Abstract
In addition to the major fusion gene PML-RARα, the t(15; 17) in acute promyelocytic leukemia (APL) produces the reciprocal fusion gene RARα-PML. To determine the scope of RARα-containing mRNA expression in APL cells, we tested PML-RARα–positive APL cells for the presence of mRNAs initiated from two distinct RARα gene promoters, α1 and α2. From the normal allele, both RARα1 and RARα2 mRNAs were expressed in all APL cases (N = 24). From the translocated allele, RARα1-PML mRNA was expressed in 77% and RARα2-PML mRNA in 28% of cases (N = 98). RARα2-PML mRNA was not observed in the absence of RARα1-PML mRNA. There was no association between RARα1-PML or RARα2-PML mRNA expression and the type of PML-RARα mRNA formed by either 5′ or 3′ breaksites in the PML gene. RARα1-PML mRNAs and RARα2-PML mRNAs from 5′ PML breaksite cases coded for full-length RARα-PML proteins but RARα2-PML mRNAs from 3′ PML breaksite cases encoded a truncated RARα2 peptide. RARα1/α2-PML mRNA expression was not associated with differences in APL cell sensitivity to all-trans retinoic acid(tRA)-induced differentiation in vitro or in clinical outcome after tRA or chemotherapy induction therapy (protocol E2491). Our analysis indicated that RARα1/α2-PML mRNA expression markedly differs from normal RARα1/α2 mRNA expression, that the difference in RARα1-PML and RARα2-PML mRNA expression frequency is primarily related to the genomic separation of the RARα1 and RARα2 coding exons, and that variations in RARα1/α2-PML mRNA expression likely have no clinically relevant function in APL cells.
THE t(15; 17) associated with acute promyelocytic leukemia (APL) results in the formation of two reciprocal fusion genes, PML-RARα and RARα-PML. Because hybrid mRNA for the PML-RARα fusion gene is expressed in virtually all APL cases, and because it encodes the most essential functional domains of both fusion gene proteins, it is regarded as the critical product for both the genesis and maintenance of the disease.1-5 RARα-PML mRNA has been reported to be expressed in 67% to 80% of APL cases.6,7 The putative proteins coded by RARα-PML contain the A-region of RARα that has ligand-independent modulatory effects on the transcriptional regulatory activity of RARα, a short common region of PML encoded by exon 7 that contains a unique casein kinase II phosphorylation site, and a variety of serine/proline-rich segments in different transcripts caused by variable PML exon splicing.6,8 Although this information content seems limited, RARα-PML has been considered to have potential biological significance, because the coding exons are consistently retained, ie, they are not deleted in the translocation process,9 and because the translational open reading frame (ORF ) is maintained in RARα-PML–positive cases.6 The possibility that RARα-PML may have selective value for the leukemic phenotype was recently stimulated by the report of a t(15; 17)-negative APL case with cryptic formation of an RARα-PML fusion gene in the absence of PML-RARα.10
The current study was performed to evaluate essentially three questions related to RARα-PML mRNA expression in APL cells. First, what is the extent of heterogeneity of RARα-PML mRNA expression, taking into consideration that two different forms of RARα (RARα1 and RARα2) have been reported in some mammalian cells11 and that previous APL cell studies only measured RARα1-PML mRNA expression?6,7,10 Normal RARα1 and RARα2 mRNA expression is governed by two independent gene promoters, and the respective proteins differ at the amino end because of alternative selection of exons encoding either the A1 or A2 region joined to a common body of the RARα molecule, the B to F regions.11 Second, is there any relationship of heterogeneity in RARα1-PML and/or RARα2-PML mRNA expression and the type of PML-RARα mRNA expressed in the same APL cells? In individual APL cases, one of three possible types of PML-RARα mRNA is expressed because of alternative break sites in the PML gene, one 5′-situated (in intron 3, the short(S)-form type) and two 3′-situated (in intron 6, the long(L)-form type, or in variable sites in exon 6, the variable(V)-form type).4,5,12 Third, is there any relationship between the heterogeneity of RARα1/α2-PML mRNA expression and important biological characteristics measured by differences in APL cell responsiveness to all-trans retinoic acid (tRA)-induced differentiation in vitro and by differences in patient pretreatment characteristics or responsiveness to therapy? The latter could be investigated because most RNA specimens were derived from APL patients entered on a randomized clinical trial comparing the effectiveness of tRA versus standard chemotherapy as remission induction therapy (Protocol INT 0129/E2491).13
MATERIALS AND METHODS
Patients and pretreatment clinical laboratory studies.Of the 98 PML-RARα–positive patients involved in this study, 72 were participants in the Eastern Cooperative Oncology Group (ECOG) clinical trial E2491. The other 26 PML-RARα–positive as well as the 38 PML-RARα–negative patients were derived from ECOG ancillary study E1485. Patient materials were obtained under an ECOG protocol-approved consent form and/or individual institutional review board approval. Baseline patient data were obtained from the ECOG central data registry and hemogram parameters, eg, white blood cell (WBC) counts, represent the presenting values before antileukemic therapy.
Cell preparation and culture.Heparinized bone marrow BM and peripheral blood (PB) specimens were received by overnight express mail from ECOG institutions at room temperature. A low-density WBC fraction (d ≤1.077 g/mL) was isolated and evaluated as previously described.12 Only low-density cell fractions with ≥75% myeloblasts/promyelocytes were used for cell culture studies and for the assessment of RARα1 and RARα2 mRNA expression. Cell culture procedures for the assessment of APL cell tRA sensitivity to tRA in vitro were as previously described.14 15
RT-PCR analysis.Total cellular RNA was prepared by a modification of the guanidine isothiocyanate extraction-cesium chloride gradient ultracentrifugation procedure, as previously described,14 or using the TRIzol reagent (Life Technologies, Inc, Gaithersburg, MD), according to the supplier's directions. Reverse transcription of RNA from random hexamer primers (Pharmacia, Piscataway, NJ) and PCR were performed essentially as previously described,14 except that amplification conditions were modified to be optimal for each primer pair used. Primer pairs and conditions for their application were selected with the assistance of Oligo™ software (National Biosciences, Inc, Plymouth, MN). The upstream and downstream primers used to amplify first strand cDNA for each gene/hybrid gene transcript were as follows: RARα1, 5′-GCCAGGCGCTCTGACCACTC-3′ and 5′-AGCCCTTGCAGCCCTCACAG-3′; RARα2, 5′-CCACCCCTAATCCCTTCCTA-3′ and 5′-AGCCCTTGCAGCCCTCACAG-3′; RARα1-PML, 5′-TCCCTACGCCTTCTTCTTCC-3′ and 5′-CCGAGCTGCTGATCACCACA-3′; and RARα2-PML, 5′-CCACCCCTAATCCCTTCCTA-3′ and 5′-CCGAGCTGCTGATCACCACA-3′. The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was used as an RNA integrity control by a competitive RT-PCR method, and primers and RT-PCR conditions for the determination of PML-RARα type were as previously described.12 The relative intensity of ethidium bromide-stained PCR gel electrophoretic bands were determined by densitometric volume measurements using a scanning laser densitometer and ImageQuant software analysis (Molecular Dynamics, Sunnyvale, CA).
DNA sequence analysis.The nucleotide sequences of the junctions between exons 2 or 3 of the A1- or A2-regions of RARα1 and RARα2 and exon 4 of the common B-region of RARα in RAR α1/α2 mRNAs and between RARα exons 2 or 3 and PML exons 4 or 7 in RARα1/α2-PML mRNAs were determined by bidirectional direct sequencing, as previously described.14
RESULTS
Expression of RARα1 and RARα2 mRNAs in APL cells.Using RT-PCR with an upstream primer specific for the A1-region of RARα1 or the A2-region of RARα2 in combination with a common downstream primer anchored in the C-region of RARα (Fig 1), constitutive expression of RARα1 and RARα2 mRNA was readily detected in pretreatment BM or PB cell preparations containing ≥75% leukemic promyelocytes or blasts in 24/24 PML-RARα–positive APL cases tested (data not shown). Experiments in which the RARα2-specific primer was competed with an equal amount of RARα1-specific primer in the same reaction tube indicated that RARα2 mRNA was expressed at a lower level than RARα1 mRNA in all of 17 cases tested (Fig 2). The mean and median ratios of RARα2 to RARα1 were, respectively, 0.58 ± 0.14 and 0.60, range 0.35 to 0.78. The RARα2 to RARα1 mRNA ratio was similar for low-density mononuclear cells from normal BM and PB. No ratio could be determined for the APL cell line NB4, because the RARα2 signal was completely competed by the RARα1 signal (Fig 2). In the absence of the RARα1 primer competitor, a low level of RARα2 mRNA was detectable in NB4 cell RNA under the same RT-PCR conditions (data not shown).
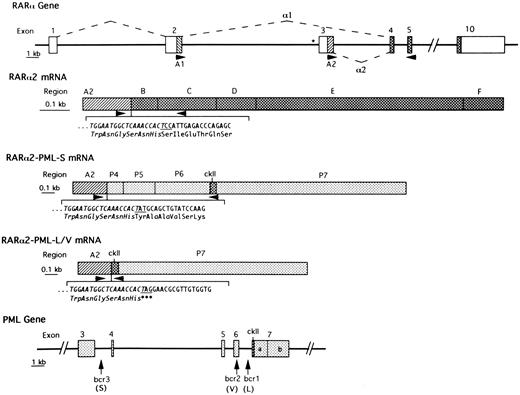
Schematic representation of the intron-exon structure of relevant parts of the RARα and PML genes and of the formation of RARα2 mRNA and of RARα2-PML mRNAs from APL cases with S-form or L/V-forms of PML-RARα mRNA. Introns are represented by heavy lines. The boxes in the top (RARα gene) and lower (PML gene) panels indicate exons. The dashed lines indicate how RARα exons are joined to form RARα1 or RARα2 mRNAs. The asterisk indicates the RARE site in the promoter region of RARα2. Open boxes indicate noncoding regions. The simple hatched areas indicate the coding regions of RARα1 (A1) and RARα2 (A2), which become fused to the variable PML carboxy-termini in RARα-PML mRNAs. Stippled boxes indicate PML exons (P4, P5, P6, P7a, and P7b), which are variably spliced in different PML-containing isoforms. Cross-hatched boxes indicate coding regions that are consistently retained in RARα- or PML-containing transcripts. ckII is the unique casein kinase-II site in constantly retained region of PML exon 7. Arrowheads indicate the site of PCR primers used in the present studies. The perijunctional nucleotide sequence and codon assignment is indicated below the schematic RARα2 and RARα2-PML mRNAs with the sequence derived from RARα exon 3 italicized and the junctional codon (derived from the last nucleotide of RARα2 exon 3 and the first two nucleotides of RARα exon 4/B-region, PML exon 4 or PML exon 7) underlined.
Schematic representation of the intron-exon structure of relevant parts of the RARα and PML genes and of the formation of RARα2 mRNA and of RARα2-PML mRNAs from APL cases with S-form or L/V-forms of PML-RARα mRNA. Introns are represented by heavy lines. The boxes in the top (RARα gene) and lower (PML gene) panels indicate exons. The dashed lines indicate how RARα exons are joined to form RARα1 or RARα2 mRNAs. The asterisk indicates the RARE site in the promoter region of RARα2. Open boxes indicate noncoding regions. The simple hatched areas indicate the coding regions of RARα1 (A1) and RARα2 (A2), which become fused to the variable PML carboxy-termini in RARα-PML mRNAs. Stippled boxes indicate PML exons (P4, P5, P6, P7a, and P7b), which are variably spliced in different PML-containing isoforms. Cross-hatched boxes indicate coding regions that are consistently retained in RARα- or PML-containing transcripts. ckII is the unique casein kinase-II site in constantly retained region of PML exon 7. Arrowheads indicate the site of PCR primers used in the present studies. The perijunctional nucleotide sequence and codon assignment is indicated below the schematic RARα2 and RARα2-PML mRNAs with the sequence derived from RARα exon 3 italicized and the junctional codon (derived from the last nucleotide of RARα2 exon 3 and the first two nucleotides of RARα exon 4/B-region, PML exon 4 or PML exon 7) underlined.
Gel electrophoretic analysis of ethidium bromide-stained DNA products synthesized in primer competition experiments from RARα2 mRNA (upper band) or RARα1 mRNA (lower band). Five picomoles each of upstream primers specific for RARα1 or RARα2 were used with 10 pmol of a common downstream primer anchored in the C-region of RARα to amplify 1 μg-RNA equivalent of cDNA, as described in Materials and Methods. The abbreviations are: M, 100-bp ladder; L1 to L3, products from 3 APL patients with L-form PML-RARα mRNA; V1, product from an APL patient with V-form PML-RARα mRNA; S1 and S2, products from 2 APL patients with S-form PML-RARα mRNA; NB4, product from APL cell line NB4 mRNA; BM, product from normal bone marrow; PB, product from normal peripheral blood. All illustrated APL RNAs were derived from specimens containing greater than 90% blasts plus promyelocytes.
Gel electrophoretic analysis of ethidium bromide-stained DNA products synthesized in primer competition experiments from RARα2 mRNA (upper band) or RARα1 mRNA (lower band). Five picomoles each of upstream primers specific for RARα1 or RARα2 were used with 10 pmol of a common downstream primer anchored in the C-region of RARα to amplify 1 μg-RNA equivalent of cDNA, as described in Materials and Methods. The abbreviations are: M, 100-bp ladder; L1 to L3, products from 3 APL patients with L-form PML-RARα mRNA; V1, product from an APL patient with V-form PML-RARα mRNA; S1 and S2, products from 2 APL patients with S-form PML-RARα mRNA; NB4, product from APL cell line NB4 mRNA; BM, product from normal bone marrow; PB, product from normal peripheral blood. All illustrated APL RNAs were derived from specimens containing greater than 90% blasts plus promyelocytes.
Expression of RARα1-PML and RARα2-PML mRNAs in APL cells.RARα1-PML and RARα2-PML are formed, using normal splicing signals by joining the 3′ terminus of RARα exon 2 or exon 3, respectively, to the 5′-terminus of either PML exon 4 (S-form PML-RARα cases) or exon 7 (L- and V-form PML-RARα cases)6 (Fig 1). In L-form and V-form cases, only one gel electrophoretic band was visualized (Fig 3), because the alternatively spliced exons are retained in the reciprocal PML-RARα product,4,12 and because the exon 6 breaksite in V-form cases lacks an appropriate splice acceptor site.6 In S-form cases, three specific gel bands were observed because of alternative splicing between PML exons 4, 5, and 6. The upper band represents full-length RARα1-PML or RARα2-PML RNAs, ie, PML exons 4 to 7 are present; the middle band lacks PML exon 5; and the lower band lacks PML exons 5 and 66 (confirmed by our sequencing results).
Gel electrophoretic analysis of ethidium bromide–stained RT-PCR products generated from mRNAs encoding RARα1-PML (left panel) or RARα2-PML (right panel) from L-form, V-form, and S-form PML-RARα mRNA type cases, including one atypical S-form case (Sv). M is Hae III-digested φX174 DNA marker; 0 is 0 RNA control.
Gel electrophoretic analysis of ethidium bromide–stained RT-PCR products generated from mRNAs encoding RARα1-PML (left panel) or RARα2-PML (right panel) from L-form, V-form, and S-form PML-RARα mRNA type cases, including one atypical S-form case (Sv). M is Hae III-digested φX174 DNA marker; 0 is 0 RNA control.
Of 98 PML-RARα–positive cases tested, RARα1-PML mRNA was detected in 75 cases (77%) and RARα2-PML mRNA in 27 cases (28%). RARα2-PML mRNA was never detected in the absence of RARα1-PML mRNA. NB4 cells, which contain the L-form of PML-RARα, expressed RARα1-PML but not RARα2-PML mRNA. The typical gel electrophoretic band pattern described above was observed in all but one S-form case (Sv) in which all three bands were coordinately reduced in size because of the absence of exon 4 (Fig 3).
RARα-PML mRNA junctional sequence analyses.Compared with the amino acid codons formed by inter-exonic splicing of normal RARα1 or PML transcripts, a novel codon is generated at the RARα1-PML junction in both S-form (ATT; asparagine) and L/V-form cases (AAG; lysine) of APL with maintenance of the translational ORF 6 (confirmed by our sequencing results). Similarly, in RARα2-PML–positive S-form APL cases, the ORF is maintained with a change in the junctional codon from the TCC (serine) of normal RARα2 to TAT (tyrosine) (Fig 1). In L- and V-form cases, however, the junctional codon changes to an amber termination codon (TAG), such that the translation product is limited to the A2 region of RARα2 (Fig 1). Also, in the single Sv noted to lack PML exon 4 in RARα1-PML mRNA (Fig 3), sequence analysis confirmed that the splicing events altered the ORF for all three RARα1-PML isoforms, each of which encodes only the A1 region of RARα1 with a short carboxy-terminus of non-PML amino acids (data not shown).
RARα1/α2-PML mRNA expression and PML-RARα fusion gene type.As summarized in Table 1, 22% to 25% of cases lacked RARα1-PML mRNA expression and, conversely, 75% to 78% of cases expressed RARα1-PML mRNA in all three PML-RARα mRNA types. In contrast to this tight range of values, the fraction of RARα2-PML mRNA expressors varied from 19% in S-form cases to 50% in V-form cases or 33% in L-form plus V-form cases, although these differences did not achieve statistical significance (P ≥ .17).
Tests for RARα1-PML mRNA in PML-RARα-negative leukemia cells.Thirty-eight leukemic cell specimens submitted to our laboratory with the suspected diagnosis of APL based on initial clinical evaluation and/or the immunophenotypic profile that, subsequently, turned out to be PML-RARα mRNA-negative were tested for the expression of RARα1-PML mRNA. All tests were negative.
Studies for possible correlation with in vitro APL tRA sensitivity.In vitro tests for tRA-induced terminal differentiation were performed in the majority of PML-RARα–positive cases analyzed for RARα1-PML and RARα2-PML mRNA expression, by the nitroblue tetrazolium dye reduction (NBT) test in 79 cases, and by the expression of CD11b, a surface membrane antigen marker for terminal myeloid cell differentiation, in 67 cases. As summarized in Table 2, no correlation was observed by either of these measures at 10−7 mol/L tRA (P ≥ .37) or by the NBT test at 10−8 mol/L tRA (data not shown). At 10−7 mol/L tRA, these values for the atypical S-form case lacking PML exon 4 in RARα1-PML mRNA were 78.5% NBT-positive and 85% CD11b-positive.
Studies for possible correlation with clinical characteristics and response to therapy.Of the 98 total APL patients analyzed, 72 were entered on protocol E2491 and were analyzed for RARα1-PML and RARα2-PML mRNA expression. The distribution of the three possible combinations of RARα1/α2-PML mRNA expression for this subset were virtually identical to those for the entire group (compare Tables 1 and 3). As shown in Table 3, there were no significant differences (P ≥ .1) in the presenting clinical characteristics of patients when analyzed for a difference in: (1) any one of the three possible RARα1-PML/RARα2-PML combinations compared with the other two; (2) RARα1-PML mRNA expressors versus nonexpressors; and (3) RARα2-PML mRNA expressors versus nonexpressors. A similar analysis of on-study hematologic characteristics (Table 4) indicted a trend toward a higher proportion of RARα1-PML mRNA nonexpressors than expressors with a presenting WBC count greater than 5,000 cells/uL (50% v 21%; P = .054).
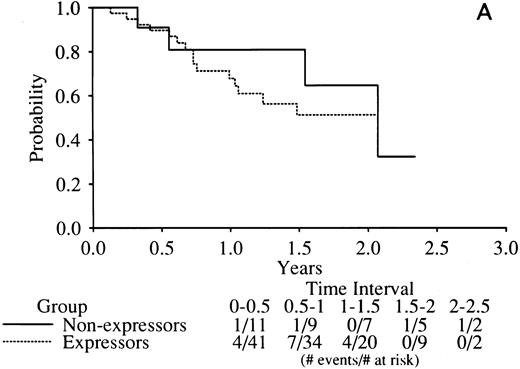
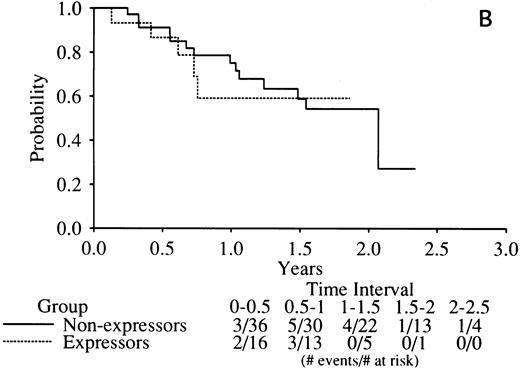
No indication of any difference was found in the complete remission (CR) rate related to RARα1/2-PML mRNA expression with the rates of all three combinations tightly clustered around the mean of 77% (Table 5). Similarly, there was no indication of any difference in DFS in RARα1-PML expressors versus nonexpressors (Fig 4A) or RARα2-PML expressors versus nonexpressors (Fig 4B).
Kaplan-Meier curves of disease-free survival in RARα1-PML mRNA (A) or RARα2-PML mRNA (B) expressors versus nonexpressors. The legend denominators indicate the number of protocol cases at risk in successive 6-month intervals up to 2.5 years of followup beginning from the time of the clinical diagnosis of complete remission. The legend numerators indicate the number of cases at risk that experienced adverse events leading to an off-protocol status (relapse or death).
Kaplan-Meier curves of disease-free survival in RARα1-PML mRNA (A) or RARα2-PML mRNA (B) expressors versus nonexpressors. The legend denominators indicate the number of protocol cases at risk in successive 6-month intervals up to 2.5 years of followup beginning from the time of the clinical diagnosis of complete remission. The legend numerators indicate the number of cases at risk that experienced adverse events leading to an off-protocol status (relapse or death).
DISCUSSION
This report documents that both RARα1 and RARα2 mRNAs are universally expressed from the nontranslocated allele in fresh APL cells. RARα2 mRNA is expressed at a lower level than RARα1 mRNA in all cases. Further studies are required to evaluate possible selective quantitative changes in the expression level of RARα2 mRNA in APL cells following tRA exposure. In murine cells, RARα1 and RARα2 can be independently regulated by tRA related to a retinoic acid response element (RARE) in the promoter region of RARα2 but not RARα1.11 On the translocated allele in APL cells, the RARα2 coding exon 3, like RARα1 coding exons 1 and 2, could be expressed as a fusion product with downstream PML exons as RARα2-PML mRNA. The incidence of RARα2-PML mRNA expression, however, was much lower than RARα1-PML mRNA with 27.6% versus 76.5% of APL cases positive, respectively. This lower incidence of RARα2-PML mRNA is not unexpected, because a complex recombinational event would be required to form RARα2-PML mRNA in cases with gene breaks between RARα exons 2 and 3. Because RARα exon 3 is situated approximately two-thirds of the distance between exon 2 and exon 4 that encodes the B-region in humans,5,15 the observed frequency of RARα2-PML mRNA expression is approximately that predicted (ie, 76.5 × 0.33 = 25.2%), if the breaksites between RARα exons 2 and 4 are equally distributed, and if no other element differentially affects the probability of forming stable RARα2-PML mRNA versus RARα1-PML mRNA. Reported mapping data of RARα “intron 2” breaksites, in fact, indicate a fairly even distribution.5,9 These considerations, as well as our observation that RARα2-PML mRNA is not expressed in the absence of RARα1-PML mRNA, suggest that the expression of these two gene products are not independently regulated in APL cells. Our finding that RARα1-PML mRNA is expressed in the absence of RARα2-PML mRNA in NB4 cells in which the RARα breaksite has been determined to lie between exons 3 and 4 without deletion9 indicates, however, that sequential regulation is not universally obligate. More generally, it is not understood why 25% of APL cases do not express RARα1-PML mRNA, although the lack of deletion suggests involvement of a transcriptional or posttranscriptional regulatory mechanism. Our calculation that the reduced incidence of RARα2-PML mRNA expression is proportionate to that for RARα1-PML mRNA also suggests involvement of a common mechanism not operative on the normal RARα allele.
In addition to confirming the previously reported incidence of RARα1-PML mRNA expression in leukemic cells from 67% to 80% of APL cases,6,7 this report establishes that its incidence is equal among all three PML-RARα fusion gene types. Although RARα2-PML mRNA expression appeared to be expressed at a somewhat lower frequency in S-form cases (7/37; 19%) compared with L-form and V-form cases (20/57; 33%), this difference was not statistically significant (P = .17; Fisher's exact test). Further, in a recent genomic mapping study, PML intron 3 breaksites, ie, S-form cases, were not selectively associated with RARα exon 3 to 4 breaksites (7/25 cases; 28%) compared with RARα exon 2 to 3 breaksites (9/36 cases; 25%).9 Thus, we conclude that there is no measurable association between RARα2-PML mRNA expression and PML-RARα type.
Most importantly, this study found no evidence for a biologically significant role of RARα1-PML mRNA or RARα2-PML mRNA expression in APL cells. At a molecular level, we found that RARα2-PML mRNA encodes a truncated RARα2 peptide restricted to the A2-region because of a translational reading frame shift at the RARα2-PML junction in all RARα2-PML–positive L- and V-form PML-RARα cases. It seems improbable that such a short peptide, even if stably produced, could have a positive functional role, although it conceivably could have a negative action by somehow interfering with its normal counterpart. Studies at the cellular level were against the latter possibility. APL cells from RARα2-PML–positive L-form cases had high-level sensitivity to tRA-induced differentiation in vitro, and, similarly, APL cells from a variant S-form PML-RARα case (Sv) that expressed only a truncated RARα1 peptide restricted to the A1-region were fully sensitive to tRA-induced differentiation in vitro. More generally, we found no correlation between the expression of either RARα1-PML or RARα2-PML mRNAs and sensitivity to tRA-induced differentiation of APL cells in vitro. At the clinical level in protocol E2491 patients, we also found no differences related to RARα1/α2-PML mRNA expression in the CR rate after randomization to either tRA or standard chemotherapy induction treatment or in the distribution of overall DFS with a median follow-up of 16 months.13 In the patient subgroup that lacked expression of either RARα1-PML or RARα2-PML mRNA, we found a trend toward a higher presenting WBC count (P = .054), a characteristic consistently associated with a poor long-term prognosis in APL.13,19,20 This weak association and limitations of statistical power related to sample size are our only reservations to the conclusion that the current study provides no indication of a clinically relevant role of the RARα1-PML or RARα2-PML fusion gene products in APL. This conclusion is analogous to that reached in a recent study of chronic myelogenous leukemia in which it was reported that the presence or absence of expression of the reciprocal product of the BCR-ABL hybrid gene associated with the t(9; 22), ie, ABL-BCR mRNA, was not correlated with clinical response to interferon-α therapy.21
ACKNOWLEDGMENT
The authors thank the physicians, nursing staffs, and data managers of the Albert Einstein Cancer Center and other ECOG affiliated institutions for their cooperation in obtaining and sending the clinical specimens used for performing these studies. We also thank Johanna Epstein for technical assistance and for performing the densitometric readings of ethidium bromide-stained PCR products after gel electrophoresis.
Supported by Public Health Service Grants No. CA56771, CA21115, CA23318, CA14985, and P30CA13330 from the National Cancer Institute, National Institutes of Health and the Department of Health and Human Services, the Chemotherapy Foundation, and the Leukaemia Research Fund of Great Britain.
This study was conducted in the majority by the Eastern Cooperative Oncology Group (Robert L. Comis, MD, Group Chair).
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Address reprint requests to Robert E. Gallagher, MD, Department of Oncology, Montefiore Medical Center, 111 210th St, Bronx, NY 10467.