Abstract
Mobilized peripheral blood progenitors (CD34+ cells) have been shown to be either in the G0 or G1 phase of the cell cycle. In this study, it is shown that they are small cells with low protein content suggestive of G0. Support for this is provided by showing that the principal E2F complex consists of hypophosphorylated p130, E2F-4, and DP-1. The E2F-4 is more highly phosphorylated than in quiescent T cells. In response to cytokines in vitro, the CD34+ cells start to enter G1 within 8 hours and enter S-phase at about 48 hours. As cells enter G1, E2F-4 is dephosphorylated to several hypophosphorylated forms and three new DNA-binding complexes appear, including one containing E2F-4, DP-1, and p107. We suggest that mobilized CD34+ cells may be maintained in G0 by p130, E2F-4, and DP-1 and the coordinate dephosphorylation of E2F-4 and hyperphosphorylation of p130 may be central to the initiation of proliferation.
THE USE OF peripheral blood progenitor cells (PBPC) to reconstitute hematopoiesis after high-dose chemo/radiotherapy has largely replaced the use of autologous bone marrow transplantation, because of the more rapid hematologic recovery that occurs with PBPC transplantation.1-3 In the steady state, there are only a very small number of stem/progenitor cells within the circulation, but the levels increase markedly during the recovery phase from chemotherapy and after the administration of a number of cytokines, especially granulocyte colony-stimulating factor (G-CSF) and granulocyte-macrophage colony-stimulating factor (GM-CSF).4 The mechanism governing the increase in circulating stem cells is ill-defined, but it is a selective phenomenon with specific subsets of progenitor cells mobilized preferentially.5 Why mobilized PBPC give rise to more rapid hematological recovery is also poorly understood. In part, it may be due to the generally higher numbers of intermediate stage progenitor cells in a PBPC collection compared with a bone marrow harvest, but in addition, there may be important functional differences between the progenitors from the two sites. It is conceivable that PBPC might be capable of more rapid proliferation after transplantation than bone marrow progenitor cells, but paradoxically far fewer PBPC appear to be cycling. A number of studies using propidium iodide staining or thymidine suicide have indicated that nearly all the circulating CD34+ cells are in G0/G1,6-9 and a recent study has suggested that these cells are in G0 on the basis of lack of Ki-67 expression.10 It is not known whether only quiescent cells are released into the circulation or whether CD34+ cells at all stages of the cell cycle are mobilized and there is then selective removal of cells as they enter S-phase either due to a change in cell size or to a change in adhesion molecule expression. If the majority of CD34+ cells were in G1, then they might be capable of rapid division giving rise to prompt engraftment.
Over the last few years, many of the molecular mechanisms that regulate entry into and then progression through the cell cycle have been elucidated. The E2F transcription factors play a pivotal role in these processes by integrating events, which occur during early cell-cycle progression, with the transcriptional machinery.11 Active E2F is a heterodimer composed of an E2F protein and a DP protein. The active E2F/DP complex can be bound and sequestered by a family of ‘pocket proteins,’ the archetype of which is the retinoblastoma protein (pRb). Phosphorylation of pRb releases the active E2F/DP complex and E2F transcriptional activity is thus controlled by cell-cycle specific kinases and phosphatases determining pocket-protein phosphorylation.12-14
There is a family of pocket proteins, which includes p107 and p130, as well as pRb. There are also families of E2F and DP proteins so that there are a large number of possible complexes with potentially different roles.15,16 The prototype complex consists of E2F-1, DP-1, and the hypophosphorylated form of pRb. When pRb becomes heavily phosphorylated in late G1, the E2F-1/DP-1 complex is released to stimulate transcription of a number of genes that are required for DNA replication, including thymidine kinase and dihydrofolate reductase.17,18 Indeed, gene deletion experiments in Drosophila or transfection of dominant negative E2F mutants into cell lines have shown that cells cannot enter S-phase without active E2F.19 20
Five E2F proteins have been identified and these have different preferences for binding pocket proteins. E2F-1, 2, and 3 bind preferentially to pRb, whereas E2F-4 binds to either pRb, p130, or p107, and E2F-5 shows specificity for p130 and p107. In addition, different E2F proteins appear to be expressed at different times during entry into the cell cycle.15,21,22 The E2Fs are not simply proteins which are regulated during cell cycle entry, they are particularly important as they can drive cells into cycle: ectopic expression of E2F-123 or of E2F-4/DP-124 causes serum-starved fibroblasts to progress through G1 into S-phase. Furthermore, E2F-1 overexpression overcomes arrest by pRb and E2F-4/DP-1 that is caused by p130.25 Thus, the balance between E2F and pocket protein levels is clearly important in determining whether a cell proliferates or not. Under normal circumstances, production of hypophosphorylated pRb and p130 in the cell cause cell cycle arrest and it has been suggested that p130 is responsible for maintaining cells in G0.25
In quiescent, serum-starved fibroblasts and in human primary T cells, which are in G0, the major E2F transcription factor is composed of E2F-4 complexed with DP-1 and p130.25 As the T cells begin to proliferate after phytohemagglutinin stimulation, E2F becomes bound to a p107/cyclinA/cdk2 complex and E2F-1, 2, and 3 are synthesized.26 We have also shown that in quiescent fully differentiated human hematopoietic cells, including B cells and monocytes, the E2F-4/DP-1/p130 complex predominates.27 All mature hematopoietic cell types are derived from CD34+ progenitors, but studies of E2F regulation in quiescent and cycling CD34+ cells have not been reported.
To understand how the proliferation of mobilized CD34+ cells is regulated, we first investigated whether these cells are in G0 or G1. Second, we determined which E2F/pocket protein complexes they contain, and we then studied how E2F activity is regulated as CD34+ progenitors enter the cell cycle in response to cytokine stimulation.
MATERIALS AND METHODS
Preparation of human primary B cells.A buffy coat from 400 mL of blood was diluted 10-fold with cation-free phosphate-buffered saline (PBS). The mononuclear cell layer was separated using Ficoll gradient-centrifugation at 800g, 20°C for 20 minutes. B cells were isolated from this by positive selection using CD19 coupled immunomagnetic beads. Approximately 3 beads per cell were added to the cells suspended in PBS and then agitated at 4°C for 45 minutes.
Purification of mobilized CD34+ cells.CD34-positive (CD34+) cells were isolated from the apheresis product of patients previously mobilized with cyclophosphamide (1.5 g/m2 ) followed by daily G-CSF (10 μg/kg/d Filgrastim or 263 μg/d Lenograstim) for 10 days. CEPRATE SC immunoaffinity columns were used for CD34+ stem cell purification.28 The antibody used on the column was monoclonal antibody (MoAb) 12.8, a CD34 class I antibody, which recognizes a sialic acid-dependent epitope.29 Purity was assessed by alkaline phosphatase antialkaline phosphatase (APAAP) staining of cytocentrifuge preparations with a CD34 antibody (HPCA-2 MoAb) different from that used for purification (purity range, 85% to 93%). Blast cells were identified by morphology after staining with May-Grünwald-Giemsa (MGG) stain. Cells were either used straight from the column or were frozen in 5 mL cryopreservative vials at −80°C and stored in liquid nitrogen. When required, the cells were thawed in a water bath at 37°C and slowly reconstituted with Iscove's Medium supplemented with 20% (vol/vol) fetal calf serum (FCS) and 2 mmol/L L-glutamine. Where indicated, CD34+ cells were grown at 1 × 105 cells/mL in Iscove's Medium supplemented with 20% (vol/vol) FCS and 2 mmol/L L-glutamine and 10 ng/mL each of stem cell factor (SCF), interleukin-3 (IL-3) and IL-6. Cells were incubated at 37°C in a fully humidified atmosphere of 5% CO2.
Cell cycle analysis.A total of 5 × 105 cells were fixed in 500 μL of −20°C 70% (vol/vol) ethanol and stored at −20°C. For DNA and protein analysis, cells were washed and incubated at 20°C for 30 minutes in 1 mL of a solution containing PBS, DNAase-free RNAase (0.5 mg/mL), propidium iodide (PI, 20 μg/mL), and fluorescein isothiocyanate (FITC, 0.05 μg/mL). The percentage of cells in different phases of the cell cycle was determined by flow cytometry (EPICS-Elite; Coulter Electronics, Luton, UK) as described previously.30
Whole cell extracts.Whole cell extracts were prepared by resuspending 2 × 107 cells in 100 μL of lysis buffer containing 20 mmol/L HEPES (N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid]), pH 7.8, 450 mmol/L NaCl, 25% (vol/vol) glycerol, 0.2 mmol/L ethylenediamine tetra-acetic acid (EDTA), 0.5 mmol/L dithiothreitol (DTT), 0.5 mmol/L phenylmethylsulfonyl fluoride (PMSF), 0.5 μg/mL leupeptin, 0.5 μg/mL Sigma protease inhibitor, 1.0 μg/mL trypsin inhibitor, 0.5 μg/mL aprotinin, 40 μg/mL bestatin, and 2 mmol/L diisofluorophosphate (DIFP). This was frozen on dry ice and thawed at 37°C three times. The samples were then spun at 13,000 rpm, 4°C for 10 minutes and the supernatant removed, aliquoted, and stored at −70°C. Protein concentration in the extracts was measured using the Bio Rad protein assay.
Western blotting.A total of 1 × 106 cells were pelleted and lysed in 100 μL of sample buffer and boiled for 10 minutes. The sample buffer contained 125 mmol/L Tris pH 7.5, 4% (wt/vol) sodium dodecyl sulfate (SDS), 0.4% (vol/vol) glycerol, 0.5 mmol/L DTT, 0.5 mmol/L PMSF, 0.5 μg/mL leupeptin, 0.5 μg/mL Sigma protease inhibitor, 1.0 μg/mL trypsin inhibitor, 0.5 μg/mL aprotinin, 40 μg/mL bestatin; the protease inhibitors being added to protect against protein degradation in the samples. Either protein from an equivalent number of cells (2 × 105 ) or 8 μg of whole cell extract boiled with sample buffer was loaded per lane, as described in each figure legend. Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using either 6%, 8%, or 10% (wt/vol) polyacrylamide gels depending on the molecular weight of the protein being probed. After transfer to nitrocellulose (Hybond C extra) by semidry blotting at 0.5 A for 40 minutes, the membranes were blocked for 1 hour with 10% (wt/vol) nonfat dried milk (Marvel) in PBS/0.05% (vol/vol) Tween 20 (PBS/T), washed, and incubated overnight at 4°C with the primary antibody in PBS/T/3% (wt/vol) bovine serum albumin (BSA). Primary antibodies used were anti-p130 antibody (C-20), anti-p107 antibody (C-18), and anti-E2F-4 antibody (C-20), all of which are rabbit polyclonal IgG antibodies and were used at 100 ng/mL. Anti-pRb antibody (PMG 3-245), a mouse monoclonal antibody, was also used at 100 ng/mL. After washing three times with PBS/T, the membranes were then incubated at room temperature for 45 minutes in the appropriate secondary antibody in PBS/T/3% BSA/3% nonfat milk. The secondary antibodies were either peroxidase conjugated antimouse or antirabbit IgG at a 1:1,000 and 1:2,500 dilution, respectively. Detection was by enhanced chemiluminescence (ECL). Antitubulin and antiactin antibodies were used at 100 ng/mL as internal controls for protein loading and integrity.
Oligonucleotides.The DNA sequences of the wild-type and mutant E2F DNA binding site oligonucleotides used in E2F DNA binding assays have been described previously.31 A total of 0.25 ng of double-stranded oligonucleotide was end-labeled with 30 μCi of [γ-32P]adenosine triphosphate (ATP) using 10 U of T4 polynucleotide kinase in T4 polynucleotide kinase buffer (70 mmol/L Tris-HCl, pH 7.6, 10 mmol/L MgCl2, 5 mmol/L DTT) in a final volume of 10 μL. After a 30-minute incubation at 37°C, the reaction was stopped by adding 2 μL 0.5 mol/L EDTA and the volume was made up to 50 μL with distilled H2O. 32P-labeled oligonucleotide was separated from free [γ-32P]ATP by centrifugation through a G25 Sephadex column at 1,100g for 5 minutes. The specific activity of the recovered probe was measured with a beta-counter following trichloroacetic acid precipitation.
Electromobility shift assays.Protein-DNA interaction was analyzed by an electrophoretic mobility shift assay (EMSA) using nondenaturing polyacrylamide gel electrophoresis. Whole cell extracts were prepared as described above and 8-μg protein samples were incubated for 10 minutes at 30°C with 300 ng of mutant oligonucleotide and 10 μL of binding buffer containing 100 mmol/L Tris pH 7.9, 30% (vol/vol) glycerol, 0.4 mmol/L EDTA, 2 mmol/L DTT, 100 mmol/L NaCl, 2 μg sheared salmon sperm DNA in a final volume of 20 μL. The samples were then incubated for a further 10 minutes at 30°C after the addition of 1 μL of labeled oligonucleotide. The amount of extract used was within the linear range of the assay (data not shown). If a supershifting antibody was used, 1 to 2 μg was added to the initial binding reaction, which was incubated for a further 1 to 2 hours at 4°C after the addition of the labeled probe. The quantity of antibody used was determined by titration to be within the range that did not cause a nonspecific supershift. For competition studies, a 100-fold molar excess of unlabeled probe was added. Protein-DNA complexes were resolved on a 4% polyacrylamide gel (acrylamide: bisacrylamide ratio 19:1) in a buffer containing 40 mmol/L Tris-acetate and 1 mmol/L EDTA. Gels were prerun for 1 hour at 150 V and, after the samples were loaded, run for 2 to 3 hours at 150 V. Gels were then dried at 80°C for 45 minutes and exposed to x-ray film (Hyperfilm) overnight at room temperature.
Phosphatase treatment.Whole cell extracts were incubated with either 500 U of lambda protein phosphatase, 10 U of T-cell protein tyrosine phosphatase, or 1 U of protein phosphatase 1 with the appropriate buffers for 1 hour at 30°C. SDS sample buffer was then added to the samples, which were then boiled for 5 minutes, separated by SDS-PAGE, and Western blotted.
Materials.The following reagents were obtained from the sources shown. Leupeptin, protease inhibitor, trypsin inhibitor, aprotinin, bestatin, DIFP, PMSF, Tween 20, EDTA, HEPES, PBS, and DTT: Sigma Chemical Co, Poole, UK. Marvel: Premier Beverages, Stafford, UK. PBS, RPMI, FCS, and Iscove's medium: GIBCO-BRL, Paisley, UK. Ficoll-Hypaque: Pharmacia, St Albans, UK. CD14 Dynabeads: Dynal, Bromborough, UK. PHA: Murex Biotech Ltd, Dartford, UK. Stem cell factor and Filgrastim were obtained from Amgen, Thousand Oaks, CA; Lenograstim was from Chugai, Tokyo, Japan. CEPRATE SC immunoaffinity columns were obtained from Cell Pro Inc, Bothwell, WA. Bio-Rad protein assay: Bio-Rad, Hemel Hempstead, UK. Hybond C extra, Hyperfilm, ECL and [γ-32 P]ATP (>3,000 Ci/mmol): Amersham International, Little Chalfont, UK. T4 polynucleotide kinase and buffer: Promega, Southampton, UK. G25 Sephadex columns: Boehringer-Mannheim, Mannheim, Germany. All phosphatases were obtained from New England Biolabs, Hitchin UK. Antibodies used for Western blotting were anti-p130 antibody (C-20), anti-p107 antibody (C-18), and anti-E2F–4 antibody (C-20) from Santa Cruz Biotechnology (Santa Cruz, CA), anti-pRb antibody (PMG 3-245) from Pharmingen (San Diego, CA), antitubulin from Boehringer-Mannheim, and antiactin from Oncogene Science (Cambridge, UK). Peroxidase conjugated secondary antimouse and antirabbit antibodies were obtained from Dakopatts (High Wycombe, UK). Supershift antibodies used in the EMSA were rabbit polyclonal anti-E2F–1 (C-20), anti-E2F–2 (L-20), anti-E2F–4 (C-20), anti-E2F–4 (C-108), anti-DP–1 (K-20), anti-p107 (C-18), and anti-Rb (C-15) antibodies and a mouse monoclonal anti-E2F–5 (MH-5) antibody. All of these and their competing peptides were obtained from Santa Cruz Biotechnology. HPCA-2 MoAb was from Becton Dickinson (Mountain View, CA). Consensus and mutant E2F oligonucleotides were obtained from Prof N. LaThangue, University of Glasgow, Glasgow, UK. A rabbit polyclonal anti-p130 supershifting antibody was a kind gift from Prof Peter Whyte, McMaster University, Ontario, Canada, and a mouse monoclonal anti-E2F–3 supershifting antibody was a kind gift from Prof Ed Harlow, Massachusetts General Hospital Cancer Center, Charlestown.
RESULTS
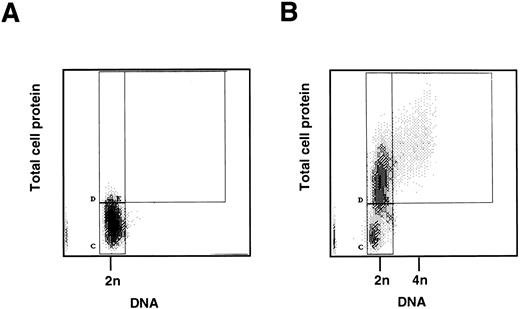
Cell cycle analysis of freshly isolated CD34+ cells.To determine whether mobilized CD34+ cells are in G0 or in G1, two parameter flow cytometric analysis of protein and DNA content was performed on the CD34+ cells immediately after purification. The majority (94.53 ± 3.95% standard deviation [SD], n = 4) had a low level of protein staining compared with proliferating CD34+ cells (see below) and a diploid (2n) DNA content, consistent with their being quiescent and in G0 (Fig 1A and B). Cytospins, stained with MGG, showed that the CD34+ cells were small compared with CD34+ cells stimulated to proliferate in vitro (see below), and there was no evidence of mitosis or differentiation.
(A) Two-color flow cytometric analysis of human CD34+ cells mobilized into the peripheral blood. CD34+ progenitor cells were isolated from peripheral blood following in vivo administration of cyclophosphamide and G-CSF. A total of 5 × 105 cells was fixed, stained, and analyzed by two-color flow cytometry for DNA and protein content as described in Materials and Methods. The quiescent cells form a homogeneous population (marked C) with 2n DNA and low protein content. (B) Two-color flow cytometric analysis of stimulated peripheral blood CD34+ cells. CD34+ progenitor cells as above were cultured in vitro with SCF, IL-3, and IL-6 for 2 days. Cells were then fixed, stained, and analyzed by two-color flow cytometry for DNA and protein content as described in Materials and Methods. The stimulated cells show two populations (marked C and D) with 2n DNA, but D has an increased protein content. This is consistent with the cells having moved from G0 (C) into the G1 phase of the cell cycle (D). Cells to the right of box D (marked E) are in S and G2/ M phases.
(A) Two-color flow cytometric analysis of human CD34+ cells mobilized into the peripheral blood. CD34+ progenitor cells were isolated from peripheral blood following in vivo administration of cyclophosphamide and G-CSF. A total of 5 × 105 cells was fixed, stained, and analyzed by two-color flow cytometry for DNA and protein content as described in Materials and Methods. The quiescent cells form a homogeneous population (marked C) with 2n DNA and low protein content. (B) Two-color flow cytometric analysis of stimulated peripheral blood CD34+ cells. CD34+ progenitor cells as above were cultured in vitro with SCF, IL-3, and IL-6 for 2 days. Cells were then fixed, stained, and analyzed by two-color flow cytometry for DNA and protein content as described in Materials and Methods. The stimulated cells show two populations (marked C and D) with 2n DNA, but D has an increased protein content. This is consistent with the cells having moved from G0 (C) into the G1 phase of the cell cycle (D). Cells to the right of box D (marked E) are in S and G2/ M phases.
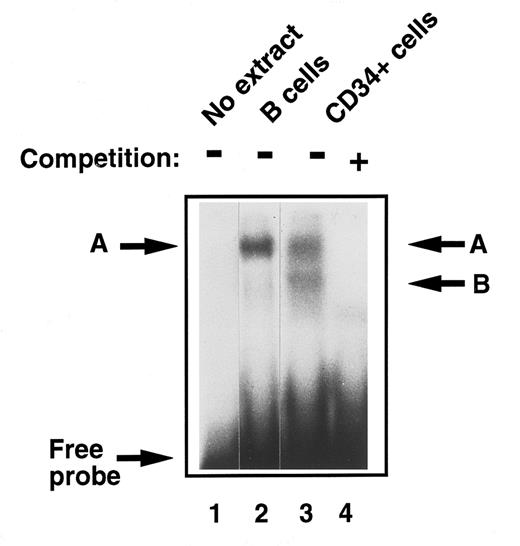
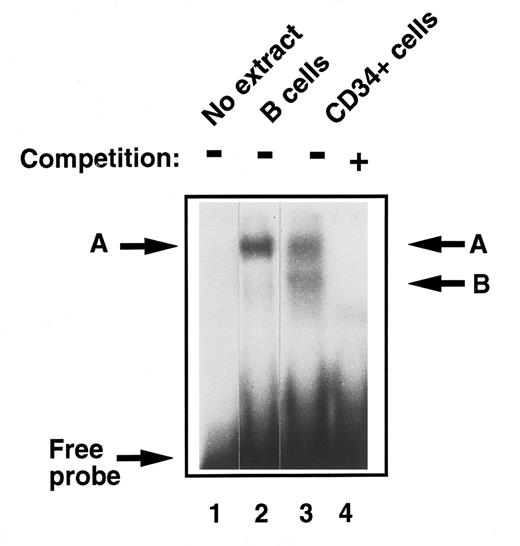
Detection of E2F in early human primary hematopoietic cells.To analyze and compare functional E2F protein-DNA complexes in these early, undifferentiated hematopoietic stem cells to those described previously in fully differentiated cells, EMSA was performed with CD34+ cell extracts. They were found to contain two E2F complexes, the slower band migrating with complex ‘A,’ which we detected in monocytes and B cells from peripheral blood27 and a lower, faster migrating band ‘B’ (Fig 2, lanes 2 and 3). The DNA binding specificity of these E2F complexes was confirmed in competition experiments where addition of excess, unlabelled wild-type E2F oligonucleotide, but not an equal amount of an oligonucleotide containing a mutant E2F site, abolished binding to the radioactive probe (Fig 2, lane 4).
Comparison of E2F-complex DNA binding activity in extracts of human primary B cells and immature CD34+ hematopoietic cells. B cells and CD34+ cells were purified and whole cell extracts prepared as described in Materials and Methods. EMSA was then performed. Equal amounts of total cell lysate (8 μg) were incubated with a 32P-labeled probe containing an E2F binding site and then separated on a 4% nondenaturing polyacrylamide gel. No whole cell extract was loaded in lane 1. Band ‘A’ was identified in quiescent B cells (lane 2) as previously described. Two bands were identified in CD34+ cells, ‘A,’ which comigrated with the band identified in B cells, and band ‘B’ (lane 3). These were confirmed as specific E2F complexes by competition with excess nonradioactive probe (lane 4).
Comparison of E2F-complex DNA binding activity in extracts of human primary B cells and immature CD34+ hematopoietic cells. B cells and CD34+ cells were purified and whole cell extracts prepared as described in Materials and Methods. EMSA was then performed. Equal amounts of total cell lysate (8 μg) were incubated with a 32P-labeled probe containing an E2F binding site and then separated on a 4% nondenaturing polyacrylamide gel. No whole cell extract was loaded in lane 1. Band ‘A’ was identified in quiescent B cells (lane 2) as previously described. Two bands were identified in CD34+ cells, ‘A,’ which comigrated with the band identified in B cells, and band ‘B’ (lane 3). These were confirmed as specific E2F complexes by competition with excess nonradioactive probe (lane 4).
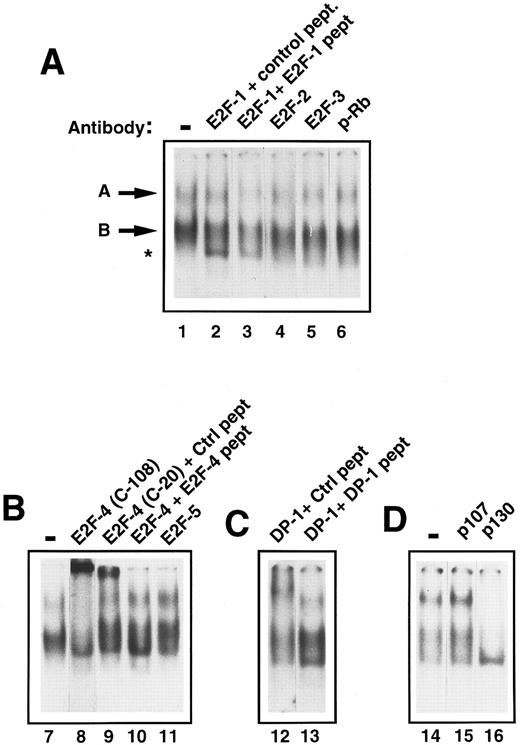
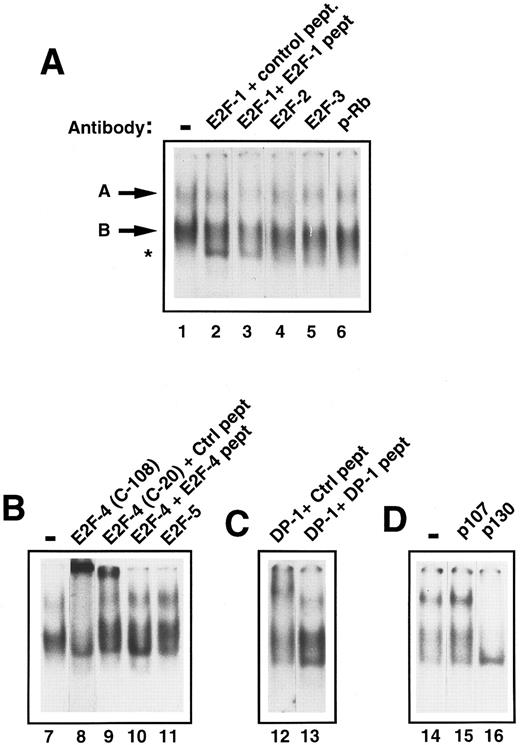
Characterization of E2F in early human primary hematopoietic cells.To determine the components of E2F complex ‘A’ and ‘B,’ supershift experiments with specific E2F antibodies were performed. Addition of anti-E2F antibodies to E2F-1, -2, -3, and -5 had no effect on band ‘A’ or ‘B’ (Fig 3A, lanes 2, 4, and 5; Fig 3B, lane 11). However, addition of an anti-E2F–4 antibody (C-108) caused a marked retardation of complex ‘A.’ There was also a significant reduction of band ‘B’ (Fig 3B, lane 8). A second, anti-E2F –4 antibody (C-20) was also used and showed a similar, if less marked, supershift of ‘A’ (Fig 3B, lane 9). Addition of the competing antigenic peptide prevented the supershift (Fig 3B, lane 10), whereas addition of a nonspecific peptide did not, showing the effect to be specific. Because E2F-4 can heterodimerize with DP-1 protein in vivo, we also investigated whether DP-1 was present in complex ‘A.’ Addition of a rabbit polyclonal DP-1 antibody led to a marked retardation of ‘A’ (Fig 3C, lane 12). Further addition to the reaction mixture of the DP-1 peptide to which the antibody was raised prevented the supershift, resulting in no movement of band ‘A’ (Fig 3C, lane 13), confirming the specificity of the above result. The p130 pocket protein has been shown to form complexes with E2F-4 and DP-1 in resting cells and, therefore, we determined whether p130 was present in complex ‘A’ or ‘B.’ Addition of an anti-p130 antibody to the assay retarded band ‘A’ and ‘B’ (Fig 3D, lane 16). In contrast, anti-p107 or anti-pRb antibodies did not affect either complex ‘A’ or ‘B’ showing that neither of these was present in either complex (Fig 3A, lane 6 and Fig 3D, lane 15). Therefore, freshly isolated CD34+ stem cells appear to contain two E2F complexes containing E2F-4, DP-1, and p130. Given that these complexes predominate in primary, quiescent hematopoietic cells, these findings are consistent with the CD34+ cells also being in the G0 phase of the cell cycle.
Identification of the components of E2F complexes in quiescent human primary CD34+ cells by supershift assays. CD34+ cells were purified and whole cell extracts prepared and EMSA was performed (see Materials and Methods). Equal amounts of CD34+ cell whole cell lysate (8 μg) were incubated with a 32P-labelled probe containing an E2F binding site and supershift assays were performed by the addition of 2 μL of the appropriate antibody (as indicated for each lane). These were then run on a 4% nondenaturing polyacrylamide gel. Excess unlabelled mutant E2F probe was added to all lanes to ensure specificity of binding. The lanes shown in each panel are from a single gel and the results are representative of four experiments. (A) The E2F complexes are arrowed as band ‘A’ and ‘B’ (lane 1). No supershifts were detected with anti-E2F–1, anti-E2F–2, anti-E2F–3, or anti-pRb antibodies (lanes 2, 4, 5, and 6). A lower complex (starred) appeared with the addition of the E2F-1 antibody and control peptide (lane 2), but was also present with the E2F-1 antibody and E2F-1 peptide (lane 3) and hence was considered to be nonspecific. (B) Both E2F complexes (lane 7) were supershifted by the addition of anti E2F–4 antibody (C-108) (lane 8). Similarly, a proportion of both complexes are retarded with a second E2F-4 antibody (C-20) in the presence of a nonspecific control peptide (lane 9), whereas the supershift is abolished by the addition of a specific E2F-4 peptide (lane 10). No supershift was detected with the addition of anti-E2F–5 antibody (lane 11). (C) A supershift of complex ‘A’ occurs with the addition of anti-DP–1 antibody with control peptide (lane 12) and is overcome by addition of specific DP-1 peptide (lane 13). (D) No supershift occurred with the addition of anti-p107 antibody (lane 15), as compared with the control lane (lane 14). Both E2F complexes were abolished with anti-p130 antibody (lane 15).
Identification of the components of E2F complexes in quiescent human primary CD34+ cells by supershift assays. CD34+ cells were purified and whole cell extracts prepared and EMSA was performed (see Materials and Methods). Equal amounts of CD34+ cell whole cell lysate (8 μg) were incubated with a 32P-labelled probe containing an E2F binding site and supershift assays were performed by the addition of 2 μL of the appropriate antibody (as indicated for each lane). These were then run on a 4% nondenaturing polyacrylamide gel. Excess unlabelled mutant E2F probe was added to all lanes to ensure specificity of binding. The lanes shown in each panel are from a single gel and the results are representative of four experiments. (A) The E2F complexes are arrowed as band ‘A’ and ‘B’ (lane 1). No supershifts were detected with anti-E2F–1, anti-E2F–2, anti-E2F–3, or anti-pRb antibodies (lanes 2, 4, 5, and 6). A lower complex (starred) appeared with the addition of the E2F-1 antibody and control peptide (lane 2), but was also present with the E2F-1 antibody and E2F-1 peptide (lane 3) and hence was considered to be nonspecific. (B) Both E2F complexes (lane 7) were supershifted by the addition of anti E2F–4 antibody (C-108) (lane 8). Similarly, a proportion of both complexes are retarded with a second E2F-4 antibody (C-20) in the presence of a nonspecific control peptide (lane 9), whereas the supershift is abolished by the addition of a specific E2F-4 peptide (lane 10). No supershift was detected with the addition of anti-E2F–5 antibody (lane 11). (C) A supershift of complex ‘A’ occurs with the addition of anti-DP–1 antibody with control peptide (lane 12) and is overcome by addition of specific DP-1 peptide (lane 13). (D) No supershift occurred with the addition of anti-p107 antibody (lane 15), as compared with the control lane (lane 14). Both E2F complexes were abolished with anti-p130 antibody (lane 15).
Cell cycle analysis and proliferation of CD34+ cells.Following selection of mobilized CD34+ cells from peripheral blood, they were cultured under the appropriate conditions and proliferation was stimulated by the addition of IL-3, IL-6, and SCF. Serial samples were taken every 12 hours and the proportion of cells in G0, G1, S, and G2/M phases was measured by two parameter flow cytometry (Fig 4A). The cells become larger as they are stimulated to enter the cell cycle from G0, as determined by protein content and forward scatter measurements. Cells in G1 can be distinguished from those in G0 by their increased size and further assessment by flow cytometry of the percentage of CD34+ cells in G0, G1, and S-phase at earlier times after cytokine stimulation showed that a small percentage of cells had entered into G1 from G0 by 8 hours (Fig 4B). Cells entering S-phase were detectable approximately 48 hours after addition of cytokines. Cell numbers doubled after approximately 72 hours and this was reflected by flow cytometric analysis, which showed an increase in the number of cells in S and G2/M (Fig 4C). Cytospins of the CD34+ cells made 4 days after the addition of the cytokines, IL-3, IL-6, and SCF and stained with MGG, showed evidence of mitosis with the appearance of mitotic figures and vacuoles. The cells remain undifferentiated at this stage with the exception of very early primary acidophilic granules.
(A) Percentage of CD34+ cells in G0, G1, S, and G2/M as assessed by flow cytometry up to 4 days following cytokine stimulation. Following mobilization with low-dose cyclophosphamide and G-CSF, CD34+ cells were isolated from peripheral blood and then resuspended in growth medium. Proliferation was stimulated by the addition of cytokines IL-3, IL- 6, and SCF. Cell samples were taken at 24-hour intervals, fixed in 70% ethanol, and flow cytometry was performed. The proportion of cells in G0 decreases with time, progressively more enter G1 by day 1 and then S and G2/M by day 3. The values plotted are % ± SD (n = 4). (B) Percentage of CD34+ cells in G0, G1, S/G2/M as assessed by flow cytometry in the first 24 hours following cytokine stimulation. Cells were stimulated as described above and samples were taken at 4-hour intervals, fixed in 70% ethanol, and flow cytometry was performed. Some cells were detected entering G1 as early as 8 hours following cytokine stimulation. The values plotted are % ± SD (n = 3). (C) Cell cycle analysis of the CD34+ cells by flow cytometry on the day of purification (day 0) and 3 days after cytokine stimulation (day 3). On day 0, there is a single (2n) DNA peak (>95%). By day 3, 60% of cells are in G1, 26% of cells are in S-phase, and 14% of cells are in G2/M with doubling of the DNA content.
(A) Percentage of CD34+ cells in G0, G1, S, and G2/M as assessed by flow cytometry up to 4 days following cytokine stimulation. Following mobilization with low-dose cyclophosphamide and G-CSF, CD34+ cells were isolated from peripheral blood and then resuspended in growth medium. Proliferation was stimulated by the addition of cytokines IL-3, IL- 6, and SCF. Cell samples were taken at 24-hour intervals, fixed in 70% ethanol, and flow cytometry was performed. The proportion of cells in G0 decreases with time, progressively more enter G1 by day 1 and then S and G2/M by day 3. The values plotted are % ± SD (n = 4). (B) Percentage of CD34+ cells in G0, G1, S/G2/M as assessed by flow cytometry in the first 24 hours following cytokine stimulation. Cells were stimulated as described above and samples were taken at 4-hour intervals, fixed in 70% ethanol, and flow cytometry was performed. Some cells were detected entering G1 as early as 8 hours following cytokine stimulation. The values plotted are % ± SD (n = 3). (C) Cell cycle analysis of the CD34+ cells by flow cytometry on the day of purification (day 0) and 3 days after cytokine stimulation (day 3). On day 0, there is a single (2n) DNA peak (>95%). By day 3, 60% of cells are in G1, 26% of cells are in S-phase, and 14% of cells are in G2/M with doubling of the DNA content.
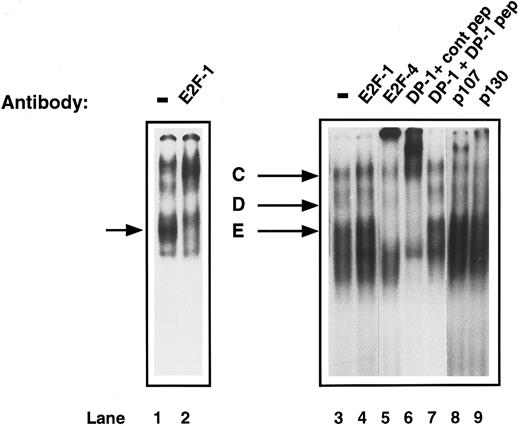
Characterization of E2F complexes in proliferating CD34+ cells.To investigate the changes that may occur in active E2F complexes as CD34+ cells proliferate, E2F-DNA binding was assessed by EMSA of whole cell extracts of CD34+ cells, which had been exposed to IL-3, IL-6, and SCF for 72 hours. A different pattern was obtained from that seen in the G0 CD34+ cells. Three bands were detected, which have been called ‘C,’ ‘D,’ and ‘E.’ Again, to determine the components of these E2F complexes, supershift experiments were performed. The addition of anti-E2F–1 antibody had no effect on any of the bands (Fig 5, lane 4). Proliferating primary T cells, which are known to contain E2F-1 that will bind DNA, were used as a control and the anti-E2F–1 antibody caused the expected supershift (Fig 5, lanes 1 and 2). Addition of either anti-E2F–4 or anti-DP–1 antibody to the proliferating CD34+ extracts caused a supershift of all three bands (Fig 5, lanes 5 and 6). Competition of the DP1 antibody with the specific peptide returned the pattern to normal (Fig 5, lane 7). Anti-p107 antibody caused a supershifted complex, which migrated above complex ‘C’; however, from which bands ‘C,’ ‘D,’ or ‘E’ it originated, was unclear (Fig 5, lane 8). A shift of bands ‘C’ and ‘D’ was also observed with anti-p130 (Fig 5, lane 9). Therefore, CD34+ cells develop a new complex containing E2F-4, DP-1, and p107 as they proliferate, although some p130 is still detectable. There is also free E2F-4/DP–1 present in the lower bands.
Identification of the components of E2F complexes in human proliferating primary CD34+ cells by supershift assays. CD34+ cells were purified and were induced to proliferate by the addition of cytokines as described in Materials and Methods. After 4 days, whole cell extracts were prepared and EMSA and supershift assays were performed as described in Fig 3. Three complexes (C, D, and E) were detected. Specific supershifted complexes were observed on addition of anti-E2F–4 antibody (lane 5) and anti-DP–1 antibody with control peptide (lane 6), but not with DP-1 peptide (lane 7). The upper complex (C) shifts with anti-p107 antibody (lane 8) and a shift was also seen with anti-p130 antibody (lane 9). An anti-E2F–1 antibody had no effect (lane 4). Proliferating T cells were run as a control (lane 1) and show a supershift with anti-E2F–1 antibody (lane 2, arrowed). The data are representative of four experiments with individual CD34+ cell isolates.
Identification of the components of E2F complexes in human proliferating primary CD34+ cells by supershift assays. CD34+ cells were purified and were induced to proliferate by the addition of cytokines as described in Materials and Methods. After 4 days, whole cell extracts were prepared and EMSA and supershift assays were performed as described in Fig 3. Three complexes (C, D, and E) were detected. Specific supershifted complexes were observed on addition of anti-E2F–4 antibody (lane 5) and anti-DP–1 antibody with control peptide (lane 6), but not with DP-1 peptide (lane 7). The upper complex (C) shifts with anti-p107 antibody (lane 8) and a shift was also seen with anti-p130 antibody (lane 9). An anti-E2F–1 antibody had no effect (lane 4). Proliferating T cells were run as a control (lane 1) and show a supershift with anti-E2F–1 antibody (lane 2, arrowed). The data are representative of four experiments with individual CD34+ cell isolates.
To determine when the development of the complex containing p107 occurs, we also analyzed extracts at time points between quiescence, when the upper complex contains E2F-4, DP-1, and p130, and proliferation. The induction of the p107 complex occurred after the cells had been cultured for 24 hours with IL-3, IL-6, and SCF. At this time, the cells are in early G1 by the criteria that they contain more protein than cells in G0, but still have a 2n DNA content.
Status of p130, p107, and pRb in quiescent and proliferating CD34+ cells.The induction of new E2F/pocket protein complexes could be due either to an increase in the expression of individual proteins or to changes in their phosphorylation state. To examine the phosphorylation states and abundance of p130, p107, and pRb, samples of CD34+ cells grown in suitable cytokine-supplemented media were taken for Western blotting and cell cycle analysis. This was done every 4 hours up to the first 20 hours, then every 24 hours thereafter until day 4 when the cells had started to proliferate. Phosphorylation of p130, p107, and pRb causes their migration to be retarded in SDS-PAGE and can thus be analyzed by Western blotting. This showed that p130 existed in G0 cells as two hypophosphorylated forms running between 126 and 128 kD (forms 1 and 2),32 but by 8 hours following cytokine stimulation, an additional, hyperphosphorylated form (form 3)32 appeared, which accumulated thereafter (Fig 6A). As described previously, two parameter cell cycle analysis at this time showed the entry of some cells into G1 from G0. Thus, phosphorylation of p130 appears to be an early event in the cell cycle progression of CD34+ cells. Hyperphosphorylated p130 predominates from approximately 20 hours onwards and correlates with the appearance of the p107/E2F complexes detected by EMSA.
(A) Expression of p130 in human primary CD34+ hematopoietic cells. Purified CD34+ cells were induced to proliferate by the addition of cytokines as previously described in Materials and Methods. Samples were taken at 0 hours, 8 hours, and every 4 hours thereafter until 20 hours and then every 12 hours up to 4 days and lysed in SDS lysis buffer. Equal amounts of total cell lysate (equivalent to 2 × 105cells) were subjected to SDS-PAGE and Western blotting. The hyperphosphorylated form of p130 (pp130) is present at 8 hours and accumulates thereafter. (B) Expression of p107 and pRb in human primary CD34+ hematopoietic cells. Purified CD34+ cells were induced to proliferate by the addition of cytokines and samples were taken at 24-hour intervals until 96 hours. Western blotting was performed using specific polyclonal antibodies that recognized p107 and pRb. The hyperphosphorylated forms of both proteins are arrowed and are detectable at 1 day and then accumulate over the next 3 days as the cells proliferate.
(A) Expression of p130 in human primary CD34+ hematopoietic cells. Purified CD34+ cells were induced to proliferate by the addition of cytokines as previously described in Materials and Methods. Samples were taken at 0 hours, 8 hours, and every 4 hours thereafter until 20 hours and then every 12 hours up to 4 days and lysed in SDS lysis buffer. Equal amounts of total cell lysate (equivalent to 2 × 105cells) were subjected to SDS-PAGE and Western blotting. The hyperphosphorylated form of p130 (pp130) is present at 8 hours and accumulates thereafter. (B) Expression of p107 and pRb in human primary CD34+ hematopoietic cells. Purified CD34+ cells were induced to proliferate by the addition of cytokines and samples were taken at 24-hour intervals until 96 hours. Western blotting was performed using specific polyclonal antibodies that recognized p107 and pRb. The hyperphosphorylated forms of both proteins are arrowed and are detectable at 1 day and then accumulate over the next 3 days as the cells proliferate.
We also examined the p107 and pRb proteins by Western blotting extracts of quiescent CD34+ cells and up to 4 days after the addition of cytokines. Hypophosphorylated forms of both p107 and pRb were expressed in G0 cells, although they were not present in the E2F complexes detected by EMSA (Fig 3). After 1 day, hyperphosphorylated p107 and pRb were detectable and these upper bands became stronger over the 4-day period, consistent with an induction of p107 and pRb with cell cycle progression (Fig 6B).
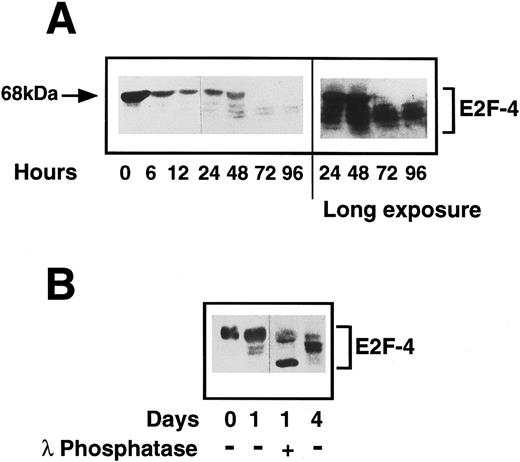
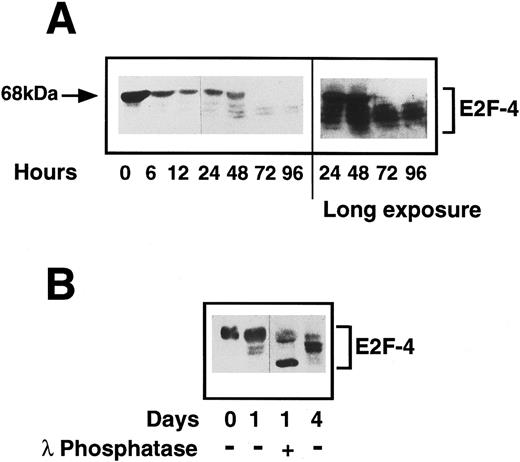
Status of E2F-4 in quiescent and proliferating CD34+ cells.We have shown E2F-4 to be present in both quiescent and proliferating CD34+ cells by EMSA and as different forms of the protein associate with p130 and p107,25 we investigated by western blotting whether the expression of the protein or its phosphorylation state changed during entry into the cell cycle. In quiescent cells, a single E2F-4 band of 68 kD was detected, at a higher molecular weight than the forms detected in fully differentiated hematopoietic cells. This protein is not an artefact of residual CD34 IgM remaining with the purified CD34+ cells, as the heavy-chain of the IgM is larger and is not detected by the anti-E2F–4 antibody (data not shown). After 24 hours, a marked decrease in expression of this form was seen and further, faster-migrating bands were detected, which became more intense as the cells progressed through cell cycle. By 3 days after cytokine stimulation, the 68-kD form had almost completely disappeared and the lower forms (48 to 65 kD) had increased to maximum intensity (Fig 7A). Flow cytometric analysis of the cells in this experiment at day 3 showed they were just entering S-phase (note that there is variability in the time it takes different CD34+ isolates to enter S-phase from 2 to 3 days).
(A) Expression of E2F-4 in human primary CD34+ hematopoietic cells. Purified CD34+ cells were induced to proliferate by the addition of cytokines as previously described in Materials and Methods. Samples were taken at the times shown and western blotting was performed using a specific anti-E2F–4 antibody. At 0 hours, the 68-kD form of E2F-4 predominates, but as the cells proliferate, the lower forms appear with the complete disappearance of the 68-kD form by 72 hours. A longer exposure of the same blot shows that there is little or no 68-kD form present in the 72- and 96-hour samples. (B) Dephosphorylation of E2F-4 in CD34+ cells by lambda phosphatase. A total of 4 μg of CD34+ whole cell lysate made 1 day after addition of cytokines was incubated for 1 hour with 500 U of lambda phosphatase. This was then added to sample buffer and separated by SDS-PAGE and probed with an anti-E2F–4 antibody. Dephosphorylation of the 68-kD form can be seen by the appearance of a lower, faster running band. Day 0 (quiescent) and day 4 (proliferating) samples are shown for comparison.
(A) Expression of E2F-4 in human primary CD34+ hematopoietic cells. Purified CD34+ cells were induced to proliferate by the addition of cytokines as previously described in Materials and Methods. Samples were taken at the times shown and western blotting was performed using a specific anti-E2F–4 antibody. At 0 hours, the 68-kD form of E2F-4 predominates, but as the cells proliferate, the lower forms appear with the complete disappearance of the 68-kD form by 72 hours. A longer exposure of the same blot shows that there is little or no 68-kD form present in the 72- and 96-hour samples. (B) Dephosphorylation of E2F-4 in CD34+ cells by lambda phosphatase. A total of 4 μg of CD34+ whole cell lysate made 1 day after addition of cytokines was incubated for 1 hour with 500 U of lambda phosphatase. This was then added to sample buffer and separated by SDS-PAGE and probed with an anti-E2F–4 antibody. Dephosphorylation of the 68-kD form can be seen by the appearance of a lower, faster running band. Day 0 (quiescent) and day 4 (proliferating) samples are shown for comparison.
Changes in electrophoretic migration of E2F-4 could be due either to changes in its phosphorylation state or to the expression of E2F-4 proteins of different sizes. To determine whether this shift represents phosphorylation of the E2F-4 protein, the 24-hour sample was incubated with a lambda protein phosphatase. Phosphatase treatment resulted in a faster migrating form and the upper, 68-kD band was diminished (Fig 7B). The same experiment repeated with the lambda phosphatase in 10x excess resulted in the complete ablation of the 68-kD form (not shown). This suggests that a hyperphosphorylated form of E2F-4 is present in quiescent CD34+ cells and that this form is lost, possibly by dephosphorylation, as the cells start to proliferate. Lambda phosphatase is a dual-specificity tyrosine and serine/threonine phosphatase. When the experiment was repeated with phosphatases specific to either phosphotyrosine or phosphoserine/threonine, only the serine/threonine-specific phosphatase caused partial dephosphorylation, and the tyrosine phosphatase had no effect (data not shown). We conclude that the novel form of E2F-4 present in quiescent CD34+ cells is hyperphosphorylated on serine/threonine and that it is dephosphorylated as cells enter G1 from G0.
DISCUSSION
CD34+ progenitor cells, which have been mobilized and then isolated from peripheral blood, are quiescent with only a very small percentage (<1%) of cells in cycle. The results presented in this study, using both the determination of cellular DNA and protein levels by flow cytometry and the molecular analysis of specific cell cycle proteins, indicates that these cells are in G0. We have shown that culturing these cells in vitro with SCF, IL-3, and IL-6 induces a normal proliferative response during which the cells increase in size and protein content before entering S-phase. In addition, we have shown changes in the expression and function of the cell cycle-dependent proteins E2F, DP-1, and the pRb family of pocket proteins that are consistent with cell cycle progression from G0 to S-phase. Further, we have shown that E2F-4 is the predominant E2F protein both in quiescent and proliferating CD34+ cells: as a p130/E2F-4/DP–1 complex in G0 and with p107 in proliferating cells. We have also shown that a novel, hyperphosphorylated form of E2F-4 exists in freshly isolated CD34+ cells, which is not present when the cells proliferate.
Several studies have suggested that CD34+ cells isolated from peripheral blood are quiescent.6-8 For example, To et al7 have reported a detailed flow cytometric analysis of progenitor cells mobilized by chemotherapy or cytokines showing that a high proportion of the cells had low CD71 expression and Rh123dull status suggestive of quiescence. Roberts and Metcalf6 extended these studies using the tritiated thymidine suicide assay for S-phase analysis. They showed that in both mice and humans peripheral blood progenitor cells are not in cycle, either in the steady state or after induction with cytokines in vivo. Recently, Jordan et al10 used flow cytometry to measure DNA content, and Ki-67 expression in CD34+ cells. They showed that CD34+ cells have 2n DNA content and approximately 75% are Ki-67 negative. Ki-67 is a nuclear antigen, which is expressed only in proliferating cells, and the lack of expression is assumed to indicate a G0 state.33,34 In our study using two color flow cytometry analysis to measure both DNA and protein levels, which similarly allows the differentiation of G0 and G1, we have found that over 95% of the mobilized peripheral blood CD34+ cells are in G0. The reason for this higher value may be due to the different technique used to define G0, but could also relate to the different mobilization procedures used. In this study, a combination of chemotherapy and G-CSF was used, whereas in the Jordan study, G-CSF alone was used.10
During the first few hours after growth factor stimulation, cells which were in G0 transit a defined cell cycle stage called G1e (entry) during which there is no increase in cell size. Thereafter, the cell cycle and growth cycle (ie, cell size) are coordinated during the progression through G1.35 We have found that after stimulation in vitro with SCF, IL-3, and IL-6, the CD34+ cells do not increase appreciably in size for about 8 hours. Subsequently, their size increases as they progress through G1, which is similar to the changes that occur when primary T cells are stimulated in vitro with PHA.30
An important factor to be taken into account in interpretation of the flow cytometry data is the heterogeneity of the CD34+ population examined. For example, whereas the majority of mobilized CD34+ cells express the CD38 antigen,36 a small proportion do not, and a recent study shows that this subpopulation of cells stay in G0 for a longer period following cytokine stimulation.10 The most primitive cells, furthermore, do not proliferate after a 2-week exposure to a cytokine cocktail37 and are presumably included among the cells which we find to still be in G0 72 hours after cytokine stimulation.
To extend these studies, we have examined specific cellular proteins that are critical for cell cycle progression. The E2F family of transcription factors play an important role in controlling cell proliferation in that they are both necessary and sufficient for cells to enter S-phase from a quiescent state. We have shown previously that human primary B cells and monocytes express a single E2F-DNA binding complex containing E2F-4, DP-1, and p130 proteins.27 Similarly, Vairo et al25 have shown this to be the major complex in quiescent T cells. All of these cells are thought to be in G0. The results presented here show that the major E2F-DNA binding complex in CD34+ cells also contains E2F-4, DP-1, and p130. It has been suggested that p130 maintains cells in G0 when in its hypophosphorylated state by suppressing E2F activity32 and our data, which show that p130 in the CD34+ cells is hypophosphorylated, is consistent with this hypothesis.
It is now well established that peripheral blood CD34+ cells when reinfused following high-dose therapy will reconstitute the bone marrow more quickly than bone marrow progenitor cells.3 Thus, they appear to be able to proliferate rapidly in vivo, despite being in G0 when freshly isolated. In addition, the ability to stimulate CD34+ cells in vitro using cytokines is now well recognized for use in retroviral gene transduction and possible ex vivo expansion. To understand the molecular mechanisms that control the proliferation of CD34+ cells, we stimulated them to proliferate in vitro with a combination of SCF, IL-3, and IL-6 and examined cell cycle-specific changes in E2F and pocket protein expression and function. Chittenden et al26 have shown previously that T cells made to proliferate by the addition of PHA and IL-2 express three E2F complexes, the principal one containing E2F-1, p107, and cyclin A. Moberg et al,15 however, showed E2F-4/p107 complexes in T cells when they were in G1. Our results show that E2F-4 also remains the predominant form of E2F in proliferating CD34+ progenitor cells. No E2F-1 was detected, although E2F-4 bound to p107 is detectable within 24 hours of cytokine stimulation. The timing is consistent with the hyperphosphorylation and reduced binding activity of p130 as the cells progress into cycle. Both pRb and a low level of p107 are present in the quiescent CD34+ cells, however, neither form detectable E2F-DNA binding complexes. p107 and pRb are both induced and phosphorylated as CD34+ cells enter S-phase from G1, which is consistent with work with other cell types.15 We detect both hyper and hypophosphorylated forms of p107 in proliferating CD34+ cells, and the hypophosphorylated form of p107 may account for formation of the p107-E2F–4 complexes detected by EMSA.38 39
We have detected a novel, heavily phosphorylated, 68-kD form of E2F-4 in quiescent CD34+ cells, which has not been described previously and which we have not detected in other quiescent hematopoietic cells including T cells, B cells, or monocytes. There is then a progressive change in its phosphorylation state, as the CD34+ cells progress from G0 into the cell cycle. Despite its dephosphorylation over the 3-day period following cytokine stimulation, E2F-4 still remains the predominant component of DNA-bound complexes. Vario et al25 have shown that certain forms of E2F-4 associate preferentially with p130 and p107. Our data show that the 68-kD form of E2F-4 also associates with p130 and binds DNA. p130 begins to be phosphorylated by about 8 hours after stimulation, which is before or during the transition of CD34+ cells from G0 into G1. We have shown that this also occurs at the same stage in primary T cells (Thomas et al, manuscript submitted) and is consistent with the conclusion that p130 complexes distinguish G0 from G1.40 Thus, in CD34+ cells, hyperphosphorylation of p130 and release of E2F-4 may be part of the molecular mechanism by which cells progress from G1e into G1.35 Dephosphorylation of the 68-kD form of E2F-4 occurs later than the phosphorylation of p130; however, it is not clear whether E2F-4 has to be dephosphorylated to allow it to become fully active.
In conclusion, we propose that hypophosphorylated p130 complexed with DP-1 and the hyperphosphorylated, 68-kD form of E2F-4 maintains CD34+ cells in a quiescent state in the peripheral blood. To proliferate in response to the appropriate cytokines, E2F-4 may need to be dephosphorylated to hypophosphorylated forms while p130 becomes hyperphosphorylated. The rapid engraftment that follows CD34+ transplantation cannot be explained by the cell cycle status of the cells.
ACKNOWLEDGMENT
We thank Profs Nic La Thangue, Peter Whyte, and Ed Harlow for providing antibodies. We would also like to thank Dr A.H. Goldstone for support.
N.S.B.T. is supported by the Kay Kendall Leukaemia Fund.
Address reprint requests to N. Shaun B. Thomas, PhD, Department of Haematology, University College London Medical School, 98 Chenies Mews, London WC1E 6HX, UK.