To the Editor:
The major histocompatibility complex class 1-like gene, HLA-H, was recently reported as being the candidate gene for the autosomal recessive disorder hereditary hemochromatosis (HH).1 There is still no formal proof that HLA-H is the HH gene, but the evidence can be compelling,1 although it may not be the only gene involved.2,3 A single base mutation, Cys282Tyr, appears to have a causative role in HH and a second variant, His63Asp, may also have a role.1 Detection of these two mutations is certainly of value in diagnosing hemochromatosis individuals.4 As a result, polymerase chain reaction (PCR) has been used for rapid detection of the mutations2,5 and a further modification for elucidation of any codon 282 mutations was recently reported6 in Blood.
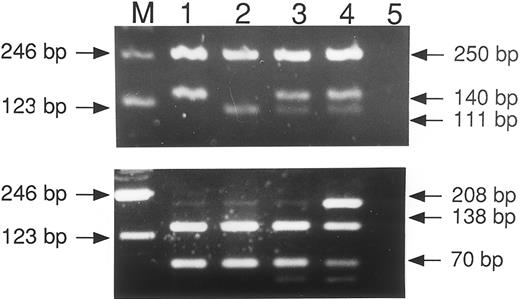
Detection of the HLA-H Cys282Tyr and His63Asp mutations by PCR-RFLP. The top panel shows the result of PCR with the codon 282 primers and restriction digest by Rsa I. The bottom panel is PCR product obtained with the codon 63 primers and digested with Bcl I. The patients are as follows: track 1, normal; track 2, homozygous mutant, codon 282; normal, codon 63; track 3, heterozygous mutant, codon 282; normal codon 63; track 4, compound heterozygote; track 5, water negative control.
Detection of the HLA-H Cys282Tyr and His63Asp mutations by PCR-RFLP. The top panel shows the result of PCR with the codon 282 primers and restriction digest by Rsa I. The bottom panel is PCR product obtained with the codon 63 primers and digested with Bcl I. The patients are as follows: track 1, normal; track 2, homozygous mutant, codon 282; normal, codon 63; track 3, heterozygous mutant, codon 282; normal codon 63; track 4, compound heterozygote; track 5, water negative control.
Beutler et al5 detect the presence of a mutation in their PCR product by time consuming allele-specific oligonucleotide hybridization. The original PCR-restriction fragment length polymorphism (PCR-RFLP) method of Jazwinska et al2 allows identification of zygosity at both alleles, but takes some time to complete and uses expensive restriction enzymes (SnaBI, ∼£36/100 U; Mbo I, ∼£43/200 U). Although the aim of Chang et al6 is rather to detect any mutation at codon 282 to emphasize the codon's importance in HLA-H function, they also use protracted cycling and are tied to the use of the expensive restriction enzyme Bcg I. The His63Asp mutation appears to increase the risk of hemochromatosis when in conjunction with the Cys282Tyr1 mutation and should also be looked for until any causative role can be disproved. Jouanolle et al7 investigate both mutations, but do not show their PCR cycling conditions and are still using Mbo I for codon 63 mutation detection. A rapid, robust, and less expensive method for PCR-RFLP detection of both HLA-H mutations has been developed for use in our routine laboratory.
Separate PCR reactions are conducted for the two mutations using the primers described by Feder et al1 for their oligonucleotide ligation assay (OLA; Table 1). DNA is extracted from 200 μL whole blood using a QIAamp Blood kit (Qiagen, Crawley, UK), eluting from the spin column with 200 μL buffer. A total PCR volume of 25 μL contains 100 ng of each primer, 1× manufacturer's PCR buffer, 200 μmol/L each dNTP, 2 μL (∼50 ng) DNA, and 0.4 U Dynazyme (Flowgen, Shenstone, UK). After 2 minutes of initial denaturation at 94°C, 35 cycles of just 1 minute at 94°C and 1 minute at 58°C are conducted in a Perkin Elmer 480 thermal cycler (Perkin Elmer, Warrington, UK). Restriction digests are performed directly in the PCR mixes by addition of 5 U Rsa I (codon 282 reactions) or Bcl I (codon 63 reactions) and incubating for 2 hours at 37°C or 50°C, respectively. The products are electrophoresed on a 3% agarose gel (Fig 1).
Amplification with the primers for codon 282 produces a PCR product of 390 bp, which in the wild-type (Cys) digests to give fragments of 250 and 140 bp and in the mutant (Tyr) fragments of 250, 111, and 29 bp (Fig 1). (The 29-bp fragment is not always visible and is not shown here.) Amplification for codon 63 gives a 208-bp product that digests in the wild-type (His) to produce 138- and 70-bp pieces but does not digest in the mutant (Asp; Fig 1).
This PCR has also been conducted successfully in my laboratory using a Techne (Cambridge, UK) Genius thermocycler and other manufacturers' DNA polymerase, including TaKaRa Taq (Severn Biotech, Kidderminster, UK) and AmpliTaq Gold (Perkin Elmer, Warrington, UK). The cycling conditions are rapid and the restriction enzymes used cheap, ie, approximately one tenth the cost of those used by Jazwinska et al (Rsa ∼£34/1,000 U; Bcl I, ∼£36/2,000 U).
The increased speed, reduced cost, and robust nature of this method make it the method of choice for the routine diagnostic laboratory.
ACKNOWLEDGMENT
I thank Dr Roger Wolff for helpful suggestions.