunc) and for luminescence detection (Microlite 2; Dynatech, Chantilly, VA) were coated with 100 μg/mL avidin (Sigma), and the free sites were saturated with bovine serum albumin. The 5′-biotinylated PCR products were incubated in microplates for 1 hour at 4°C and then denatured with NaOH (0.25 mol/L) for 15 minutes at room temperature. The captured strand was hybridized with a digoxigenin-labeled probe (0.2 pmol/well) in 0.5× sodium chloride sodium phosphate EDTA buffer for 2 hours at 42°C. The bound probe was detected using an anti-digoxigenin alkaline phosphatase-coupled antibody (1/7,500; anti-DIG-AP; Boerhinger) for 1 hour at room temperature. Two methods of revelation were used according to the experiment. For colorimetric detection, paranitrophenylphosphate substrate (1 mg/mL Sigma) was added in 0.1 mol/L diethanolamine buffer. The optical density (OD) at 405 nm was measured after incubation overnight at 37°C. For luminescence detection, which enhances the sensitivity of the method, 2.5 mmol/L of 1,2 dioxetane chemiluminescent substrate for alkaline phosphatase (CSPD; Tropix, Bedford, MA), and 0.1 mg/mL of luminescence enhancer (Sapphire; Tropix) were added in 0.1 mmol/L diethanolamine buffer. Luminescence was measured after incubation for 1 hour at room temperature using Micro Beta Plus (Wallac, Turku, Finland).
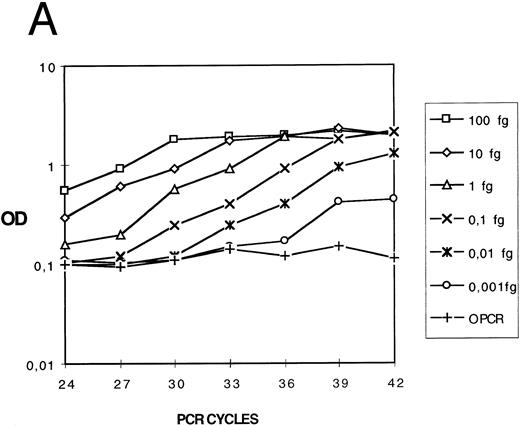
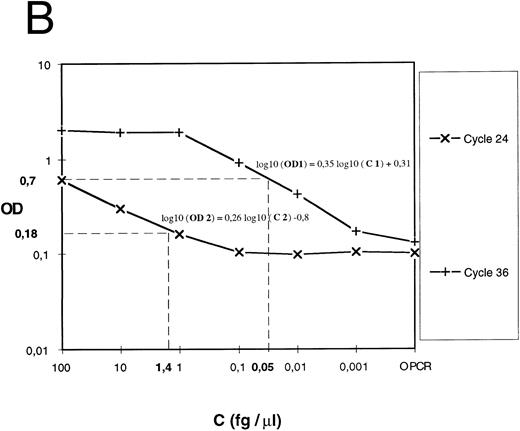
Expression of the results.For cDNA quantification, samples were amplified and PCR products were quantified at regular intervals during PCR amplification. For each experimental sample, the linear region of the curve (signal value [OD or luminescence values] v PCR cycles) was compared with the linear region of standard curves generated by amplifying an external DNA scale in the same experiment (Fig 1A). The DNA scale (femtograms per microliter) was made of serially diluted PCR products that were previously gel-purified (QIAEX gel extraction, Quiagen), and quantitated by two methods: UV absorbance and comparison to molecular weight markers after ethidium bromide staining.
Quantification of IL-12 p40 mRNA. (A) A IL-12 p40 cDNA scale constituted from gel-purified, quantified, and serially diluted PCR product was amplified. PCR products were sampled from every well at the indicated cycle and submitted to colorimetric detection as described in the Materials and Methods. (B) For each indicated cycle, the linear region of the curve (optical density versus standard cDNA amount) was used to estimate experimental cDNA amount (eg, cycles 24 and 36 and 2 experimental samples at 0.05 and 1.4 fg ).
Quantification of IL-12 p40 mRNA. (A) A IL-12 p40 cDNA scale constituted from gel-purified, quantified, and serially diluted PCR product was amplified. PCR products were sampled from every well at the indicated cycle and submitted to colorimetric detection as described in the Materials and Methods. (B) For each indicated cycle, the linear region of the curve (optical density versus standard cDNA amount) was used to estimate experimental cDNA amount (eg, cycles 24 and 36 and 2 experimental samples at 0.05 and 1.4 fg ).
For each cycle, the linear region of the curve (signal value [OD] v standard DNA concentration [Cst]) was used for computing the slope (a) and intercept (yo) of the following linear regression: log10 (OD) = a log10 (C)+ yo. a and yo parameters were used for determining experimental cDNA concentration (femtogram/per microliters) (Fig 1B) according to the following equation: C exp (fg/μL) = 10([log 10 (OD)−yo]/a). For a given sample, if signal values obtained at several successive samplings were in the linear region, cDNA amounts computed for each one were averaged.
To correct for variability in RNA recovery and efficiency of reverse transcription, GAPDH cDNA was amplified and quantified for each cDNA preparation. The results were expressed as relative mRNA units (computed as the ratio of gene of interest over GAPDH, both in femtograms per microliter).
Cytokine assays.Quantitation of cytokines were performed using commercially available specific ELISA assays (human IL-12 p40 [Medgenix Diagnostics, Fleurus, Belgium], human IL-12 p70 heterodimer [Quantikine; human IL-12 immunoassay; R&D Systems],28 and human IL-10 [IL-10-Easia; Medgenix Diagnostics]) according to the manufacturer's instructions.
RESULTS
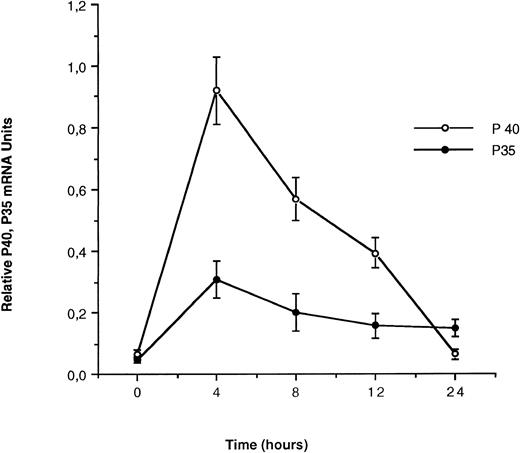
Expression of IL-12 p40 and p35 mRNA by SAC-activated monocytes.Using a quantitative PCR assay, we first studied the kinetics of IL-12 p35 and p40 mRNA levels in monocytes in response to SAC, which is a potent inducing agent of IL-12 secretion.5 As shown in Fig 2, nonactivated monocytes express both IL-12 p40 and p35 mRNA subunits at very low levels. SAC induces IL-12 subunits mRNA accumulation that reaches a peak at 4 hours of 0.919 ± 0.11 relative mRNA units for p40 and 0.307 ± 0.059 for the p35 subunit. This peak is followed by a rapid decrease in the level of IL-12 p40 transcripts that return at baseline level after approximately 24 hours: 0.063 ± 0.015 relative mRNA units. p35 mRNA is expressed more weakly, and the level of p35 transcripts decreases more slowly than that of the p40 subunit: 0.149 ± 0.039 mRNA relative units for p35 at 24 hours after SAC activation. Because monocyte-enriched adherent cells contained a minor proportion of B cells (5% to 10% according to CD19 staining), we determined the capacity of these cells to produce IL-12 in response to SAC. Interestingly, in highly purified B cells (>98%, obtained by positive selection using anti-CD19–coated magnetic beads), SAC induces a significant p35 mRNA expression after 4 or 24 hours of activation but no p40 mRNA expression or free p40 protein or bioactive p70 secretion in culture medium (data not shown).
Kinetics of IL-12 p35 and p40 mRNA accumulation in monocytes. Monocyte-enriched adherent cells were cultured with SAC. Cells were harvested for mRNA assay at various times after the beginning of the culture. mRNA is expressed as normalized femtograms of IL-12 p35 and IL-12 p40 cDNA. For normalization, the IL-12 p35 and p40 mRNA amount in femtograms was divided by femtograms of GAPDH. Each point plotted represents the mean ± standard error of mean (SEM) of quadruplicate culture samples. This experience is representative of three independent experiments.
Kinetics of IL-12 p35 and p40 mRNA accumulation in monocytes. Monocyte-enriched adherent cells were cultured with SAC. Cells were harvested for mRNA assay at various times after the beginning of the culture. mRNA is expressed as normalized femtograms of IL-12 p35 and IL-12 p40 cDNA. For normalization, the IL-12 p35 and p40 mRNA amount in femtograms was divided by femtograms of GAPDH. Each point plotted represents the mean ± standard error of mean (SEM) of quadruplicate culture samples. This experience is representative of three independent experiments.
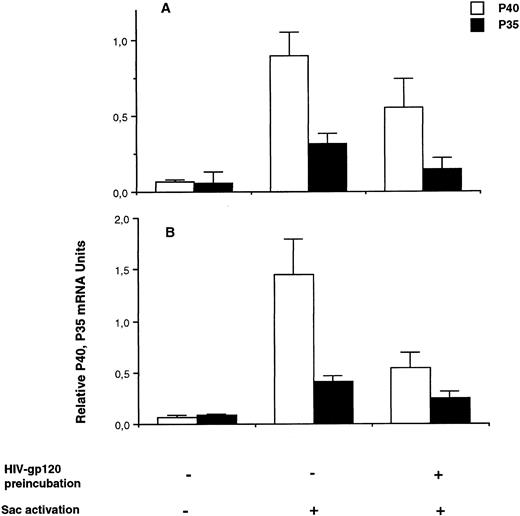
HIV-gp120 preincubation inhibits IL-12 mRNA expression and protein secretion by SAC-activated monocytes and macrophages.We studied the effect of 24 hours of preincubation with HIV recombinant gp120 (rgp120) on IL-12 production by SAC-activated freshly isolated monocytes. Cells were preincubated for 24 hours with rgp120 (3 μg/mL) and then activated by SAC until the peak of IL-12 mRNA subunit expression, which was 4 hours, as determined in the previous experiments. In these conditions, as shown in Fig 3A, rgp120 induces a significant decrease of both p35 and p40 steady state mRNA levels: for p40, 0.550 ± 0.190 relative mRNA units after rgp120 preincubation and SAC activation versus 0.900 ± 0.150 after SAC alone; for p35, the values are, respectively, 0.150 ± 0.071 versus 0.320 ± 0.060. In the same conditions, we tested the effect of rgp120 preincubation on macrophages cultured for 7 days. As shown in Fig 3B, rgp120 preincubation reduces more drastically p40 mRNA expression in macrophages compared with freshly isolated monocytes: 0.540 ± 0.150 relative mRNA units after gp120 preincubation and SAC activation versus 1.350 ± 0.350 after SAC activation only; for p35, respectively, 0.250 ± 0.065 versus 0.410 ± 0.050.
Effect of HIV recombinant gp120 on IL-12 p35 and p40 mRNA accumulation in monocytes and macrophages. (A) Monocyte-enriched adherent cells were preincubated or not with HIV recombinant gp120 for 24 hours and then activated with SAC for 4 hours. Cells were harvested for mRNA assay. Each value represents the mean ± SEM of quadruplicate culture samples. Similar results were obtained in two other experiments. (B) Seven-day macrophages (obtained as described in the Materials and Methods) pretreated or not with HIV recombinant gp120 for 24 hours were activated by SAC for 4 hours. Each value represents the mean ± SEM of duplicate culture samples.
Effect of HIV recombinant gp120 on IL-12 p35 and p40 mRNA accumulation in monocytes and macrophages. (A) Monocyte-enriched adherent cells were preincubated or not with HIV recombinant gp120 for 24 hours and then activated with SAC for 4 hours. Cells were harvested for mRNA assay. Each value represents the mean ± SEM of quadruplicate culture samples. Similar results were obtained in two other experiments. (B) Seven-day macrophages (obtained as described in the Materials and Methods) pretreated or not with HIV recombinant gp120 for 24 hours were activated by SAC for 4 hours. Each value represents the mean ± SEM of duplicate culture samples.
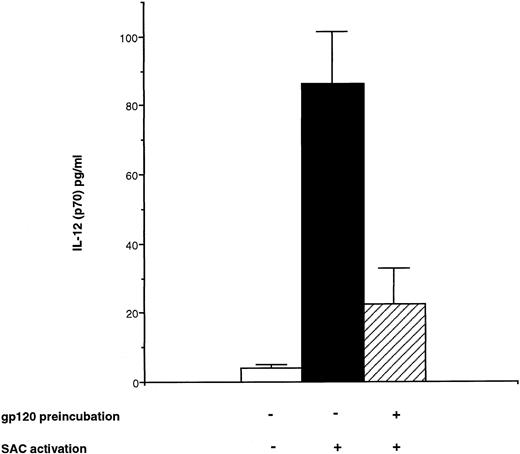
To extend the data obtained with PCR at the protein level, bioactive p70 protein and p40 protein were measured using specific ELISA28 in the supernatant of monocyte cultures pretreated with rgp120 for 24 hours and then activated by SAC for 24 hours. In these conditions, it was observed a significant decrease for IL-12 heterodimer secretion: 22.5 ± 10.5 pg/mL after rgp120 preincubation and SAC activation versus 84.3 ± 16 pg/mL with SAC alone (Fig 4). Similarly, p40 subunit protein was decreased to 825 ± 167 pg/mL after rgp120 preincubation versus a level of 1,664 ± 262 pg/mL after SAC alone (Fig 7).
Effect of HIV recombinant gp120 on IL-12 (p70) production by monocytes. Monocyte-enriched adherent cells were pretreated or not with HIV recombinant gp120 for 24 hours and then activated with SAC for 24 hours. Supernatants were collected and tested for IL-12 p70 secretion. Each value represents the mean ± SEM of quadruplicate culture samples.
Effect of HIV recombinant gp120 on IL-12 (p70) production by monocytes. Monocyte-enriched adherent cells were pretreated or not with HIV recombinant gp120 for 24 hours and then activated with SAC for 24 hours. Supernatants were collected and tested for IL-12 p70 secretion. Each value represents the mean ± SEM of quadruplicate culture samples.
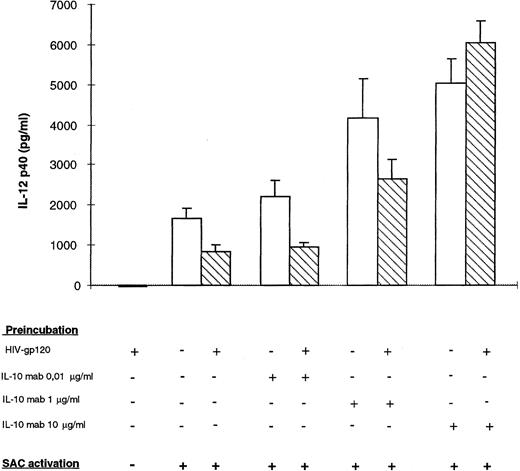
Effect of neutralizing anti–IL-10 on HIV recombinant gp120-inhibited IL-12 p40 secretion by monocytes. Monocyte-enriched adherent cells were pretreated with HIV recombinant gp120 and different concentrations of monoclonal neutralizing anti–IL-10 for 24 hours and then activated by SAC for 24 hours. Supernatants were harvested for IL-12 p40 assay. Each value represents the mean ± SEM of triplicate culture samples.
Effect of neutralizing anti–IL-10 on HIV recombinant gp120-inhibited IL-12 p40 secretion by monocytes. Monocyte-enriched adherent cells were pretreated with HIV recombinant gp120 and different concentrations of monoclonal neutralizing anti–IL-10 for 24 hours and then activated by SAC for 24 hours. Supernatants were harvested for IL-12 p40 assay. Each value represents the mean ± SEM of triplicate culture samples.
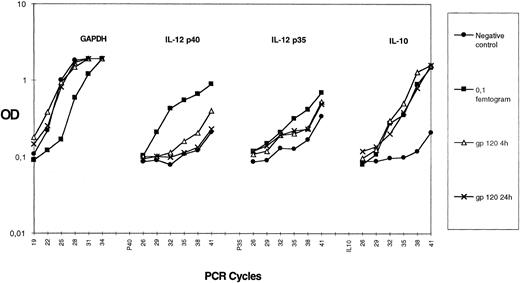
HIV gp120 induces IL-10 but not IL-12 production by purified monocytes.Several reports have shown that monocytes/macrophages can produce different cytokines in response to HIV infection or after exposure to HIV-gp120.29 30 We studied the capacity of HIVgp120 to induce IL-10 and IL-12 expression in a highly purified monocyte population. We prepared greater than 95% pure monocytes using an elutriation method (as described in the Materials and Methods). Purified monocytes were treated with rgp120 (3 μg/mL) for 4 or 24 hours and semiquantitative PCRs for IL-10; IL-12 p40; IL-12 p35; and the house keeping gene, GAPDH, were performed on cDNA. As shown in Fig 5, there is a similar amount of GAPDH in all cDNA preparations. rgp120 induces IL-10 mRNA accumulation, which occurs from 4 hours and persists at 24 hours. Expression of IL-12 p35 mRNA is slightly enhanced in contrast to p40 mRNA, which is not induced after exposure to rgp120. The level of rgp120-induced IL-10 protein in culture medium after 24 hours was 60 ± 15 pg/mL (mean ± standard error or mean of quadruplicate culture samples), whereas purified monocytes cultured in the medium alone did not produce detectable IL-10 (sensitivity of the assay <5 pg). Furthermore, no IL-12 p40 protein or bioactive p70 was detectable after 24 hours of monocyte culture with rgp120 with the ELISA kits used.
Effect of HIV recombinant gp120 on IL-10 mRNA accumulation in monocytes. Elutriated monocytes were cultured with HIV recombinant gp120. Cells were harvested for IL-10, p35, p40, and GAPDH mRNA assay at 4 hours or 24 hours after the beginning of the culture. For each gene, the 0.1-fg-point of purified PCR product amplified in the same experiment is shown. Each point plotted represents the mean of duplicate culture samples. Similar results were obtained in another experiment.
Effect of HIV recombinant gp120 on IL-10 mRNA accumulation in monocytes. Elutriated monocytes were cultured with HIV recombinant gp120. Cells were harvested for IL-10, p35, p40, and GAPDH mRNA assay at 4 hours or 24 hours after the beginning of the culture. For each gene, the 0.1-fg-point of purified PCR product amplified in the same experiment is shown. Each point plotted represents the mean of duplicate culture samples. Similar results were obtained in another experiment.
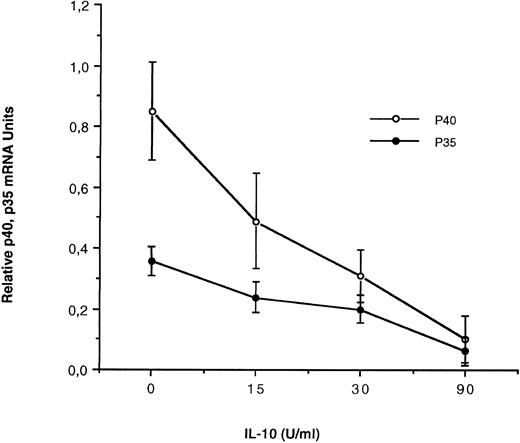
IL-10 inhibits IL-12 subunit expression at the RNA level by SAC-activated monocytes.It has been previously shown that IL-10 inhibits IL-12 production by SAC-activated peripheral blood mononuclear cells.31 We have studied, precisely, in our experimental conditions, the effect of IL-10 on IL-12 subunit mRNA expression in SAC-activated monocytes using quantitative PCR. For this purpose, monocytes were preincubated 4 hours with increased concentrations of human recombinant IL-10 (0, 15, 30, or 90 U/mL), then activated by SAC 4 hours. mRNA were extracted and p35 and p40 mRNA levels were evaluated using quantitative PCR. In these conditions, as shown in Fig 6, IL-10 induces a dose-dependent inhibitory effect on both IL-12 p40 and to a lesser extent p35 mRNA levels in response to SAC.
Effect of IL-10 on IL-12 p35 and p40 mRNA accumulation in monocytes. Monocyte-enriched adherent cells were preincubated for 4 hours with recombinant IL-10 at 0, 15, 30, and 90 U/mL; activated with SAC for 4 hours and finally harvested for mRNA assay. Each value represents the mean ± SEM of triplicate culture samples.
Effect of IL-10 on IL-12 p35 and p40 mRNA accumulation in monocytes. Monocyte-enriched adherent cells were preincubated for 4 hours with recombinant IL-10 at 0, 15, 30, and 90 U/mL; activated with SAC for 4 hours and finally harvested for mRNA assay. Each value represents the mean ± SEM of triplicate culture samples.
IL-10 neutralizing antibody reconstitutes IL-12 p40 production by gp120-treated monocytes.To determine the role of IL-10 in the rgp120-inhibited IL-12 production mechanism, IL-10 neutralizing monoclonal antibody was added at different concentrations (0.01, 1, and 10 μg/mL), with or without rgp120, 24 hours before monocyte-SAC activation. As shown in Fig 7, by neutralization of endogenous IL-10, p40 production was enhanced in both supernatants of gp120-treated or nontreated cells. However, the level of p40 was significantly lower in gp120-treated samples at nonsaturating concentrations of IL-10 monoclonal antibody (0.01 and 1 μg/mL). At high concentrations of anti–IL-10 (10 μg/mL), there was no significant difference between gp120-treated cells or non–gp120-treated cells. These data show that IL-10 production was higher in gp120-treated cells and that gp120-induced IL-10 production is involved in p40 downregulation.
DISCUSSION
The defect of IL-12 production by SAC-activated monocytes observed in vivo in HIV-infected patients and in vitro after HIV infection of monocytes remains incompletely explained.21-23 However, it seems very probable that this deficit is not directly related to monocyte infection but rather to soluble factors such as cellular or viral products produced during the HIV infection.21 Based on this hypothesis, we have studied the effect of monocyte-preincubation with HIV-gp120 on IL-12 expression at mRNA and protein level by SAC-activated monocytes. To develop optimal conditions for this study, we have first determined kinetics of IL-12 p35 and p40 mRNA accumulation in monocytes in response to SAC. Our results show that SAC-activation induces a rapid accumulation of IL-12 p40 transcripts, which reaches a peak after 4 hours (approximately a 20-fold increase over baseline). In contrast with other investigators who report that p35 is constitutive and minimally regulated,5 our results show that IL-12 p35 exhibits kinetics similar to those of p40 but with a weaker level of expression (approximately a sevenfold increase over baseline, (Fig 2). This quantitative differential expression of the two mRNA subunits, in agreement with the fact that monocytes produce an excess of p40 protein over the p70 heterodimer, probably has a biological significance. It may induce, at translation steps, favorable conditions for heterodimer formation. Alternatively, p40 subunit can exhibit a biological activity distinct from the IL-12 heterodimer. In this regard, it has recently been shown in the murine system that IL-12 p40 homodimer is a potent IL-12 heterodimer antagonist, which blocks binding of IL-12 to IL-12 receptor and could modulate its biological activity.32 We show that monocytes or macrophages pretreated with HIV-gp120 for 24 hours and then activated by SAC until the peak of mRNA accumulation (4 hours) have a decreased ability to express both p35 or p40 mRNA and to produce IL-12 proteins (Figs 3, 4, and 7). This inhibitory effect can be the consequence of a direct action of the retroviral glycoprotein or an indirect effect mediated by one or several inhibitory factors produced during the preincubation step with HIV-gp120. It has recently been shown, using PCR and quantification of PCR products by densitometric analysis of gels, that a synthetic retroviral peptide partially homologous to HIV-gp41 could inhibit IL-12 p40 mRNA accumulation when it was added simultaneously with staphylococcal enterotoxin A.33 These observations suggest a direct effect of this viral peptide on IL-12 p40 mRNA accumulation.33 In contrast, in this work, by adding simultaneously rgp120 and SAC to monocytes or macrophage cultures, no inhibitory effect has been observed on either IL-12 p35 or p40 mRNA accumulation or p40 protein and p70 heterodimer production (data not shown). Thus, our observations suggest rather an indirect effect of HIV-gp120 mediated by one or several inhibitory factors produced by monocytes during the gp120 preincubation step. Several cytokines are produced after interaction between HIV-gp120 and monocytes/macrophages.29,30 IL-10 is a likely candidate because it is an important negative regulator of monokine secretion.34 IL-10 is produced relatively late after SAC or LPS activation of monocytes and downregulates the production of numerous proinflammatory cytokines such as IL-1, IL-6, IL-8, and tumor necrosis factor α (TNFα).34 It has been previously shown that IL-10 inhibits IL-12 expression by PBMC in response to SAC.31 Confirming and extending these data, we show, using quantitative PCR, that 4 hours of preincubation with IL-10 induces in SAC-activated monocytes a dose-dependent inhibition in both IL-12 p35 and p40 mRNA accumulation (Fig 6). These results, which were obtained after 4 hours of SAC activation, suggest an inhibitory effect of IL-10 on IL-12 subunit rate transcription. Several studies have shown that IL-10 is induced in vivo and in vitro during HIV infection and suggest that IL-10 plays an important role in immune dysregulation in HIV-infected patients.35,36 The neutralization of endogenous IL-10 production can restore certain defective antigen-specific T-helper function of PBMC from HIV+ individuals.36 In a recent publication, IL-10 mRNA was detected in all lymph nodes from AIDS patients examined but was absent in normal lymph node biopsies.35 The cells that produced the IL-10 within the patient's lymph nodes were not identified, but it has been shown that biologically active IL-10 is produced by HIV-infected monocytes or macrophages as early as 12 hours after the infection.35 In our experimental conditions and in agreement with another previously published study,30 we show that HIV-gp120 clearly induces on highly purified monocytes a consequent IL-10 mRNA expression (Fig 5) and a significant but weak level of proteins compared with other macrophage stimuli, such as SAC or lipopolysaccharide (LPS), probably because SAC and LPS induce IL-10 mRNA expression and an efficient translation and secretion of IL-10 mRNA and proteins. In these experiments, no significant IL-12 p40 mRNA expression, IL-12 p40 protein, or bioactive p70 secretion were detected. Further experiments are necessary to clarify the exact molecular mechanism involved in the HIV-gp120 induction of IL-10. It may be the consequence of a direct effect of HIV-gp120 on the regulation of IL-10 mRNA transcription or an indirect effect through the induction of other cytokines, such as TNFα, a known potent inducer of IL-10 mRNA accumulation in monocytes/macrophages.37 By using a neutralizing IL-10 antibody, we reconstitute IL-12 production by gp120-treated monocytes, confirming the role of IL-10 in the rgp120-inhibited IL-12 production mechanism (Fig 7). Our data and that of others suggest that HIV-gp120 acts on monocytes/macrophages by first inducing the expression of several monokines such as IL-6, IL-1, TNFα,29,38,39 and IL-10 but not IL-12, probably because IL-12 p40 gene expression requires other signals. HIV-gp120–induced IL-10 acts secondly by downregulating IL-12 at the RNA level during the subsequent SAC-activation step. In addition, SAC can provide an efficient signal to translate gp120-induced IL-10 mRNA, which upregulates the IL-10 level in gp120-treated supernatants. Despite the fact that neutralization of endogenously produced IL-10 enhances TNFα and IL-6 production, we did not find, in our experimental conditions, significant inhibition for these inflammatory cytokines in gp120-treated preparations (data not shown); however, the regulation of IL-6 or TNFα production by monocytes includes many components such as TNFα itself, which is induced by HIV-gp120 and which upregulated IL-6 and its own production.37
In vivo, productive HIV infection of lymphocytes and macrophages, in particular in lymph nodes, can provide a continual source of gp120 that induces permanent local production of IL-10, which downregulates IL-12 production by surrounding infected or uninfected macrophages. This local cytokine disregulation can alter T-helper development and then the efficiency of immune response. In this regard, additional experiments are being conducted to study the effect of HIV infection and HIV-gp120 on IL-12 production of macrophages via CD40-CD40 ligand interaction. On the other hand, clinical trials with IL-12 are in progress in HIV-infected patients. It would be interesting to analyze immune functions, including T-helper functions, before and after IL-12 administration.
Supported by ANRS-Sidaction-Fondation pour la Recherche Médicale.
Address reprint requests to Y. Taoufik, PhD, Laboratoire Virus, Neurone et Immunité (CR INSERM 4 U012B, Pr. M Tardieu), Faculté de médecine de Bicêtre, Université Paris-Sud, 63, rue Gabriel Peri, Le Kremlin-Bicêtre Cedex, 94276, France.