Abstract
Glucocorticoids (GC) have long been used as the most effective agents for the treatment of allergic diseases accompanied by eosinophilia such as chronic asthma and atopic dermatitis. The development of chronic eosinophilic inflammation is dependent on interleukin-5 (IL-5), a selective eosinophil-activating factor, produced by helper T cells. To delineate the regulatory mechanisms of human IL-5 synthesis, we established allergen-specific CD4+ T-cell clones from asthmatic patients. GC efficiently suppressed IL-5 synthesis of T-cell clones activated via either T-cell receptor (TCR) or IL-2 receptor (IL-2R). Induction of IL-5 mRNA upon TCR and IL-2R stimulation was totally inhibited by dexamethasone. Human IL-5 promoter/enhancer-luciferase gene construct transfected to T-cell clones was transcribed on either TCR or IL-2R stimulation and was clearly downregulated by dexamethasone, indicating that the approximately 500-bp human IL-5 gene segment located 5′ upstream of the coding region contains activation-inducible enhancer elements responsible for the regulation by GC. Electrophoretic mobility shift assay analysis suggested that AP-1 and NF-κB are among the possible targets of GC actions on TCR-stimulated T cells. NF-AT and NF-κB were not significantly induced by IL-2 stimulation. Our results showing that GC suppressed IL-5 production by human CD4+ T cells activated by two distinct stimuli, TCR and IL-2R stimulation, underscore the efficacy of GC in the treatment of allergic diseases via suppression of T-cell IL-5 synthesis.
INTERLEUKIN-5 (IL-5) is a potent selective growth factor, differentiation factor, activating factor, and chemotactic factor for human eosinophils.1-3 Several animal studies have shown that tissue infiltration of eosinophils is dependent on IL-5 produced by T cells.4,5 Persistent eosinophilic inflammation of the bronchial mucosa is a characteristic pathologic feature of bronchial asthma.6-8 The number of CD4+ T cells expressing IL-5 mRNA is increased in the bronchial mucosa of symptomatic asthmatics,9 is correlated with the number of activated eosinophils, and is further increased upon allergen inhalation challenge.10,11 L-5 is the predominant eosinophil-active cytokine present in the broncho-alveolar lavage fluids obtained during allergen-induced late-phase reaction.12 Oral prednisolone therapy reduced serum IL-5 concentration13 and the number of IL-5 mRNA-expressing cells in broncho-alveolar lavage fluid,14 in parallel with clinical improvement. Control of T-cell IL-5 production seems to be an effective strategy for the management of allergic diseases characterized by eosinophilic inflammation.
Glucocorticoids (GC) have long been considered the most effective treatment for eosinophilic disorders, including chronic asthma.15 The efficacy of GC is ascribed to their multiple pharmacologic actions, one of which is the suppression of inflammatory cytokine production.16,17 We and others reported that IL-5 synthesis by peripheral blood mononuclear cells was inhibited by GC in vitro.18,19 Because of the mixed cell preparations used in those studies, the question of whether GC act directly on T cells or not still remains to be answered. It is well known that GC induce non-T cells to produce anti-inflammatory mediators, eg, lipocortins, which exhibit immunosuppressive effects on T cells.20-24 To address this issue, we used allergen-specific human T-cell clones in the present study and showed that GC directly act on T cells to suppress IL-5 synthesis at the level of gene transcription.
We have recently reported that human helper T-cell clones produce IL-5 in response to IL-2 as well as to antigenic stimulation.25 Several investigators have reported that the production of not only Th2 cytokines but also IL-2 (a Th1 cytokine) by infiltrating T cells is enhanced at the sites of allergic inflammation.26-29 Locally produced IL-2 may facilitate the development of eosinophilic inflammation through inducing IL-5 production by T cells. IL-2 exerts its effects after binding with its receptor, which is composed of three subunits, IL-2 receptor (IL-2R) α, β, and γ chain.30,31 The IL-2R signal activates multiple signal transduction pathways, eg, protein tyrosine kinases,32,33 PI-3 kinase,34 and proto-oncogenes.35-37 The signal transduced by IL-2R is reported to be very distinct from the signal transduced by T-cell receptor (TCR) in that (1) TCR signal induces Ca2+ influx but IL-2R signal does not,38,39 (2) TCR signal induces PI turnover but IL-2R signal does not,40 (3) TCR signal induces protein kinase C (PKC) activation but IL-2R signal does not,41,42 and (4) TCR signal is sensitive to FK506 and cyclosporin, but IL-2R signal is not.43-45 It was also reported that TCR-induced cytokine production was inhibited by GC at quite low concentrations, but, in contrast, the proliferation of murine cytotoxic T cells induced by IL-2 was affected only marginally by GC,46 suggesting that the TCR signal is mediated by quite distinct pathways from the IL-2R signal. So it is now an intriguing question as to whether IL-2–induced IL-5 synthesis is inhibited by GC as effectively as TCR-stimulated IL-5 synthesis. We showed here for the first time that GC inhibited IL-2–induced cytokine synthesis and proliferation of helper T cells. Our results further supported the efficacy of GC for the treatment of atopic diseases by acting as IL-5 synthesis inhibitors.
MATERIALS AND METHODS
Reagents.Hydrocortisone, prednisolone, and dexamethasone were purchased from Sigma Chemicals (St Louis, MO) and dissolved in ethanol at a concentration of 2 mmol/L as stock solutions. Anti-CD3 (Leu 4), anti-CD4 (Leu 2), and anti-CD8 (Leu 3) monoclonal antibodies (MoAbs) were purchased from Becton Dickinson (San Jose, CA). Anti-CD3 MoAb (OKT3) was from Ortho (Raritan, NJ). Phorbol 12-myristate 13-acetate was from Sigma and ionomycin was from Calbiochem (La Jolla, CA). Recombinant human IL-2 (rIL-2) was kindly provided by Shionogi Pharmaceutical Co (Osaka, Japan). The specific activity of the recombinant material for IL-2 was approximately 1 U/ng protein. Recombinant Der f II (rDer f II) was directly expressed in Escherichia coli using the Der f II cDNA clone and prepared as previously described.47 AIM-V medium (GIBCO BRL, Gaitherburg, MD) was used for T-cell cultures. RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 U/mL penicillin was used for the transient transfection experiments.
Establishment of antigen-specific T-cell clones. Der f II-specific human T-cell clones were derived from peripheral blood mononuclear cells (PBMC) of atopic asthmatic patients by antigen stimulation followed by the limiting dilution method, as described previously.48
Briefly, PBMC (2 × 106/mL) were cultured with rDer f II protein (1 μg/mL) for 10 days in 24-well culture plates and nonadherent cells were recovered. Then,102 to 104 live cells were cultured in 96-well round-bottom culture plates (Nunc, Roskilde, Denmark) with antigen and 2,500-rad–irradiated autologous PBMC (5 × 104 cells). Fresh medium containing 10 U/mL rIL-2 was added once weekly. When less than 1 of 10 wells contained proliferating cells, the resulting cell lines were considered to have originated from a single clone. To ensure their clonality, these T-cell clones were further subcloned by limiting dilution using irradiated autologous PBMC and antigen. After 10 to 14 days, expanding cultures were transferred to 24-well culture plates (Becton Dickinson). T-cell clones were maintained by antigenic stimulation with irradiated autologous PBMC (2 × 106/well) and rDer f II protein every 2 to 3 weeks.
Stimulation of T-cell clones.T cells were harvested at least 10 days after the last antigenic stimulation, layered onto Ficoll-Paque, and centrifuged. The interface was recovered, washed twice, and resuspended in fresh medium. The resulting preparation usually consisted of more than 98% CD3+ cells, as determined by flow cytometry. Cells (105/well) were cultured in triplicate with various stimuli in 96-well round-bottom culture plates for 24 hours, and then supernatants were harvested and kept frozen at −70°C until use. In some cultures, wells were preincubated with 10 μg/mL anti-CD3 MoAb (OKT3) in 0.05 mol/L carbonate-bicarbonate buffer (pH 9.6) at 4°C overnight. For proliferation analysis, cells were cultured for 72 hours. 3H-Thymidine (0.5 mCi/well) was pulsed for the last 16 hours. To obtain cytoplasmic RNA, T cells (4 × 106/well) were cultured in 24-well culture plates with various stimulants for the designated time periods.
Quantitation of cytokines.IL-5 was measured by a sandwich enzyme-linked immunosorbent assay (ELISA) using monoclonal antihuman IL-5 (D138) as the capture antibody and biotinylated purified rabbit antihuman IL-5 as the second antibody, as described previously.49 The linear portion of the standard curve was between 3.9 and 500 pg/mL. IL-2 and IL-4 were measured by specific ELISA (Quantikine ELISA kits; R&D Systems, Minneapolis, MN) according to the manufacturer's directions.
Reverse transcription-polymerase chain reaction (RT-PCR) assay.IL-5 gene expression was analyzed using the RT-PCR method as reported previously.19 Briefly, RNA was extracted from the pelleted cells essentially following the one-step acid guanidinium isothiocyanate/phenol-chloroform extraction method of Chomczynski and Sacchi50 using Isogene (Nippongene, Tokyo, Japan). cDNA was synthesized from 1 μg of cytoplasmic RNA using random primers and murine Moloney leukemia virus reverse transcriptase (GIBCO BRL). PCRs were performed using the following primers (Clontech, Palo Alto, CA): IL-5: sense strand primer, 5′-GCTTCTGCATTTGAGTTTGCTAGCT-3′, and antisense strand primer, 5′-TGGCCGTCAATGTATTTCTTTATTAAG-3′; β-actin: sense strand primer, 5′-ATGGATGATGATATCGCCGCG-3′, and antisense strand primer, 5′-CTAGAAGCATTTGCGGTGGACGATGGGGGCC-3′. To 50 μL (final volume) of amplification solution (50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 8.3, 2 mmol/L MgCl2 , 0.01% [wt/vol] gelatin, 0.2 mmol/L of each deoxynucleotide triphosphate; Toyobo, Osaka, Japan), 2 μL of cDNA (corresponding to about 250 ng of starting RNA material), 0.4 μmol/L of each primer, and 2 U of Taq DNA polymerase (AmpliTaq; Perkin Elmer Cetus, Norwalk, CT) were added. The mixture was heated at 95°C for 5 minutes, followed by 25 cycles, each consisting of incubations for 1 minute at 95°C, 2 minutes at 60°C, and 3 minutes at 72°C. The PCR products were analyzed by agarose gel electrophoresis in the presence of ethidium bromide.
Plasmid constructs.Human genomic DNA isolated from HeLa cells was used as a template. IL-5 promoter/enhancer gene (−511 to +4 relative to the transcription initiation site) was PCR amplified using a sense strand primer (5′-ATACTCGAGGGATCCTAATCAAGACCC-3′) and an antisense strand primer (5′-TGCAAGCTTTGCATAGTACAAGACTGC-3′) according to the DNA sequence reported by Tanabe et al.51 This 515-bp PCR product was inserted into the Xho I-HindIII site of the pGL2 basic vector (Promega, Madison, WI) to construct pIL-5(-511)Luc. Identity of the promoter/enhancer sequence with the originally reported DNA sequence was confirmed by chain termination sequencing using Sequenase 2.0 (Strategene, La Jolla, CA). pCMV-β-gal control vector (Riken DNA Bank, Tokyo, Japan) was used as a transfection control.
Transient transfection, luciferase assay, and β-galactosidase assay.T-cell clones were transfected by electroporation using a Gene Pulser (Bio-Rad, Richmond, CA). Briefly, 2.5 × 107 T-cell clones in 500 μL RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 U/mL rIL-2 in a 0.4-cm electroporation cuvette were incubated with 50 μg of pIL-5(-511)Luc and 10 μg of pCMV-β-gal and electroporated at 270 V, 960 μF with a mean time constant of 19 milliseconds. After transfected cells were cultured in a fresh medium with or without stimulation for 24 hours, protein extracts were prepared and luciferase activity was assayed using the Luciferase Assay System (Promega). Relative luciferase unit (RLU) was calculated by the following formula: RLU = Luciferase Activity in Cell Lysate/Protein Content (in milligrams per milliliter). β-Galactosidase activity was assayed using chlorophenol red-β-D-galactopyranoside (CPRG) as the substrate. First, 150 μL of cell lysates diluted with Z buffer (0.1 mol/L Na2HPO4 -NaH2PO4 , pH 7.5, 10 mmol/L KCl, 1 mmol/L MgSO4 , 50 mmol/L 2-mercaptoethanol) and 30 μL of 15 mmol/L CPRG were incubated at 37°C for 30 minutes. The reaction was then stopped by the addition of 75 μL of 1 mol/L Na2CO3 and OD574 was measured.
Preparation of nuclear extracts.Crude nuclear and cytoplasmic extracts were prepared from unstimulated or stimulated cells as described by Schreiber et al,52 with modifications. Cells were washed in ice-cold phosphate-buffered saline, suspended at 5 × 107cells/mL in ice-cold buffer A (10 mmol/L HEPES-KOH, pH 7.9, 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.1 mmol/L EGTA, 1 mmol/L dithiothreitol [DTT], and 0.5 mmol/L phenylmethyl sulfonyl fluoride [PMSF ]) and kept on ice for 15 minutes. Then 1/16 vol of 10% Nonidet P-40 was added and the mixture was vigorously vortexed. After centrifugation at 12,000 rpm for 30 seconds at 4°C, the cytoplasmic supernatant was retained on ice and the nuclear pellet was washed with the same buffer (buffer A containing NP-40). The pellet was next incubated with 3 vol of ice-cold buffer C (10 mmol/L HEPES-KOH, pH 7.9, 400 mmol/L NaCl, 10 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L DTT, 1 mmol/L PMSF ) at 5 × 108 nuclei/mL for 15 minutes and centrifuged at 15,000 rpm for 15 minutes at 4°C. The nuclear and cytoplasmic supernatants were kept frozen in aliquots at −70°C. For some experiments, they were dialyzed against 500 vol of dialysis buffer (10 mmol/L HEPES-KOH, pH 7.9, 50 mmol/L NaCl, 50% glycerol [vol/vol], 1 mmol/L DTT, and 1 mmol/L MgCl2 ) for 12 hours at 4°C.
Electrophoretic mobility shift assay (EMSA).The oligonucleotides used for AP-1, NF-κB and Oct-1 mobility shift assays were purchased from Promega: AP-1 (5′-CGCTTGATGAGTCAGCCGGAA-3′),53 NF-κB (5′-AGTTGAGGGGACTTTCCCAGGC-3′),54 and Oct-1 (5′-TGTCGAATGCAAATCACTA GAA-3′).55 NF-AT oligonucleotides were from Sawady Technology (Tokyo, Japan); NF-AT (5′-GGAGGAAAAACTGTTTCATACAGAAGGCGT-3′) from distal NF-AT site of human IL-2 gene.56 Pairs of synthetic high-performance liquid chromatography-purified oligonucleotides containing complementary sequences were annealed by boiling equimolar concentrations of each strand for 10 minutes and allowing the mixture to slowly cool in a water bath to room temperature. Then, 3.5 pmol of annealed oligonucleotides was incubated in a 10-μL reaction mixture containing 70 mmol/L Tris-HCl, pH 7.6, 10 mmol/L MgCl2 , 5 mmol/L DTT, and 10 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham, Arlington Heights, IL) with 10 U of T4 polynucleotide kinase (Takara, Otsu, Japan) for 30 minutes at 37°C. The reaction was stopped by adding 1 μL of 0.5 mol/L EDTA and 89 μL TE buffer (10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA). Gel-shift analysis was performed according to the manufacturer's protocol (Gel Shift Assay Systems; Promega) with slight modifications. Thirty-five femtomoles of 32P end-labeled oligonucleotides was incubated in a 10-μL reaction mixture containing 10 mmol/L Tris-HCl, pH 7.5, 0.5 mmol/L EDTA, 0.5 mmol/L DTT, 4% glycerol, 50 mmol/L NaCl, 1 mmol/L MgCl2 , and 0.5 μg poly (dI-dC)⋅poly (dI-dC) with 4 μg of nuclear extracts for 30 minutes at room temperature. In competition experiments, a 20 to 200 molar excess of unlabeled oligonucleotides, relative to the probe, was added to the binding reaction. After incubation, bromophenol blue and xylene cyanol were added to 0.02% and the resulting complexes were resolved on 4% polyacrylamide gel (acrylamide:bisacrylamide, 30:1 wt/wt) by electrophoresis at 100 V in 0.5× TBE buffer (1× TBE: 89 mmol/L Tris-HCl, pH 8.0, 89 mmol/L boric acid, and 2 mmol/L EDTA) at room temperature. The gel was subsequently dried and exposed to RX film (Fuji Photo Film, Tokyo, Japan) at −70°C.
Protein assay.Protein concentrations were determined using bicinchoninic acid protein assay reagent (Pierce, Rockford, IL) according to the manufacturer's directions.
Statistical analysis.Statistical analysis was performed using the Student's t-test. A value of P < .05 was considered to be statistically significant. Responses are presented as the mean ± standard error of the mean (SEM).
RESULTS
GC suppressed cytokine production of human T-cell clones stimulated via T-cell receptor complex.The first experiment was performed to test whether IL-5 production of human helper T-cell clones is suppressed by GC. T-cell clones were used for experiments at least 10 days after the last antigenic stimulation. As described in the Materials and Methods, T cells obtained from the interface of Ficoll-Paque density gradient consisted of more than 98% pure CD3+ CD4+ cells. They were washed three times, resuspended in fresh medium, and stimulated via T-cell receptor using immobilized anti-CD3 MoAb. Culture supernatants were harvested after 24 hours. As shown in Fig 1, all of the three agents, hydrocortisone (A), prednisolone (B), and dexamethasone (C), suppressed the production of IL-5 by activated T-cell clones in a dose-dependent manner. The concentrations of each agent required for half maximal inhibition (IC50 ) were ∼100 nmol/L, ∼20 nmol/L, and ∼1 nmol/L, respectively. All three agents did not alter the viability of cells after 24 hours of incubation at the concentrations used in these experiments, thereby excluding nonspecific toxicity of the agents (data not shown). The results clearly indicate that GC directly act on T cells to suppress IL-5 synthesis.
TCR-induced IL-5 production was suppressed by glucocorticoids. A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2) were cultured in 96-well round-bottom culture plates pretreated with OKT3 MoAb (10 μg/mL). Designated concentrations of hydrocortisone (A), prednisolone (B), and dexamethasone (C) were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL).
TCR-induced IL-5 production was suppressed by glucocorticoids. A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2) were cultured in 96-well round-bottom culture plates pretreated with OKT3 MoAb (10 μg/mL). Designated concentrations of hydrocortisone (A), prednisolone (B), and dexamethasone (C) were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL).
Dexamethasone suppressed IL-5 production induced by IL-2.We have recently reported that T-cell clones that produce IL-5 upon antigenic stimulation also produce IL-5 in response to IL-2 and that IL-5 production of peripheral T cells is dependent on IL-2.25 PI turnover, PKC activation, and Ca2+ influx were not observed in T cells stimulated with IL-2,38-42 although these early biochemical events are the hallmarks of TCR-mediated T-cell activation.57 TCR-induced proliferation of T cells is inhibited by FK506, but IL-2–induced proliferation is not.43-45 Therefore, the effects of GC on IL-2–induced IL-5 production were next examined. T-cell clones HK5, YA5, HK2, and YA8 were stimulated by recombinant human IL-2 (100 U/mL) for 24 hours in the presence or absence of dexamethasone. As shown in Fig 2, dexamethasone clearly suppressed IL-5 production (A) and proliferation (B) of these T-cell clones in a dose-dependent manner, indicating that IL-2 signals leading to cytokine production and proliferation of helper T cells are both sensitive to GC. Essentially the same results were obtained in five other T-cell clones, indicating that this observation was not restricted to these particular clones (data not shown).
IL-2–induced IL-5 production and proliferation of T-cell clones were suppressed by dexamethasone. (A) A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2; [•] YA8) were cultured in 96-well round-bottom culture plates with human rIL-2 (100 U/mL). Designated concentrations of dexamethasone were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL). (B) For proliferation assays, cells were cultured for 72 hours. 3H-Thymidine (0.5 μCi/well) was pulsed for the last 16 hours. Data are expressed as the mean of triplicate cultures ± SEM. 3H-Thymidine incorporation of unstimulated HK5, YA5, HK2, and YA8 cells was 362 ± 154, 526 ± 193, 759 ± 227, and 668 ± 149, respectively.
IL-2–induced IL-5 production and proliferation of T-cell clones were suppressed by dexamethasone. (A) A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2; [•] YA8) were cultured in 96-well round-bottom culture plates with human rIL-2 (100 U/mL). Designated concentrations of dexamethasone were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL). (B) For proliferation assays, cells were cultured for 72 hours. 3H-Thymidine (0.5 μCi/well) was pulsed for the last 16 hours. Data are expressed as the mean of triplicate cultures ± SEM. 3H-Thymidine incorporation of unstimulated HK5, YA5, HK2, and YA8 cells was 362 ± 154, 526 ± 193, 759 ± 227, and 668 ± 149, respectively.
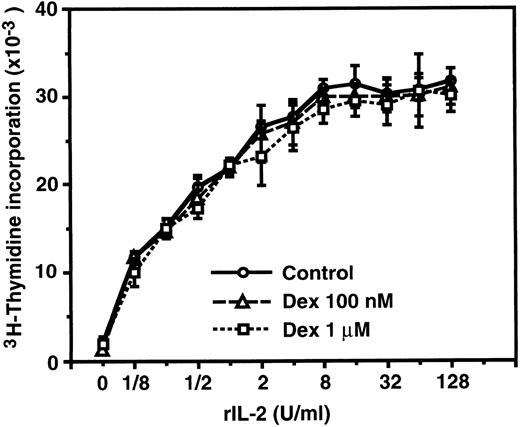
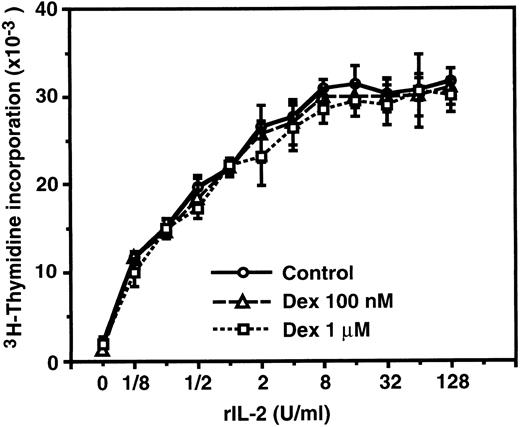
IL-2–induced proliferation of CTLL-2 cells is not affected by dexamethasone.Because dexamethasone-resistant proliferation of murine CD8+ T cells stimulated with IL-2 was reported by previous investigators,46 we confirmed the inability of dexamethasone to suppress IL-2–induced proliferation employing a murine cytotoxic T-cell line, CTLL-2, which is a well-characterized IL-2–dependent cell line commonly used for the bioassay of human IL-2, murine IL-2, and murine IL-4.58 As shown in Fig 3, CTLL-2 cells proliferated in response to exogenous rIL-2 in a dose-dependent manner. Dexamethasone at concentrations of 100 nmol/L and 1 μmol/L did not significantly affect the proliferative response of CTLL-2 cells, suggesting that the IL-2R signal leading to the proliferation of CD8+ (cytotoxic) T cells is transduced by GC-resistant signaling pathway(s) that is distinct from the IL-2R signal of CD4+ T cells.
IL-2–induced proliferation of CTLL-2 cells was not affected by dexamethasone. CTLL-2 cells (104/well) were incubated with the designated concentrations of rIL-2 for 24 hours. Either 100 nmol/L or 1 μmol/L dexamethasone was included from the start of some cultures. 3H-Thymidine (0.5 μCi/well) was pulsed for the last 6 hours. Data are expressed as the mean of triplicate cultures ± SEM.
IL-2–induced proliferation of CTLL-2 cells was not affected by dexamethasone. CTLL-2 cells (104/well) were incubated with the designated concentrations of rIL-2 for 24 hours. Either 100 nmol/L or 1 μmol/L dexamethasone was included from the start of some cultures. 3H-Thymidine (0.5 μCi/well) was pulsed for the last 6 hours. Data are expressed as the mean of triplicate cultures ± SEM.
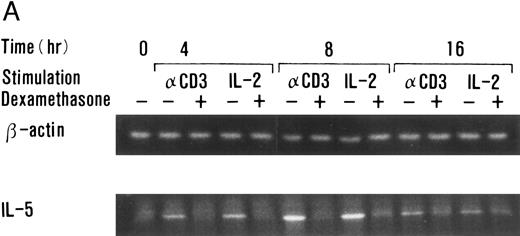
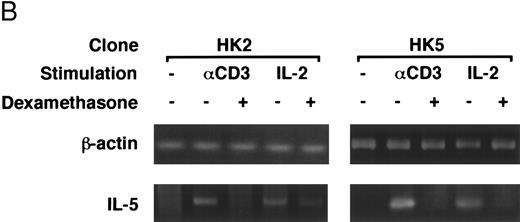
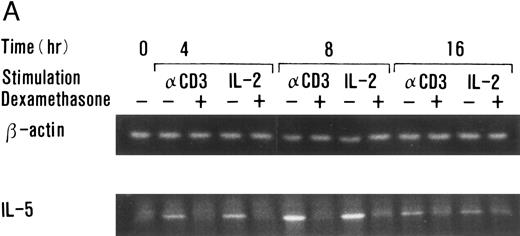
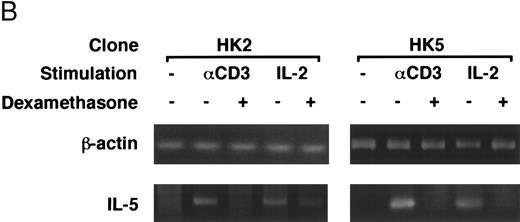
Dexamethasone suppressed IL-5 gene expressions.We next examined the effects of dexamethasone on IL-5 gene expression using RT-PCR analysis. T-cell clone (YA5) was stimulated with immobilized anti-CD3 MoAb or rIL-2 (100 U/mL) for the designated time periods and total cytoplasmic RNA was extracted. RNA was then reverse transcribed and amplified by PCR using primer sets specific for human IL-5 and β-actin. The number of cycles was titrated before the experiment and reactions were repeated for 25 cycles, because the PCR products were amplified exponentially between 23 and 30 cycles. IL-5 mRNA was induced in T-cell clones stimulated with either immobilized anti-CD3 antibody or rIL-2 and reached maximum after 6 to 9 hours, as reported previously.25 48 A representative result is shown in Fig 4. IL-5 mRNA induced by either stimulus was totally downregulated by dexamethasone. Essentially the same results were obtained when PCR was performed for 23 and 27 cycles, because the PCR reaction was linear between 23 and 30 cycles (data not shown), indicating that the amounts of PCR products at 25 cycles well reflect the relative quantity of mRNA in the original RNA preparations.
Dexamethasone suppressed IL-5 gene expression of human T-cell clones stimulated via TCR and IL-2R. (A) T-cell clone (YA5, 2 × 106/well) was cultured in 24-well culture plates for the designated time periods. Some wells were pretreated with OKT3 MoAb (10 μg/mL), as indicated. rIL-2 (100 U/mL) was added to the designated wells. Dexamethasone (100 nmol/L) was included throughout the culture period in some wells, as indicated. Cells were harvested after 0, 4, 8, and 16 hours. Total RNA was then extracted, reverse transcribed, and amplified by PCR. The 279- and 1,128-bp products correspond with the expected size of IL-5 and β-actin amplification products, respectively. (B) T-cell clone (HK2 and HK5) was stimulated for 8 hours.
Dexamethasone suppressed IL-5 gene expression of human T-cell clones stimulated via TCR and IL-2R. (A) T-cell clone (YA5, 2 × 106/well) was cultured in 24-well culture plates for the designated time periods. Some wells were pretreated with OKT3 MoAb (10 μg/mL), as indicated. rIL-2 (100 U/mL) was added to the designated wells. Dexamethasone (100 nmol/L) was included throughout the culture period in some wells, as indicated. Cells were harvested after 0, 4, 8, and 16 hours. Total RNA was then extracted, reverse transcribed, and amplified by PCR. The 279- and 1,128-bp products correspond with the expected size of IL-5 and β-actin amplification products, respectively. (B) T-cell clone (HK2 and HK5) was stimulated for 8 hours.
Dexamethasone suppressed the transcriptional activity mediated by ∼500-bp human IL-5 promoter/enhancer.It has been shown that dexamethasone inhibits the induction of IL-2 gene transcription mainly by interfering with AP-1 activity.59 60 We used a transient transfection system to analyze whether dexamethasone suppresses human IL-5 transcriptional activity. The 511-bp human IL-5 promoter/enhancer-luciferase gene construct, pIL-5(-511)Luc, was transiently transfected to T-cell clones by electroporation. The transfected cells were then stimulated with immobilized anti-CD3 MoAb or rIL-2 for 24 hours. As shown in Table 1, luciferase activity was induced upon stimulation and was clearly downregulated by dexamethasone, indicating that the effect of GC on IL-5 synthesis is exerted at the level of gene transcription. β-Galactosidase activity derived from pCMV-β-gal, a transfection control, did not differ significantly either between stimulated and unstimulated cells or between GC-treated and GC-nontreated cells. GC-sensitive transcription of pIL-5Luc was confirmed in two other T-cell clones (HK2 and HK5).
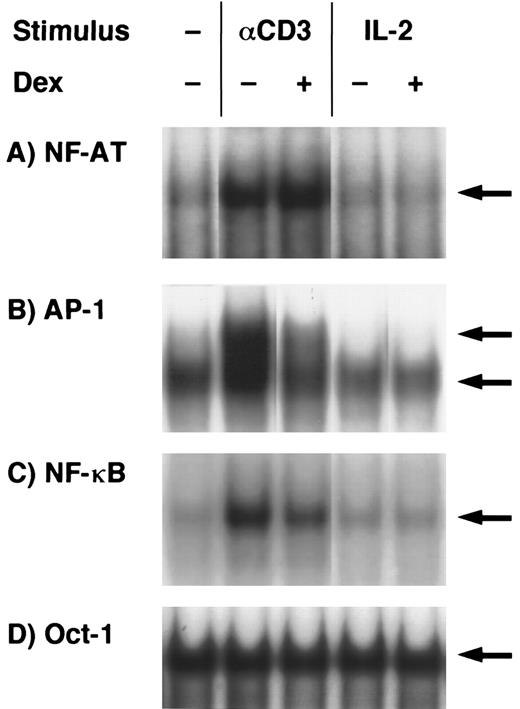
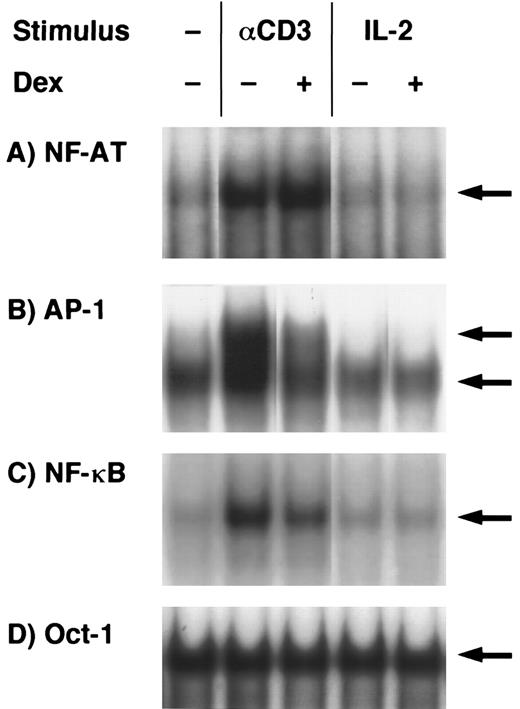
EMSA analysis of dexamethasone-treated T-cell clones.The findings given above that both activation signals mediated through TCR- and IL-2R–induced GC-sensitive gene transcription and protein synthesis of IL-5 prompted us to examine whether AP-1 and NF-κB, GC-sensitive transcription factors, were induced by either TCR or IL-2R stimulation and were downregulated by dexamethasone. Representative results are shown in Fig 5. Each band indicated by the arrowhead shows the specific binding that was diminished by the addition of excess amounts of unlabeled probes in the preliminary experiments (data not shown). NF-AT, AP-1, and NF-κB binding activities were clearly upregulated in the nuclear protein preparations upon stimulation with immobilized anti-CD3 MoAb. Oct-1 binding activities were constitutively present in the stimulated and unstimulated nuclear proteins and were not affected by dexamethasone. AP-1 and NF-κB binding activities were clearly downregulated by dexamethasone, suggesting that these transcription factors are the targets of GC action on IL-5 synthesis. NF-AT binding was not affected by dexamethasone. In contrast to the TCR stimulation, neither NF-AT nor NF-κB was significantly induced in the nuclear extracts of IL-2–stimulated T cells. Essentially the same results were obtained in three other T-cell clones (data not shown). Although AP-1 and NF-AT are critical transcription factors for the induction of IL-2 gene,57 these findings suggest that IL-5 gene transcription using human helper T cells may occur in the absence of apparent induction of AP-1 and NF-AT and still in a manner sensitive to GC.
Effects of dexamethasone on NF-AT, AP-1, NF-κB, and Oct-1 binding activities. T-cell clones (YA5) were stimulated with immobilized anti-CD3 MoAb or rIL-2 (100 U/mL) for 2 hours. Dexamethasone (1 μmol/L) was added to some cultures throughout the culture period, as indicated. Nuclear proteins were then extracted and DNA binding activity was measured by EMSA. Arrows indicate specific binding to NF-AT (A), AP-1 (B), NF-κB (C), and Oct-1 (D) oligonucleotides.
Effects of dexamethasone on NF-AT, AP-1, NF-κB, and Oct-1 binding activities. T-cell clones (YA5) were stimulated with immobilized anti-CD3 MoAb or rIL-2 (100 U/mL) for 2 hours. Dexamethasone (1 μmol/L) was added to some cultures throughout the culture period, as indicated. Nuclear proteins were then extracted and DNA binding activity was measured by EMSA. Arrows indicate specific binding to NF-AT (A), AP-1 (B), NF-κB (C), and Oct-1 (D) oligonucleotides.
DISCUSSION
In our present study, allergen-specific human T-cell clones established from atopic asthmatic donors were used to delineate the action of GC on IL-5 synthesis. It was clearly shown that IL-5 synthesis of human helper T cells induced by two distinct stimuli mediated through TCR and IL-2R was completely suppressed by GC (Figs 1 and 2). IL-5 mRNA expression induced by either stimulus was clearly downregulated by dexamethasone (Fig 4). Dexamethasone suppressed the transcriptional activity mediated through the ∼500-bp human IL-5 promoter/enhancer gene segment located 5′ upstream of the coding region (Table 1), indicating that the inhibition of IL-5 synthesis by GC was exerted at the level of gene transcription.
It has been shown that GC inhibit the production of various cytokines, such as IL-2, IL-3, IL-4, IL-6, IL-8, interferon-γ, and granulocyte-macrophage colony-stimulating factor.46,61-66 The mechanisms of action of GC have been elucidated since the molecular identification of the GC receptor (GR).67,68 GC exerts its effects through binding with its receptor existing in the cytoplasm. GC-GR complex can translocate into the nucleus and bind with the specific enhancer gene segment called glucocorticoid responsive element (GRE; GGTACANNNTGTTCT), either to enhance or suppress the target gene transcription. In some instances, GC-GR complex interferes with the transcriptional activity of AP-1, a ubiquitous transcription factor that is composed of Jun and Fos families (c-Jun, Jun-B, Jun-D, c-Fos, FosB, Fra1, and Fra2) through direct protein-protein interaction.69-71 The inhibition of IL-2 enhancer by GC maps to the locus containing AP-1 element.59,60 It is also reported that GC interfere with NF-κB activity through induction of IκB synthesis.72 Our present results are consistent with the previous reports that AP-1 and NF-κB are the targets of GC actions. The possible location of AP-1 and NF-κB elements responsible for the GC sensitivity of human IL-5 gene warrants further investigation.
Because the CD3 molecule is a component of the T-cell receptor complex, the activation signal transduced by the cross-linkage of CD3 molecules is considered to mimic antigenic stimuli delivered through TCR under physiologic conditions.57 We and others reported that IL-5 synthesis of human PBMC was suppressed by GC.18,19 Because GC induce anti-inflammatory second messengers, including lipocortins, in various cell populations,20-24 it is still unclear whether the suppressive effect of GC on IL-5 production is a direct action on T cells or an indirect one. Our present findings indicate that GC can exert their effects directly on T cells, because the T-cell preparations used in our study consisted of more than 98% pure T-cell populations.
In addition to the activating signal through T-cell receptor complex, exogenous IL-2 is sufficient to induce IL-5 synthesis by human allergen-specific T-cell clones.25 Although the roles of Th2 cytokines in atopic diseases have been emphasized,11,73 it has been reported that local IL-2 production was also enhanced during the late phase reaction after allergen exposure,26-29 suggesting the involvement of IL-2 (Th1 cytokine) in addition to Th2 cytokines in the pathogenesis of atopic diseases. So it is an intriguing question as to whether IL-2–induced IL-5 production is also suppressed by GC. IL-2 induces PTK activation and PI-3 kinase activation, but neither Ca2+ mobilization, PI turnover, nor PKC activation occurs in T cells in response to IL-2 stimulation,32-34,38-42 whereas these early biochemical events are the hallmarks of TCR-mediated T-cell activation.57 TCR signal is sensitive to FK506 and cyclosporin A, but IL-2R signal is not,43-45 indicating that cytokine synthesis induced by TCR signal is transduced by a quite different pathway from that induced by IL-2R signal. It was also reported that TCR-induced cytokine production was inhibited by quite low concentrations of GC, but, in contrast, the proliferation of murine cytotoxic T cells induced by IL-2 was affected only marginally by GC,46 suggesting that the signals mediating cytokine synthesis use different pathways from those mediating IL-2–induced cell proliferation in T cells. It was clearly shown here for the first time that not only the proliferation but also IL-5 synthesis of human helper T cells induced by IL-2 is totally suppressed by GC. These findings further support the efficacy of GC in the treatment of eosinophilic inflammation acting as IL-5 synthesis inhibitors. GC suppressed the expression of IL-5 mRNA induced by IL-2 (Fig 4) and at the same time suppressed the gene transcription mediated by the human IL-5 promoter/enhancer gene located 5′ upstream of the coding region (Table 1), suggesting that GC exert their action on T cells at the transcriptional level.
EMSA analysis indicated that AP-1 and NF-κB complex were among the possible targets of GC action on TCR-stimulated helper T cells (Fig 5), which is consistent with previous reports.59,60,72 Whether the effect of GC on IL-5 gene specifically maps to AP-1 or NF-κB element within human IL-5 promoter/enhancer gene should further be determined by the transfection analysis using mutant promoter/enhancer plasmids as performed in IL-2 gene.60 AP-1, NF-κB, and NF-AT binding was not detectable after IL-2R stimulation, in contrast to TCR stimulation, despite the fact that endogenous IL-5 gene expression (Fig 4) and transcriptional activity as determined by pIL-5(-511)Luc (Table 1) induced by either TCR or IL-2R stimulation were quite comparable, suggesting that AP-1 and NF-κB activities seem to be important for IL-5 synthesis induced by TCR stimulation, but are not essential for IL-5 synthesis induced by IL-2. Because the induction of NF-AT, AP-1, and NF-κB complexes in IL-2–stimulated T cells may follow different kinetics than in TCR-stimulated cells, we examined NF-AT, AP-1, and NF-κB binding activities at different time points after IL-2 and TCR stimulation. No significant binding activities of NF-AT, AP-1, and NF-κB were detected at 0.5, 1, and 4 hours after IL-2 stimulation, in accordance with the results obtained at 2 hours (shown in Fig 5), although those binding activities were clearly induced by TCR stimulation (data not shown). The present findings suggest that GC may exert their effects through affecting a still unknown transcription factor(s) other than AP-1, NF-AT, NF-κB, and Oct-1, and a GC-sensitive unique transcription factor(s) might regulate human IL-5 gene activation in allergic diseases associated with eosinophilic inflammation such as asthma and atopic dermatitis.
To date, several transcription elements, including AP-1, NF-κB, NF-AT, Oct-1, GATA, and conserved lymphokine element 0 (CLE0), have been indicated by the EMSA and homology search analyses.74-78 NF-AT binding activity was induced in an activated murine mast cell line and the human IL-5 sequence between −120 and −98 corresponded to the activity.76 Both GATA binding element located −82 to −62 and CLE0 element located −56 to −42 are essential for the spontaneous, not inducible, transcriptional activity expressed in an adult T-cell leukemia virus-transformed T-cell line.77 Two octamer binding sites located −244 to −237 and somewhere between −67 and −30 were also identified.78 Our results showing that human IL-5 promoter/enhancer-luciferase gene construct transfected to T-cell clones was transcribed upon stimulation and was downregulated by dexamethasone (Table 1) indicate that the approximately 500-bp human IL-5 gene segment located 5′ upstream of the coding region contains an activation-inducible enhancer element(s) regulated by GC. Because the function of transcription factors is context-dependent, it is essential to determine (1) whether those possible elements homologous to the consensus binding sequences of known transcription factors such as NF-AT, AP-1, and NF-κB really bind their corresponding transcription factors and (2) whether those elements and transcription factors are functionally active in human helper T cells and regulated by GC. The studies necessary to answer these questions are currently being undertaken using transient transfection systems, EMSA, and footprint analyses using genuine human IL-5 sequences. The further identification of IL-2–responsive and GC-sensitive transcriptional elements in the human IL-5 promoter/enhancer gene will improve understanding of the molecular mechanisms of human T-cell IL-5 synthesis and may facilitate future therapeutic intervention for severe atopic diseases by the development of selective IL-5 synthesis inhibitors.
ACKNOWLEDGMENT
The authors thank Dr S. Morimura for helpful discussion, Dr W.A. Gray for reviewing this manuscript, and M. Komatsu and Y. Shirai for technical assistance.
Address reprint requests to Hirokazu Okudaira, MD, Phd, Associate Professor, Department of Medicine and Physical Therapy, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113, Japan.

![Fig. 1. TCR-induced IL-5 production was suppressed by glucocorticoids. A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2) were cultured in 96-well round-bottom culture plates pretreated with OKT3 MoAb (10 μg/mL). Designated concentrations of hydrocortisone (A), prednisolone (B), and dexamethasone (C) were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2891/4/m_bl_0032f1c.jpeg?Expires=1765901051&Signature=3WxKN1yLZ-7XmF8M93Aa3WRsn1OYgeaFN5b~FDp9vSaRapFUPj6J-86KtD7D8nXe3209PKwIJ0j85AH7Ku97EaR~lb8qtvxInahGh6ZECID9Y1aQkTTj8RJapcZ~xKf6IIrgXCLMqOFOinHtGNgqWJn3RavrEZL4flpg4ecmpUB81DLPJfQRRcfUgVSpRswMdgZ2dvWCSXTgv1k0MhyUZTVVV9J~EW44ABSJJzc9p4NlavYIVbYtWalaP0kaDeQhOh5aMPESEL~cTOjezNRsP886oPp~3kWyf-1T3nTZ9cmOoEdKxuCu4zkQrSlhtf2qK2YQgm8oW7G~7-MBftpD4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. TCR-induced IL-5 production was suppressed by glucocorticoids. A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2) were cultured in 96-well round-bottom culture plates pretreated with OKT3 MoAb (10 μg/mL). Designated concentrations of hydrocortisone (A), prednisolone (B), and dexamethasone (C) were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2891/4/m_bl_0032f1a.jpeg?Expires=1765901052&Signature=0x3ZJWZJnLb4zuZrjPleK~Y~AyJsV8Sq2qOj1jKcvqkgbUgEt54b8LjxWKNC3pMyUi9ySCCtz0n9fetxm-8-IwFLdOt394nLNpMpMc0XEpyKpCP1eBXXal2nYjnYYzK34ilCKYsxK5JUQrb81cQKYErsayu1wdBcHWkoJLxfKwtgcrIMuMj-Wp2hcgaB9qivO87gy18fcddnfMnNeHeng-kKQ~2gtQivmoxtgT5b-9elZslmiESCrZ1IY4-GOgi5zAkbdItRMf-wS678YZ75dajWyecT2G2VHqD-XHPCkptjv~kndg6KTFAjTzx-qzhEPJXtaYbd~Nw5qYR6qu5OUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. IL-2–induced IL-5 production and proliferation of T-cell clones were suppressed by dexamethasone. (A) A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2; [•] YA8) were cultured in 96-well round-bottom culture plates with human rIL-2 (100 U/mL). Designated concentrations of dexamethasone were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL). (B) For proliferation assays, cells were cultured for 72 hours. 3H-Thymidine (0.5 μCi/well) was pulsed for the last 16 hours. Data are expressed as the mean of triplicate cultures ± SEM. 3H-Thymidine incorporation of unstimulated HK5, YA5, HK2, and YA8 cells was 362 ± 154, 526 ± 193, 759 ± 227, and 668 ± 149, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2891/4/m_bl_0032f2.jpeg?Expires=1765901052&Signature=kGN-WYuGa4-J-x2uWFGo~vuA-x5SO5~BVBM79DeBvkCtQo9zcDtZkTLBZLQJ6yDK1mUNTfxJMUjeSr7cEC3bPbb8LYacaRf38vtBfKoMTk9sc8CyBVCShIRTQvIuo~McHVl~40ryQ2x5BMfP4W4tQIYDocBPFy-O2mC7E8pFHhcrmi2epHGK0pAgwXUq0p4FhbBPGmjfbzgT~hHnbrZf7FjfPDIwLPb9JppmDnogn1H5ARBORc7dcx7P-F8Dv~qjiHhf8qXGPQFozumb3sERv37RavM4yG5qFbT1RWs989UY1xeXiz9nyDH1m-GhDxVCDb~bRIADYxSHofKQW8io2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 1. TCR-induced IL-5 production was suppressed by glucocorticoids. A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2) were cultured in 96-well round-bottom culture plates pretreated with OKT3 MoAb (10 μg/mL). Designated concentrations of hydrocortisone (A), prednisolone (B), and dexamethasone (C) were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2891/4/m_bl_0032f1c.jpeg?Expires=1765966564&Signature=Kj9KxlN12jj5t~qoXek8ccFU~ZWrwCcG6CVfS-XU59uPK9lUjH6wRrPQWEUpiMMu~s00mB8rqHCkA8Sr~t0-Pr386xDqcfk~eHESGBSh7MikclPfwkRMrap9d9Q7ngg-fQdcDFrywLj-NotsMOhj0cppegK5rX-5tlKnwnrWQgpTQQ7BHWME-L28dkBWaw0IoRemlK47q7eH~r39~ZubRdIpfPPr~XY5wpL8XGt1PF3LIuo4G3QAyEveTSypPH3~OXW3aMCWDYWn9rlfi61gLiCnRBDyhCAVzua9BhgsllXstI7dpWq-Kza-m4F8eqkIYTOIxlzA09fqWSifqyqIbg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. TCR-induced IL-5 production was suppressed by glucocorticoids. A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2) were cultured in 96-well round-bottom culture plates pretreated with OKT3 MoAb (10 μg/mL). Designated concentrations of hydrocortisone (A), prednisolone (B), and dexamethasone (C) were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2891/4/m_bl_0032f1a.jpeg?Expires=1765966564&Signature=qEAQaii6-J7vx9hlVHJIgoJ15B2GfA1tHsCUhRgLbSsmerTMr6bWAMpOpAumAVgD2EYbSgk7ntqRTNAmkEjENOwHskdN9sIhroxn8Qo-M4uWRhW0a-brqYPvbMuXPmbPWckddA33KG8ARPciceoX9-BWJrc-Da7GvNoHR~8YSBaGFIt5hIrMCSp9UhXE-8HO03F6Wge0~WjOYaBS-LUyfcdiI-g-KoGVHMqLhMD03g08Ggi6dxvGqol27ObQ4mOwnYPed8xPX9I4qKgCL2sv7PSulRy3S~Gm-RiYgETPlqkLKfVu2tFjfSRCpIMN-eaPH8p44GLvsXHREkDrobIpBw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. IL-2–induced IL-5 production and proliferation of T-cell clones were suppressed by dexamethasone. (A) A total of 105 T-cell clones ([○] HK5; [▵] YA5; [□] HK2; [•] YA8) were cultured in 96-well round-bottom culture plates with human rIL-2 (100 U/mL). Designated concentrations of dexamethasone were included from the start of some cultures. Culture supernatants were harvested after 24 hours and assayed for IL-5 by a specific ELISA. Data are expressed as the mean of triplicate cultures ± SEM. IL-5 production in the unstimulated cultures was always below the detection limit of the ELISA system (<1 pg/mL). (B) For proliferation assays, cells were cultured for 72 hours. 3H-Thymidine (0.5 μCi/well) was pulsed for the last 16 hours. Data are expressed as the mean of triplicate cultures ± SEM. 3H-Thymidine incorporation of unstimulated HK5, YA5, HK2, and YA8 cells was 362 ± 154, 526 ± 193, 759 ± 227, and 668 ± 149, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2891/4/m_bl_0032f2.jpeg?Expires=1765966564&Signature=1veZiRuyDB999wOnrGhrZpNvbd0F8DOwNGbWspzW~BmgDSaUOxjbZM1JGSpG0FboARvHMD6WvNvbTAAssJALJ5JOX4ijVj15ZCrfWCiqh8s4DlkqwF1bPuxlNNatW8XGNQfrOwqMCF6uKLQRQRKcka~6CXdGm2ZgpxZP6641lQdmFhRx42nb5PUrGxexlkc2xpjMn7qVTb~K6oCwWmRY6RcvOdghYT-NyWD6iNq7tl6gRi0DczLMU4SWUPXrYuvGayYZUBnvbYK4XdJuC-XZjjnVx41DMTecikRsxjYhZLdBVuF2UHMz-f5Q~zxh0npoImp1KZhfvw5QtKL7pTJ1Rw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)