Abstract
Fanconi anemia (FA) is a genetically and phenotypically heterogeneous disorder defined by cellular hypersensitivity to DNA cross-linking agents; mutations in the gene defective in FA complementation group C, FAC, are responsible for the syndrome in a subset of patients. We have performed an analysis of the clinical effects of specific mutations in the FAC gene. Using the amplification refractory mutation system assays that we developed to rapidly detect FAC mutations, at least one mutated copy of the FAC gene was identified in 59 FA patients from the International Fanconi Anemia Registry (IFAR). This represents 15% of the 397 FA patients tested. FA-C patients were divided into three subgroups based on results of a genotype-phenotype analysis using the Cox proportional hazards model: (1) patients with the IVS4 mutation (n = 26); (2) patients with at least one exon 14 mutation (R548X or L554P) (n = 16); and (3) patients with at least one exon 1 mutation (322delG or Q13X) and no known exon 14 mutation (n = 17). Kaplan-Meier analysis shows that IVS4 or exon 14 mutations define poor risk subgroups, as they are associated with significantly earlier onset of hematologic abnormalities and poorer survival compared to exon 1 patients and to the non-FA-C IFAR population. There was no direct correlation between the degree of cellular hypersensitivity to the clastogenic effect of diepoxybutane and severity of clinical phenotype. Sixteen of the 59 FA-C patients (27%) have developed acute myelogenous leukemia. Thirteen of these patients have died; AML was the cause of death in 46% of the expired FA-C patients. This study enables us to define this clinically heterogeneous disorder genotypically to better predict clinical outcome and aid decision-making regarding major therapeutic modalities for a subset of FA patients.
FANCONI anemia (FA) is a genetically and phenotypically heterogenous disorder defined by cellular hypersensitivity to DNA cross-linking agents such as diepoxybutane (DEB) and mitomycin C.1 Patients may be severely affected, with multiple congenital anomalies, or may have a mild phenotype with no major malformations.2-4 FA patients develop progressive bone marrow failure manifested by pancytopenia and are predisposed to acute myelogenous leukemia (AML); the age of onset of hematologic disease is variable.5 FA represents the most frequent cause of inherited aplastic anemia and is an important cause of childhood AML.2 6
FA patients constitute at least five different complementation groups (FA-A to FA-E) as defined by cell fusion experiments7; the gene for group C (FAC) has been cloned by functional complementation.8 The coding sequence of the FAC gene is comprised of 14 exons9 and encodes a protein of 558 amino acid residues with a molecular mass of 60 kD.10 Cytoplasmic localization has been shown to be essential for the intracellular activity of the FAC protein, but the mechanism of action of the protein is still unknown.10 FAC mutations have been identified in affected individuals by single-strand conformation polymorphism (SSCP) analysis and by chemical mismatch cleavage analysis.11-15 The pathogenicity of several of these have been shown by functional assays, which test the ability of mutated FAC cDNAs to alter the cellular phenotype of FA-C or normal cells.16-18 Mutation analysis of all 14 exons has failed to detect the alteration on the second allele of some compound heterozygotes.11 15
The most common FAC mutation is IVS4 +4 A>T, a splice site mutation in intron 4 found in patients of Ashkenazi Jewish ancestry.11,14,19 Less common mutations include 322delG and Q13X in exon 1, R185X in exon 6, and R548X and L554P in exon 14.11-13 15 We have developed amplification refractory mutation system (ARMS) assays for these six FAC mutations19 and have used these assays to screen the patients in the International Fanconi Anemia Registry (IFAR) database. Preliminary analysis suggests that the FAC genotype affects the phenotype and allows segregation of patients into risk groups.11 In this paper, we report the incidence of FAC mutated alleles in 59 patients enrolled in the IFAR and analyze phenotypic expression and clinical outcome by FAC genotype.
MATERIALS AND METHODS
Registry characteristics.The IFAR was established at The Rockefeller University in 1982 to collect clinical and genetic information from a large number of FA patients. Registration into the IFAR is usually at the time of diagnosis and information is collected initially on hematologic and congenital abnormalities. Comprehensive attempts to obtain follow-up data are made yearly. All reporting is voluntary and obtained with the consent of the patients or guardian. Diagnosis of FA is confirmed by study chromosomal breakage induced by DEB in peripheral blood (PB) lymphocytes. The protocol used for DEB testing is described in detail in Current Protocols in Human Genetics.20
Hematologic abnormalities.Onset of hematologic abnormalities is defined as the time at which one of the following laboratory parameters was observed; platelet count less than 100 × 109/L, hemoglobin (Hb) less than 10 g/dL or absolute neutrophil count (ANC) less than 1 × 109/L.5 Less severe decreases in blood cells and alterations in red blood cell mean corpuscular and fetal Hb (HbF ) are often noted first, but are not defined as hematologic abnormalities in determining age of hematologic onset. AML is defined as greater than 30% myeloid blasts in the bone marrow (BM) or greater than 20% myeloid blasts in the blood.
Congenital malformations.Patients were scored for the presence of major congenital malformations, defined as structural alterations that occur during embryogenesis that have medical and social consequences. Minor anomalies, which are unusual morphologic traits that are of no serious medical or cosmetic consequence to the patient, and abnormalities in growth parameters, were omitted in the scoring system used in this study. The spectrum of major congenital malformations found associated with FA patients in this study included abnormalities of the kidney and urinary tract, genitalia, heart, gastrointestinal system, central nervous system, and other skeletal system abnormalities in addition to defects of the radius and thumb.
FAC mutation analysis.DNA samples were obtained from the PB of patients enrolled in the IFAR. ARMS assays for six known FAC mutations were performed as previously described.19 Patients lacking all six of these FAC mutations were designated non-FA-C.
Statistical analysis.All computations were performed using SPSS 6.1.3 (SPSS, Inc, Chicago, IL). Correlation of mutation with DEB-sensitivity was calculated using analysis of variance. Correlation of mutation with age of hematologic onset and with survival for the patients with mutations in FAC was calculated with the Cox proportional hazards model using backward selection of the likelihood-ratio statistic based on the maximum partial likelihood estimates.21 Survival curves were calculated using the Kaplan-Meier method22; the interval was from birth to the date of the relevant event (hematologic onset, leukemia, or death) or to the last review. The univariate comparison between survival curves was performed using the log-rank test. The study closing date was May 1996 for the patients with mutations in FAC. The median age of onset of hematologic abnormalities and the estimated median survival for the 338 non-C patients were based on data collected for a previous study5; all patients from that study were subsequently tested for FAC mutations.
RESULTS
Incidence of FAC mutated alleles.Genomic DNA isolated from 397 FA patients in IFAR were screened by ARMS assays for six known FAC mutations. At least one mutated FAC allele was identified in 59 FA patients. This represents 14.9% of the patients tested. The distribution of FAC mutant alleles is shown in Table 1.
The most frequent allele identified is IVS4 +4 A>T. Twenty-five patients are homozygous for this allele and one patient is heterozygous with an unknown second mutant allele. The second most common allele identified is 322delG, a deletion of a single nucleotide in the first exon of the coding sequence. One patient is homozygous for 322delG. The second mutant allele is Q13X in 2 patients, R185X in 4 patients, R548X in 3 patients, and unknown in 7 patients. Q13X, another exon 1 mutation, is homozygous in one patient, associated with 322delG in two patients, and with an unknown mutant allele in two patients. R548X is the most common FAC mutation observed in exon 14. We identified 4 patients homozygous for this allele, and 3 patients heterozygous with a 322delG allele (as described above) and 5 patients heterozygous with an unknown second mutant allele. L554P, another exon 14 mutation was found in 4 patients, all with an unknown second mutant allele.
Genotype-phenotype correlations.To analyze genotype-phenotype correlations, we first determined the correlation of the six different FAC mutations with hematologic onset and with survival using the Cox proportional hazards model. Patients were scored for the presence or absence of mutations in exon 1 (322delG, Q13X), IVS4 (IVS4 +4 A>T), exon 6 (R185X), and exon 14 (R548X, L554P). Mutations in exon 14 and IVS4 correlated negatively with survival (exon 14, P = .009; IVS4, P = .0370) and with age of hematologic onset (exon 14, P < .00005; IVS4, P < .00005). FAC patients were divided into three groups based on these results: patients with at least one exon 14 mutation (n = 16); patients with at least one IVS4 mutation (n = 26); and patients with at least one exon 1 mutation, but no mutations in exon 14 or IVS4 (n = 17).
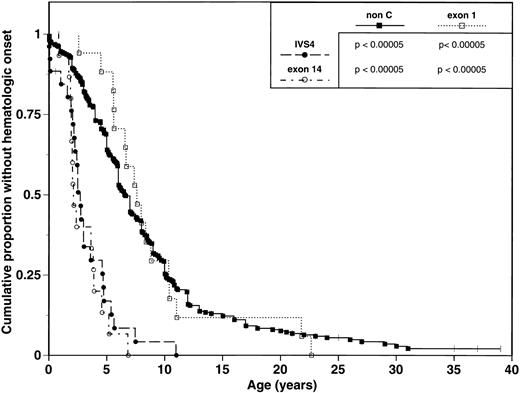
We analyzed and compared the clinical phenotypes of the three FAC subgroups, as defined above (Table 2). IVS4 and exon 14 subgroups are associated with a severe phenotype manifested by multiple major congenital malformations and early onset of hematologic disease. The median age of hematologic disease onset is a 2.7 years (range 0 to 11 years) in the IVS4 group and 2.1 years (range 0.8 to 5.2 years) in the exon 14 group (Table 2, Fig 1). In comparison, the exon 1 subgroup displays a mild phenotype with few major congenital malformations and a later onset of hematologic disease. The median age of onset of hematologic disease is 7.6 years (range 2.6 to 21.8 years) (Table 2, Fig 1). Pairwise comparisons of Kaplan-Meier curves for age of hematologic onset found significant differences between exon 14 patients and exon 1 patients (P < .00005; Fig 1); similar results were obtained when comparing IVS4 patients to exon 1 patients (P < .00005; Fig 1).
Comparison of hematologic onset in FA patients grouped by mutation. Tick marks represent patients without hematologic onset. P values represent results from univariate comparison between pairs of survival curves using the log-rank test.
Comparison of hematologic onset in FA patients grouped by mutation. Tick marks represent patients without hematologic onset. P values represent results from univariate comparison between pairs of survival curves using the log-rank test.
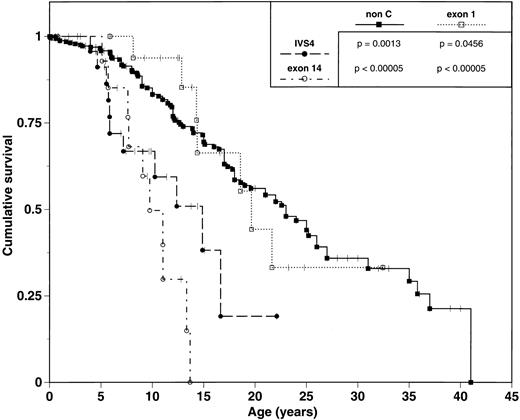
Earlier hematologic disease onset and a greater number of congenital malformations is associated with poorer survival. IVS4 and exon 14 subgroups experienced median survivals of 14.9 and 9.7 years, respectively. In contrast, exon 1 patients had a median survival of 19.7 years (Table 2, Fig 2) and were significantly different from both IVS4 (P = .0456) and exon 14 patients (P < .00005).
Comparison of survival of FA patients grouped by mutation. Tick marks represent patients still alive. P values represent results from univariate comparison between pairs of survival curves using the log-rank test.
Comparison of survival of FA patients grouped by mutation. Tick marks represent patients still alive. P values represent results from univariate comparison between pairs of survival curves using the log-rank test.
Phenotypic analysis of 338 non-C FA patients reveals a median age of onset of hematologic abnormalities of 6.6 years (range 0 to 31 years) and an estimated median survival of 23.0 years. These data are similar to those observed in the exon 1 subgroup, and contrasts to the early hematologic disease onset and poor survival of the IVS4 and exon 14 subgroups (Table 2; Figs 1 and 2).
Sixteen of the 59 FAC patients (27%) have developed AML (Table 3). The incidence of leukemia in each of the FAC subgroups ranges from 19% to 37%. The median age at diagnosis of leukemia is younger in the IVS4 and exon 14 groups compared to the exon 1 group (10.8 and 15.9 years v 21.9 years). Twenty-eight of the 59 FAC patients have died. Leukemia was the cause of death in 13 of the 28 (46%). Three patients with a history of leukemia survive; one patient was recently diagnosed, and two are in remission post-bone marrow transplant (BMT).
DEB sensitivity.Sensitivity to DEB-induced chromosomal breakage was analyzed in cultured PB lymphocytes from the patients in each of the three FAC subgroups. The mean chromosome breaks/cell was 12.6 (S.D. = 4.81) for the exon 1 subgroup, 7.41 (S.D. = 6.09) for IVS4 subgroup, and 5.99 (S.D. = 4.68) for exon 14 subgroup. Analysis of variance revealed a negative correlation between genotype and DEB-induced chromosomal breakage. Patients with exon 1 mutations exhibited a significantly higher frequency of DEB-induced chromosomal breakage than either IVS4 or exon 14 patients (P = .0031). IVS4 and exon 14 patients were not significantly different from each other. This did not change with the square root transformation to approximate normality.
DISCUSSION
FA is a clinically heterogeneous disorder. The heterogeneity of disease severity, particularly the great variability in age of hematologic onset and survival, complicate the clinical care of the FA patient. Major issues for the clinician are surveillance of disease progression and optimal timing of major therapeutic interventions, such as BMT. With the knowledge that genotype determines phenotype, we attempted to define risk groups in the FAC population, by first separating patients into three genotypic subgroups and then comparing the clinical phenotypes of these subgroups.
Two of the subgroups, IVS4 and exon 14, are associated with a poor prognosis (“poor risk”) manifest by onset of hematologic disease before 3 years of age and median survival of 10 to 15 years. These two subgroups have a significantly worse prognosis than the non-C IFAR population, who experience a median onset of hematologic disease at 6.6 years and a median survival of 23 years. Exon 1 patients have a better prognosis than the other two FAC subgroups with a later onset of hematologic disease and a survival comparable to the non-C IFAR group.
BMT remains the only curative therapy for FA-related hematologic disease. Recent analysis of 151 HLA-matched sibling transplants for FA from the multicenter International Bone Marrow Registry (IBMTR) and 18 HLA-matched sibling transplants from Cincinnati show increased survival is associated with younger age, less severe hematologic disease, and absence of malignant transformation.23 24 Therefore, optimal BMT results require careful monitoring of hematologic progression and advance donor selection. Although most clinicians HLA type family members of all newly diagnosed FA patients, it is imperative that typing be performed as soon as possible in the patients in the poor risk groups defined in this study. Since patients with HLA-matched siblings are typically transplanted early in the progression of hematologic disease, most of these patients will require transplant before the age of 5 years.
Early knowledge of prognosis is important for these poor risk patients, as they need time to explore treatment options if they lack an HLA-matched sibling for transplant. Many families in this situation will pursue future pregnancies in hopes of having a non-affected HLA-matched sibling. Alternatively, searches for unrelated bone marrow or cord blood donors are often conducted. Although the results of unrelated BMTs are inferior to those with HLA-matched sibling,23,25 and the experience is small with unrelated cord blood transplants, they may be the only option for the patient with severe hematologic disease or evidence of early malignant transformation. Unrelated searches to locate a suitable donor can take months to years, and should be considered at diagnosis or early in the course of the disease in the high-risk patients.
In addition to the identification of two poor risk groups in the FAC population, this study clearly shows that leukemia is a significant complication and cause of morbidity in the FAC patients. IVS4 and exon 14 patients develop leukemia at a younger age than exon 1 patients. We have previously shown that the actuarial risk of developing leukemia is very high for FA patients in the IFAR5; our present data is the first to show the risk in a specific complementation group. We are currently studying the risk of leukemia in FAA patients. The careful surveillance of bone marrow for evidence of clonal disease is extremely important in FA. We recommend yearly bone marrow aspirations for morphologic examination and cytogenetic analysis. Patients with hematologic disease, who are at higher risk for leukemia, should have bone marrow examinations every 6 months. These frequent observations will allow future study of the evolution of AML in FA and also are important in decisions regarding the timing of transplant especially in those patients without a matched sibling donor.
Our data support the results of a recent study by Yamashita et al26 correlating FAC protein expression with the frequency of congenital malformations in patients.26 While the function of the FAC protein remains unknown, the FAC gene encodes a cytoplasmically localized protein of 60 kD that inhibits the induction of DNA cross-links, and that binds to several other cytosolic proteins.10, 27-29 One of these FAC-related proteins, the 50-kD protein FRP-50, is actually an amino terminal truncated FAC protein resulting from an internal translation initiation at methionine 55. Yamashita et al26 showed that FRP-50 is expressed in normal cell lines as well as cell lines with the 322delG (exon 1) mutation, but not expressed in IVS4 cell lines. Expression of FRP-50 was correlated with phenotype as defined by the number of physical stigmata: patients with either one (compound heterozygotes) or two copies of the 322delG mutation had a mild phenotype while IVS4 patients had a severe phenotype. Exon 14 patients were not included in their study and the relationship of genotype to the severity of hematologic disease was also not analyzed. Like Yamashita et al, we found that compound heterozygotes with a single copy of 322delG, such as our patients with 322delG/R548X (exon 1/exon 14), are generally mild with respect to congenital malformations (data not shown). However, our data show that these patients share the severe hematologic phenotype of an R548X homozygote, indicating that the presence of the FRP-50 protein is not sufficient to mitigate the severe hematologic phenotype associated with mutations in other domains of the FAC protein.
Our data show that there is an inverse correlation between disease severity and the degree of sensitivity to DNA damage as measured by the DEB assay. Patients with exon 14 and IVS4 mutations, which define poor risk subgroups associated with early age of hematologic onset and with poor survival, exhibited less sensitivity to the clastogenic effect of DEB than patients with mutations in exon 1. Thus, while the presence of the FRP-50 protein in these patients may affect clinical phenotype,26 it does not appear to have a mitigating effect on DEB sensitivity. Yamashita et al26 state that they have unpublished results showing the cells from patients with either 322delG or IVS4 mutations exhibit similar sensitivity to MMC in vitro.26 Our data indicate that cross-link hypersensitivity, which defines the FA cellular phenotype, may be a secondary effect of an abnormal FA protein, consistent with data showing that the FAC protein is localized to the cytoplasm.27-29 We suggest that the various domains of the FAC gene have diverse functions, resulting in the lack of correlation between clinical and cellular phenotype. The formal possibility also exists that the clinical phenotype is affected by closely linked modifying genes in linkage disequilibrium with the different mutant FAC alleles. Since the high frequency of the IVS4 mutation in the Ashkenazi Jewish population is due to a founder effect,30 it is likely to be in linkage disequilibrium with particular variants in other closely linked genes.
Unfortunately, some alleles are still missing from this analysis. The second mutation is undefined in all four patients with L554P and in 5 of the 12 patients with R548X. The second allele in at least two of the L554P patients is thought to represent a deletion in the promoter region of the gene, apparently resulting in a null allele (M. Buchwald, personal communication, March 1995). This type of mutation may be representative of the alleles that have gone undetected both by SSCP and by chemical mismatch cleavage, and could explain the dominance of the severe phenotype in these compound heterozygotes.
Because only 10% to 15% of FA patients in the IFAR are in FA-C, these results affect only a small portion of the FA population. However, the FA-A gene has recently been mapped to chromosome 16q24.3,31,32 and has been isolated by two independent approaches, based on positional cloning and on expression cloning.33,34 Mutations in FAA account for 60% to 65% of FA cases35-36; mutation screening for this gene and FAC will therefore allow detection of the defect in about 75% of FA families. FAA mutation analysis is likely to yield genotype-phenotype differences in this heterogeneous population, again allowing subgroup segregation and prognostication.
Further identification of mutant FAC alleles may allow more precise genotype segregation and perhaps more clinical homogeneity between subgroups. Nevertheless, this study enables us to define this clinically heterogeneous disorder genotypically to better predict clinical outcome and aid decision-making regarding therapeutic modalities for a subset of FA patients.
ACKNOWLEDGMENT
We gratefully acknowledge the contribution of the many physicians who referred patients to the IFAR.
Supported in part by Grant No. HL32987 from the National Institutes of Health, Bethesda, MD, to A.D.A. and by General Clinical Research Center Grant No. RR00102 from the National Insitutes of Health to The Rockefeller University Hospital.
Address reprint requests to Arleen D. Auerbach, PhD, Laboratory of Human Genetics and Hematology, The Rockefeller University, 1230 York Ave, Box 178, New York, NY 10021-6399.