Abstract
Evaluation of candidate genes for stem cell gene therapy for acquired immunodeficiency syndrome (AIDS) has been limited by the difficulty of supporting in vitro T-cell differentiation of genetically modified hematopoietic progenitor cells. Using a novel thymic stromal culture technique, we evaluated the ability of a hairpin ribozyme specific for simian immunodeficiency virus (SIV) and human immunodeficiency virus type 2 (HIV-2) to inhibit viral replication in T lymphocytes derived from transduced CD34+ progenitor cells. Retroviral transduction of rhesus macaque CD34+ progenitor cells with a retroviral vector (p9456t) encoding the SIV-specific ribozyme and the selectable marker neomycin phosphotransferase in the presence of bone marrow stroma and in the absence of exogenous cytokines resulted in efficient transduction of both colony-forming units and long-term culture-initiating cells, with transduction efficiencies ranging between 21% and 56%. After transduction, CD34+ cells were cultured on rhesus thymic stromal culture (to support in vitro differentiation of T cells) or in the presence of cytokines (to support differentiation of macrophage-like cells). After expansion and selection with the neomycin analog G418, cells derived from transduced progenitor cells were challenged with SIV. CD4+ T cells derived from CD34+ hematopoietic cells transduced with the ribozyme vector p9456t were highly resistant to challenge with SIV, exhibiting up to a 500-fold decrease in SIV replication, even after high multiplicities of infection. Macrophages derived from CD34+ cells transduced with the 9456 ribozyme exhibited a comparable level of inhibition of SIV replication. These results show that a hairpin ribozyme introduced into CD34+ hematopoietic progenitor cells can retain the ability to inhibit AIDS virus replication after T-cell differentiation and support the feasibility of intracellular immunization of hematopoietic stem cells against infection with HIV and SIV. Protection of multiple hematopoietic lineages with the SIV-specific ribozyme should permit analysis of stem cell gene therapy for AIDS in the SIV/macaque model.
GENETIC MODIFICATION of hematopoietic stem cells has received increasing attention as a therapeutic strategy for a variety of disorders affecting hematopoietic cells, including acquired immunodeficiency syndrome (AIDS). The capacity of hematopoietic stem cells for self renewal and their ability to differentiate into multiple hematopoietic lineages offer the potential for a life-long supply of cells carrying a therapeutic gene. Moreover, because of the multiple rounds of cell division that occur during hematopoiesis,1,2 stable introduction of a gene into even a small number of hematopoietic stem cells may result in a significant expansion of cells derived from transduced stem cells during the course of lineage-specific differentiation. This expansion of genetically modified cells may be even more pronounced in the case of gene therapy for human immunodeficiency virus (HIV) disease. In light of evidence suggesting a relatively rapid turnover of T cells in HIV-infected individuals,3,4 CD4+ T cells carrying a gene that inhibits HIV-1 replication should have a survival advantage in vivo, a characteristic that may compensate in part for the relatively low levels of genetically modified cells that have been observed in most studies of transduced hematopoietic stem cells in large animals and human clinical trials.5-7
A wide variety of different approaches have been devised to inhibit HIV replication, including ribozymes, RNA decoys, antisense molecules, transdominant proteins, intracellular antibodies, and suicide genes (reviewed in Yu et al8 and Sarver and Rossi9 ). Although all of these approaches have been able to inhibit viral replication in vitro, there are several considerations that favor the potential use of ribozymes as therapeutic agents. Ribozymes are small RNA molecules that have catalytic activity and can be designed to cleave specific nucleic acid sequences.10 Ribozymes may be expressed by RNA polymerase III promoters, resulting in relatively high levels of expression in multiple cell types. Unlike foreign proteins, ribozymes are unlikely to serve as targets for immune responses. In addition, because ribozymes can be designed to target specific sequences, they can be directed at relatively conserved regions of the viral genome or at sequences contained both in genomic viral RNA (thereby preventing infection of a cell) and in viral mRNA (thus inhibiting viral replication in an already infected cell). Ribozymes have the potential for widespread use in gene therapy and have been shown to inhibit expression of oncogenes,11,12 to repair defective host mRNA molecules,13 and to inhibit replication of a variety of viruses, including HIV.14-16
Despite considerable interest in stem cell gene therapy for AIDS and the availability of multiple genetic approaches to inhibit HIV replication, there has been relatively little information on the ability of genes to inhibit viral replication in hematopoietic progeny derived from transduced CD34+ hematopoietic cells. Yu et al8 were the first to show inhibition of HIV-1 replication in macrophage-like cells derived from genetically modified human CD34+ hematopoietic progenitor cells. In this study, CD34+ cells were transduced with a retroviral vector encoding the hairpin ribozyme MJT, which cleaves a conserved sequence contained in the HIV-1 LTR. Similar observations have been recently reported by others who have shown inhibition of HIV-1 replication in myelomonocytic cells derived from CD34+ cells transduced with retroviral vectors encoding a rev-responsive element decoy, a transdominant Rev protein, or a double hammerhead ribozyme.17-19 However, until recently, no data were available on whether genes designed to inhibit HIV-1 or simian immunodeficiency virus (SIV) replication could be introduced into CD34+ progenitor cells and remain active after T-cell differentiation. The lack of information on this critical issue arises in part from the fact that T-cell differentiation normally occurs in the unique three-dimensional environment of the thymus20 21 and has been difficult to model using in vitro culture systems.
Because properties of genes designed to inhibit HIV-1/SIV replication may differ between macrophages and T cells, both with respect to expression, toxicity, and efficacy, it is important to analyze the ability of candidate therapeutic genes for stem cell therapy for AIDS for their ability to inhibit viral replication in T cells as well as macrophages derived from transduced progenitor cells. In the present study, we evaluate the ability of an SIV-specific hairpin ribozyme22 to inhibit viral replication in T cells and macrophages derived from transduced CD34+ progenitor cells. Infection of macaques with SIV is generally considered to be the leading animal model for the study of AIDS.23 In addition, the close phylogenetic relationship between humans and macaques and their widespread use in preclinical studies for stem cell gene therapy24 suggest that results in the rhesus macaque model may yield valuable information for efforts to use genetically modified human stem cells as a therapy for HIV-infected people. The SIV-specific ribozyme 9456 recognizes a highly conserved sequence spanning the U3/R region of SIV and HIV-2 strains that is contained in the 3′ LTR of both genomic viral RNA and all SIV viral mRNA transcripts.22 Using a recently described thymic culture system that supports in vitro T-cell differentiation of CD34+ hematopoietic progenitor cells,25 we analyzed T cells derived from rhesus CD34+ progenitor cells transduced with a retroviral vector encoding the 9456 ribozyme. Our results now demonstrate that both T cells and macrophage-like cells derived from rhesus macaque CD34+ progenitor cells transduced with the 9456 ribozyme are highly resistant to SIV infection.
MATERIALS AND METHODS
Bone marrow harvest and immunomagnetic bead CD34 cell isolation. Rhesus monkeys (Macaca mulatta) used in this study were normal, colony-born animals maintained in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the Guide for the Care and Use of Laboratory Animals (DHHS Publication No. [NIH] 85-23, revised 1985). Bone marrow aspirates were obtained from anesthetized macaques and CD34+ cells purified by immunomagnetic beads (Dynal, Lake Success, NY), as previously described.25 Rhesus CD34+ cells obtained in this fashion were greater than 90% CD34+ and contained less than 1% residual CD3+ T cells. Purified CD34+ cells were subsequently further depleted of CD3+ cells by incubation with a CD3-specific antibody 6G12 (kindly provided by J. Wong, Massachusetts General Hospital, Boston, MA),26 followed by incubation with sheep antimouse magnetic beads (Dynal) and depletion of cells bound to beads using a magnetic separation device (Dynal).
Flow cytometric analysis and cell sorting. Antibodies used for immunophenotyping of rhesus cells included anti-CD3 (6G12), anti-CD4 (OKT4; obtained from the American Type Culture Collection, Rockville, MD), anti-CD8 (Leu-2a; Becton Dickinson, San Jose, CA), anti-CD14 (MOP9; Becton Dickinson), and anti-CD34 (Qbend-10; Immunotech, Westbrook, ME). Cells were stained in the presence of phosphate-buffered saline with 2% mouse serum. Cells were harvested from in vitro T-cell cultures and incubated with antibodies to CD3, CD4, and CD8. After staining, cells were sorted into CD3+CD4+CD8− and CD3+CD4−CD8+ subsets using a Becton Dickinson FACS Vantage. Fluorescence was excited using an argon Coherent Enterprise laser (Mountain View, CA) generating 90 mW of 488 nm light. Fluorescein isothiocyanate emission was detected through a 530/30-nm band pass filter and phycoerythrin emission was detected through a 575/26-nm band pass filter. Fluorescence was directed to the appropriate detectors using a 560-nm short pass dichroic.
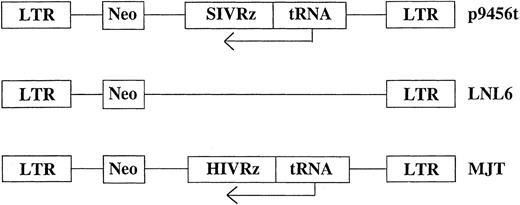
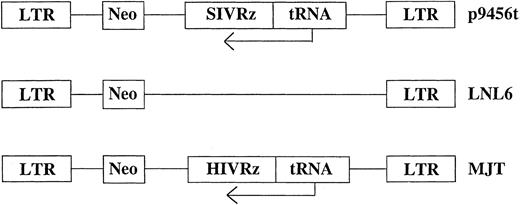
Retroviral vectors and transduction. Retroviral vectors used in these studies are diagrammed in Fig 1. The vector p9456t encodes an SIV-specific hairpin ribozyme 945622 expressed under the control of a tRNAVal promoter in the retroviral vector LNL6.27 Two control retroviral vectors were used: MJT, which encodes an HIV-1–specific ribozyme that does not recognize SIV,15,22 and the unmodified vector LNL6. All retroviral vectors were produced in the murine amphotropic packaging cell line PA317.28 Retroviral packaging cell lines were grown in an artificial capillary system (ACS; Cellco, Germantown, MD). All transductions were performed using frozen supernatants with titers ranging from 1.0 to 1.6 × 106 colony-forming units (CFU)/mL as determined by end point dilution on 3T3 cells.29 No replication competent retrovirus was detected in retroviral supernatants as assessed using the S+L− assay employing PG4 cells.30 31 Rhesus CD34+ bone marrow cells were transduced for 3 days with cell-free retroviral vector supernatants. Transductions were performed in the presence of irradiated allogeneic bone marrow stroma. Rhesus bone marrow stromal cultures were established in long-term culture media (Stem Cell Technologies, Vancouver, British Columbia, Canada) plus 10−6 mol/L hydrocortisone (Sigma, St Louis, MO) and γ irradiated (15 Gy) after 7 to 14 days of culture before replating. Nonadherent cells were removed after 3 days of transduction, counted, and used to establish in vitro T lymphopoiesis or macrophage cultures as described below.
Schematic representation of the retroviral vectors used for transduction of rhesus CD34+ cells. Ribozymes were cloned into the retroviral vector LNL627 as previously described under the control of the tRNAVal promoter.15 neo, neomycin phosphotransferase gene.
T-cell differentiation of CD34+ bone marrow cells. To support differentiation into T cells, transduced CD34+ cells were cultured on a thymic stromal monolayer as previously described.25 Briefly, thymic stromal monolayers were established in 24-well plates from cryopreserved fetal rhesus thymus cell suspensions and nonadherent cells were vigorously washed away after 3 days. After 7 days, thymic stromal monolayers were inoculated with 2 × 105 CD34+ cells/well and cultured in the presence of RPMI with 10% fetal calf serum (FCS). After 14 days, samples were removed for flow cytometry and polymerase chain reaction (PCR) analysis, and the remaining cells were expanded by stimulation with Concanavalin A (ConA; 5 μg/mL) in the presence of recombinant interleukin-2 (rIL-2; 20 U/mL; provided by Dr M. Gately, Hoffman-La Roche, Nutley, NJ) and irradiated human peripheral blood mononuclear cells (PBMC). Aliquots of T cells were analyzed by three-color flow cytometry and CD3+CD4+CD8− cells were obtained by cell sorting as described above. Purified CD4+ T cells were then restimulated with ConA, IL-2, and irradiated PBMC and selected in G418 (200 μg/mL active concentration; GIBCO BRL, Gaithersburg, MD) for 7 days before restimulation and SIV infection.
Colony-forming assays and differentiation of macrophage-like cells from CD34+ cells. CFU assays were established by culturing CD34+ cells at a concentration of 1 × 104 cells/mL in complete methylcellulose media (Stem Cell Technologies) containing human erythropoietin (10 ng/mL; Stem Cell Technologies), granulocyte-macrophage colony-stimulating factor (GM-CSF; 30 ng/mL; Genzyme, Cambridge, MA), human IL-3 (20 ng/mL; Genzyme), and human stem cell factor (SCF; 50 ng/mL; Genzyme) in the presence and absence of G418 (400 μg/mL active concentration; GIBCO BRL). Hematopoietic colonies were scored at 14 and 21 days based on standard criteria.32 Transduction efficiency was calculated by dividing the number of G418-resistant CFU by the total number of CFU for each culture condition examined. Previous experiments have shown that greater than 95% of G418-resistant CFU obtained under these conditions contain neo vector DNA.33 Individual myelomonocytic colonies were pooled, counted, and cultured in 24-well plates in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and supplemented with GM-CSF (20 ng/mL) and macrophage colony-stimulating factor (M-CSF; 40 U/mL; Genzyme) to induce differentiation of macrophage-like cells. After 7 days of selection, the resulting cells were infected with SIV as described below.
Proliferation assays. T cells derived from CD34+ cells were washed and resuspended in RPMI with 10% FCS at a concentration of 106 cells/mL. One hundred microliters (105 cells) were added to each well of a 96-well plate. Cells were stimulated with either ConA (5 μg/mL) or monoclonal antibodies to CD3 (6G12) and CD28 (CD28.2; Immunotech, Larrabee, ME) (0.15 μg/mL) in the presence of IL-2 (20 U/mL) and irradiated human PBMC (105 cells/well in 100 μL of RPMI with 10% FCS). Purified goat antimouse F(ab)′2 fragments (Kirkegard and Perry Laboratories, Gaithersburg, MD) were used as a cross-linking agent for the stimulation with the monoclonal antibodies to CD3 and CD28. Wells were pretreated with 1.25 μg/mL of goat antimouse antibody for 45 minutes at 37°C and washed three times before the addition of monoclonal antibodies to CD3 and CD28. After 7 days in culture at 37°C, each well was pulsed with 1 μCi of methyl-3H-thymidine (DuPont, NEN, Boston, MA) for 16 hours and harvested onto a glass fiber filter (Harvester 96; Tomtec, Orange, CT) and the incorporation of 3H-thymidine was determined using a liquid scintillation counter (Micro Beta Plus; Wallac, Gaithersburg, MD). Five replicate wells were assayed for each condition.
PCR analysis of retroviral genes in T cells. Cellular DNA was extracted using the Puregene DNA isolation kit (Gentra Systems, Inc, Minneapolis, MN). DNA template (100 ng, corresponding to approximately 104 cell equivalents) was added to each PCR reaction (50 μL) containing 50 ng each of the sense and antisense PCR primer, 1.0 mmol/L MgCl2 , 200 μmol/L of each dNTP, and 2.5 U of AmpliTaq (Perkin Elmer, Norwalk, CT). The neo primers34 (5′ primer, listed 5′ to 3′) TCC ATC ATG GCT GAT GCA ATG CGG C and (3′ primer) GAT AGA AGG CGA TGC GCT GCG AAT CG yield a 416-bp product; the 5′ PCR primer was end-labeled with 33P-dATP (DuPont, NEN). Amplification was performed at 92°C for 2 minutes, followed by 35 cycles of 92°C for 30 seconds, 65°C for 30 seconds, and 72°C for 1 minute and a final extension at 72°C for 10 minutes using a PTC 200 DNA Engine (M.J. Research, Watertown, MA). To facilitate semiquantitative analysis of samples, each PCR amplification included amplification of a series of standards consisting of 10-fold dilutions (100%, 10%, 1%, and 0.1%) of the CEMx174 cell line stably transduced with the LN retroviral vector diluted in genomic rhesus DNA. PCR products were separated on a 4% native polyacrylamide gel and then visualized by autoradiography using Biomax MR Film (Eastman Kodak, Rochester, NY).
SIV infection of T cells and macrophage-like cells. Rhesus CD4+ T-cell progeny and macrophage-like cells derived from CD34+ cells transduced with the ribozyme or the control vectors were infected with SIVmac239 (T cells) or SIVmac316 (macrophage-like cells) for 4 hours at 37°C. SIVmac239 is a pathogenic molecular clone of SIV that replicates efficiently in T cells.35 SIVmac316 is derived from SIVmac239, differing in 8 amino acids in the envelope sequence, and replicates well in macrophages.36 T cells were challenged with SIVmac239 at multiplicities of infection (MOI) ranging from 0.001 to 1.0 TCID50 /cell; macrophage-like cells were challenged with SIVmac316 at MOI of 0.001 to 0.01 TCID50 /cell. The titer of virus stocks was performed by endpoint dilution on CEMx174 cells performed in 6 replicate wells as previously described for HIV-1.37 Free virus was removed by vigorously washing three times with phosphate-buffered saline. Infections were performed in duplicate and the results were presented as the mean of each well. Viral replication was assessed by enzyme-linked immunosorbent assay determination of SIV p27 antigen (Coulter, Hialeah, FL) according to the manufacturer's protocol. The lower limit detection for SIV p27 is 50 pg/mL.
RESULTS
Transduction of rhesus CD34+ progenitor cells with retroviral vectors encoding the SIV-specific ribozyme 9456. We first evaluated retroviral transduction efficiency of CD34+ progenitor cells as assessed by outgrowth of CFU resistant to the neomycin analog G418. G418-resistant erythroid and myelomonocytic CFU were assessed after transduction with a retroviral vector encoding an SIV-specific ribozyme (p9456t) and the neomycin phosphotransferase gene neo or the LNL6 vector encoding neo alone. Based on the observation that stimulation with exogenous cytokines inhibits the ability of CD34+ cells to undergo T-cell differentiation (Rosenzweig et al, manuscript in preparation), CD34+ cells were transduced by three successive incubations with cell-free retroviral supernatant in the presence of bone marrow stroma and the absence of exogenous cytokines. Using this approach, transduction efficiencies of 21% to 35% were obtained using the p9456t vector and 30% to 45% using the control LNL6 vector (Table 1). Similar numbers of myelomonocytic (colony-forming unit–granulocyte-macrophage [CFU-GM]) and erythroid (burst-forming unit-erythroid [BFU-E]) colonies were obtained after transduction with the p9456t and LNL6 vectors, suggesting that 9456 ribozyme did not interfere with differentiation of myelomonocytic or erythroid lineages.
To assess transduction of more primitive hematopoietic cells, we also analyzed the yield of G418-resistant long-term culture-initiating cells (LTC-IC) obtained from transduced CD34+ cells. Rhesus CD34+ progenitor cells were transduced as described above, cultured on irradiated bone marrow stroma for 5 weeks, and then replated in methylcellulose in the presence and absence of G418. After transduction with the p9456t retroviral vector, we observed between 30% and 56% transduction of LTC-IC, and a similar transduction efficiency with the LNL6 vector (Table 2).
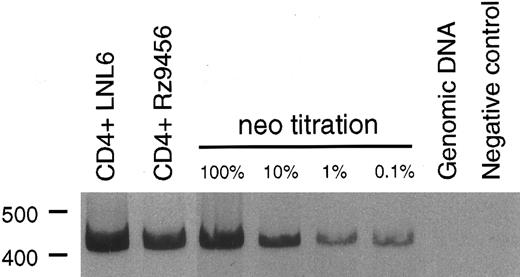
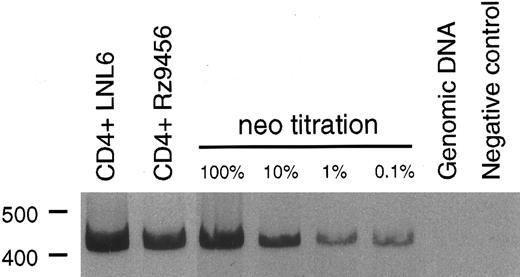
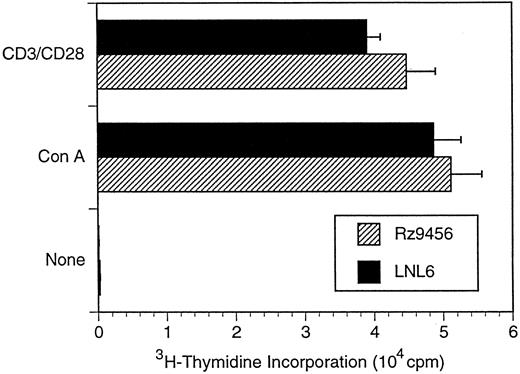
T-cell differentiation of CD34+ hematopoietic cells transduced with the 9456 ribozyme. Analysis of the ability of genes designed to inhibit HIV or SIV replication in T cells derived from transduced CD34+ cells has been limited by the difficulty in supporting T-cell differentiation in vitro. We used cultures of rhesus thymic stroma to support T-cell differentiation of rhesus CD34+ progenitor cells transduced with retroviral vectors. T-cell differentiation occurring in this system phenotypically mirrors that of normal T-cell ontogeny and results in a polyclonal population of CD4+ and CD8+ T cells.25 CD34+ cells were transduced as described above with three successive incubations with retroviral vector and then cultured on rhesus thymic stroma. T cells derived from CD34+ cells transduced with either the p9456t or LNL6 retroviral vectors were phenotypically similar to untransduced cells, with comparable levels of CD4+ and CD8+ cells (Table 3). After 14 to 21 days of differentiation on thymic monolayers, T cells were expanded by stimulation with the lectin ConA, rIL-2, and irradiated human PBMC. Purified CD4+ T cells were then obtained by cell sorting, reexpanded with ConA, and then incubated with the neomycin analog G418 to select for transduced cells. PCR analysis using primers specific for the neo gene showed that a high percentage of G418-selected T cells derived from p9456t- and LNL6-transduced cells contained retroviral vector DNA (Fig 2). Flow cytometric analysis of CD4+ T cells derived from transduced progenitor cells showed persistent levels of CD4 expression after G418 selection (Table 3). In addition, T cells containing the 9456 ribozyme had proliferative responses to the mitogen ConA or to stimulation with CD3 and CD28 antibodies similar to that of control T cells from LNL6-transduced cells (Fig 3).
PCR analysis of vector DNA in T cells derived from transduced CD34+ progenitor cells. Rhesus CD34+ cells were transduced with either the LNL6 or p9456t retroviral vectors and then cultured on thymic stroma to support T-cell differentiation. T cells derived from transduced progenitor cells were expanded in the presence of G418 and then 100 ng of DNA was analyzed for the presence of vector DNA by PCR using primers specific for the neo gene. The neo titration samples consisted of the indicated percentage of CEMx174 cells stably transduced with the retroviral vector LN diluted in rhesus genomic DNA.
PCR analysis of vector DNA in T cells derived from transduced CD34+ progenitor cells. Rhesus CD34+ cells were transduced with either the LNL6 or p9456t retroviral vectors and then cultured on thymic stroma to support T-cell differentiation. T cells derived from transduced progenitor cells were expanded in the presence of G418 and then 100 ng of DNA was analyzed for the presence of vector DNA by PCR using primers specific for the neo gene. The neo titration samples consisted of the indicated percentage of CEMx174 cells stably transduced with the retroviral vector LN diluted in rhesus genomic DNA.
Proliferative responses of rhesus CD4+ T cells derived from transduced CD34+ cells. CD4+ T cells were derived from transduced or untransduced CD34+ cells as described in the legend to Fig 2. T cells were stimulated with lectin (5 μg/mL ConA) and irradiated PBMC in the presence of IL-2 (20 U/mL). Data are plotted as the mean of five replicate wells ± standard deviation and are representative of two independent experiments.
Proliferative responses of rhesus CD4+ T cells derived from transduced CD34+ cells. CD4+ T cells were derived from transduced or untransduced CD34+ cells as described in the legend to Fig 2. T cells were stimulated with lectin (5 μg/mL ConA) and irradiated PBMC in the presence of IL-2 (20 U/mL). Data are plotted as the mean of five replicate wells ± standard deviation and are representative of two independent experiments.
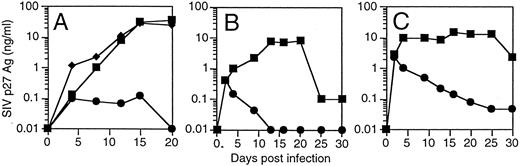
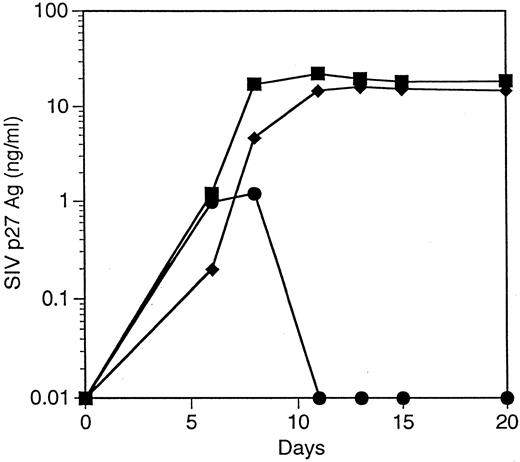
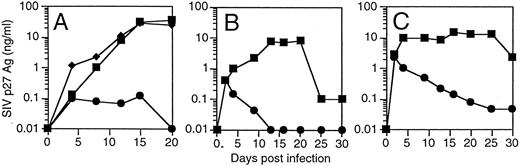
T cells derived from CD34+ cells transduced with an SIV-specific ribozyme are protected from SIV infection. T cells derived from transduced CD34+ cells were restimulated and then challenged with a SIVmac239, a strain of SIV that replicates well in T cells.35 T cells containing the SIV-specific ribozyme 9456 were highly resistant to SIV infection after an MOI of 0.01 TCID50 /cell, with only small amounts of detectable SIV replication not exceeding 100 pg/mL (Fig 4A). In contrast, T cells containing the control vector LNL6 or the HIV-1–specific ribozyme MJT,15 which does not recognize SIV, were efficiently infected with SIV, with peak viral replication exceeding 30 ng/mL. Similar levels of viral inhibition were observed in three independent experiments (data not shown). T cells containing the 9456 ribozyme maintained viability after SIV infection (viability, 85% ± 6%; n = 3), whereas viability in LN or MJT control T cells was less than 30%.
Inhibition of SIV replication in CD4+ T cells derived from CD34+ cells transduced with the 9456 ribozyme. CD34+ cells were transduced with a retroviral vector containing either the 9456 ribozyme (•) or, as controls, the LNL6 vector (▪) or the HIV-1–specific ribozyme MJT (♦), which does not recognize SIV. Transduced CD34+ cells were then cultured on thymic stroma to support T-cell differentiation, expanded, selected in G418, and then infected with SIVmac239 at an MOI of 0.01 TCID50 (A), 0.1 TCID50 (B), or 1 TCID50 /cell (C).
Inhibition of SIV replication in CD4+ T cells derived from CD34+ cells transduced with the 9456 ribozyme. CD34+ cells were transduced with a retroviral vector containing either the 9456 ribozyme (•) or, as controls, the LNL6 vector (▪) or the HIV-1–specific ribozyme MJT (♦), which does not recognize SIV. Transduced CD34+ cells were then cultured on thymic stroma to support T-cell differentiation, expanded, selected in G418, and then infected with SIVmac239 at an MOI of 0.01 TCID50 (A), 0.1 TCID50 (B), or 1 TCID50 /cell (C).
The ability of candidate therapeutic genes to inhibit HIV or SIV replication has frequently been attenuated at higher multiplicities of infection. We wished to examine the ability of T cells containing the 9456 ribozyme to resist infection after a relatively high MOI. We therefore infected T cells containing either the 9456 ribozyme or LN control with an MOI of 0.1 and 1 TCID50 /cell. Although a dose-dependent increase in viral replication was observed in the p9456t-containing T cells, viral replication progressively decreased below the level of detection of the SIV p27 antigen assay (Fig 4B and C). As observed at lower MOIs, T cells containing the 9456 ribozyme maintained viability (data not shown). In contrast, LNL6-transduced cells supported relatively high levels of SIV replication. Thus, transduction of CD34+ progenitor cells with the SIV-specific ribozyme 9456 results in T cells that are highly resistant to SIV infection, even after a relatively high MOI.
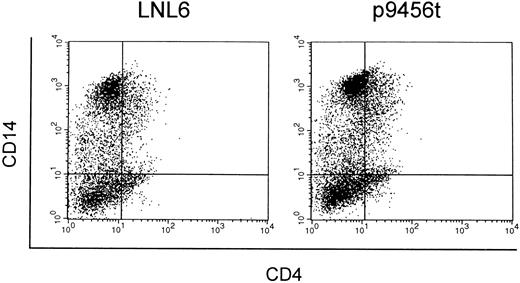
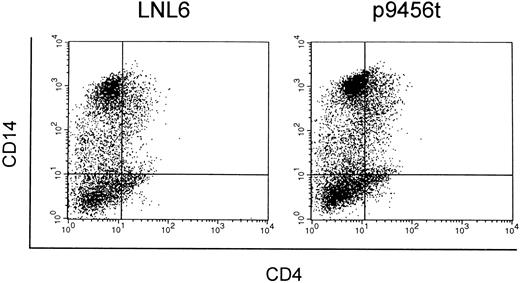
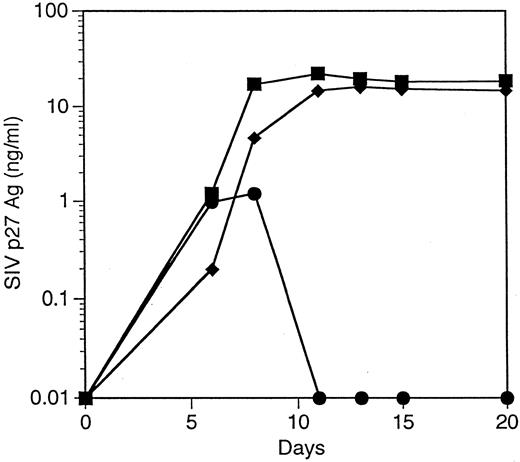
Macrophage-like cells derived from CD34+ cells transduced with the 9456 ribozyme are protected from SIV infection. We next evaluated the ability of the 9456 ribozyme to inhibit SIV replication in macrophage-like cells derived from transduced CD34+ cells. Rhesus CD34+ cells were transduced as described above and then cultured in methylcellulose in the presence of IL-3, SCF, GM-CSF, and G418. Individual colonies were then picked, pooled, and cultured in GM-CSF, M-CSF, and G418. Macrophage-like cells derived from CD34+ cells transduced with the 9456 ribozyme expressed similar levels of CD4 and CD14 as cells derived from LNL6-transduced cells (Fig 5). Cells obtained in this fashion were then infected with SIVmac316, a strain of SIV known to replicate in macrophages.36 Macrophage-like cells containing the 9456 ribozyme were highly resistant to infection with SIVmac316, whereas control macrophage-like cells supported efficient replication of SIV (Fig 6), with comparable levels of inhibition observed in two independent experiments. Thus, the 9456 ribozyme is able to inhibit SIV replication in both macrophage-like cells and T cells derived from transduced CD34+ progenitor cells.
Expression of CD4 and CD14 on macrophage-like cells derived from transduced CD34+ cells. CD34+ cells transduced with either the LNL6 (A) or ribozyme 9456 (B) retroviral vectors were cultured in methylcellulose in the presence of G418 and then replated in liquid culture supplemented with GM-CSF, M-CSF, and G418. Macrophage-like cells derived from these cultures were then analyzed for expression of CD4 and CD14 by flow cytometry.
Expression of CD4 and CD14 on macrophage-like cells derived from transduced CD34+ cells. CD34+ cells transduced with either the LNL6 (A) or ribozyme 9456 (B) retroviral vectors were cultured in methylcellulose in the presence of G418 and then replated in liquid culture supplemented with GM-CSF, M-CSF, and G418. Macrophage-like cells derived from these cultures were then analyzed for expression of CD4 and CD14 by flow cytometry.
Inhibition of SIV replication in macrophage-like cells derived from CD34+ cells transduced with the 9456 ribozyme. CD34+ cells were transduced with a retroviral vector containing either the 9456 ribozyme (•) or, as controls, the LNL6 vector (▪) or the HIV-1–specific ribozyme MJT (♦). After transduction, the macrophage-like cells were derived as described in the legend for Fig 5 and challenged with SIVmac316 using an MOI of 10−2 TCID50 /cell.
Inhibition of SIV replication in macrophage-like cells derived from CD34+ cells transduced with the 9456 ribozyme. CD34+ cells were transduced with a retroviral vector containing either the 9456 ribozyme (•) or, as controls, the LNL6 vector (▪) or the HIV-1–specific ribozyme MJT (♦). After transduction, the macrophage-like cells were derived as described in the legend for Fig 5 and challenged with SIVmac316 using an MOI of 10−2 TCID50 /cell.
DISCUSSION
It has been almost 10 years since the strategy of intracellular immunization of CD34+ progenitor cells against AIDS virus infection was first proposed.38 Since that time, despite multiple reports demonstrating the ability of genes to inhibit HIV or SIV replication in transformed cell lines,39 40 there have been relatively few publications that have analyzed inhibition of replication in the progeny of transduced hematopoietic progenitor cells and most of these have analyzed myelomonocytic rather than T cells. In this study, we examined the ability of an SIV-specific hairpin ribozyme to inhibit viral replication in hematopoietic cells derived from transduced CD34+ progenitor cells. Our data show that the 9456 ribozyme is quite effective in inhibiting SIV replication in both T cells and macrophage-like cells derived from transduced progenitor cells. We observed a greater than 500-fold reduction in viral replication as compared with controls, even after a relatively high MOI of 1 TCID50 /cell. In addition, we observed no evident toxic effects on T-cell differentiation, cellular proliferation, or expression of CD4.
Our results add to the expanding body of evidence showing the efficacy of ribozymes for gene therapy and also emphasize their potential value for stem cell gene therapy. The hairpin ribozyme MJT, which cleaves a conserved sequence in the U5 region of the HIV-1 LTR,41 is able to inhibit replication of HIV-1 in a variety of cell types, including transformed cell lines,42 primary human T cells,43 and macrophage-like cells derived from transduced CD34+ bone marrow cells.8,14,15,43 Efficient expression of ribozymes by RNA polymerase III promoters, which are active in diverse cell types, and the absence of demonstrable toxicity of ribozymes represent distinct advantages of these molecules for stem cell gene therapy, where it is desirable to achieve sustained expression in multiple different hematopoietic lineages without adverse effects. Our results with the SIV-specific 9456 ribozyme suggest that comparable hairpin ribozymes specific for HIV-18,15 may have a similar capacity to inhibit viral replication in T cells derived from transduced progenitor cells. Possible disadvantages of ribozymes include the potential for the emergence of virus variants with mutations in the target sequence recognized by the ribozyme, a particular concern given the relatively high mutation rate of HIV and the relatively rapid rates of virion turnover in vivo.44 However, the use of ribozymes in combination with other ribozymes or other gene therapy approaches45 and the choice of target sequences in highly conserved regions of the viral genome should limit the emergence of resistance.
Several considerations suggest that evaluation of genes designed to inhibit HIV replication in T cells derived from transduced progenitor cells may offer significant information not available from the analysis of myelomonocytic progeny. T cells are the dominant population of HIV-infected cells in vivo46 and are estimated to account for greater than 99% of viral production on a daily basis.47 Differentiation of T cells from progenitor cells differs considerably from that of myelomonocytic cells. T-cell differentiation involves a complex series of developmental stages resulting in the rearrangement of T-cell receptor genes and the production of a diverse population of T cells able to recognize foreign antigenic peptides presented by self major histocompatibility complex molecules.20 A large number of genes are expressed during the different stages of T-cell development and interference with these genes may adversely influence T-cell production.48 The potential for toxicity of foreign genes expressed in hematopoietic cells derived from transduced CD34+ progenitor cells may therefore be greater in T cells and their progenitors than in macrophages. In addition, expression of genes in T cells may differ from that in macrophages. Our current data show that introduction of the 9456 ribozyme into CD34+ progenitor cells does not appear to interfere with T-cell differentiation and that the 9456 ribozyme is expressed in mature CD4+ T cells at sufficient levels to inhibit SIV replication. We have also recently shown that a polymeric transactivation response (TAR) element decoy linked to an antisense tat molecule expressed under the control of a modified HIV-1 LTR is able to inhibit both HIV-1 and SIV replication in T cells derived from transduced CD34+ progenitor cells.33 Similar results have been subsequently reported using a retroviral vector encoding a transdominant mutant HIV-1 RevM10 protein.49 Taken together, these results demonstrate that candidate therapeutic genes introduced into CD34+ hematopoietic progenitor cells can retain the ability to inhibit AIDS virus replication after T-cell differentiation.
Definition of the optimal conditions for retroviral transduction that maintain the pluripotency of hematopoietic stem cells has been difficult to achieve. Because retroviral vectors based on Moloney murine leukemia virus require cell cycling for integration of vector DNA,50 most investigators have used stimulation with hematopoietic growth factors to enhance retroviral transduction of hematopoietic progenitor cells.34,51-53 Although cytokine stimulation can significantly enhance retroviral transduction, it may also lead to a loss of pluripotency. Stimulation of murine hematopoietic cells with cytokines significantly decreases their ability to reconstitute irradiated hosts54,55 and to differentiate into B cells.56 We have observed that the cytokines IL-3 and SCF, which are commonly used to enhance retroviral transduction,34,52 significantly inhibit the T-cell differentiation capacity of CD34+ progenitor cells (Rosenzweig et al, manuscript in preparation). Because bone marrow stroma has previously been shown to enhance retroviral transduction efficiency57 and to maintain the long-term engraftment of transduced CD34+ human cells in immunodeficient mice,58 in these studies we performed retroviral transduction in the presence of bone marrow stroma and the absence of exogenous cytokines. Using these conditions and relatively high titer retroviral supernatant, we were able to obtain efficient transduction of CFU and LTC-IC and maintain T-cell differentiation capacity of CD34+ cells. Even in the absence of transduction of hematopoietic stem cells, transduction of T-cell progenitors may well lead to in vivo differentiation of genetically modified T cells that may persist for several years. Thus, retroviral transduction conditions of hematopoietic stem and progenitor cells for use in human gene therapy protocols for HIV disease should preferably be designed so as to preserve T-cell differentiation capacity. In vivo studies in macaques should permit examination of the efficiency of retroviral transduction of true hematopoietic stem cells under the conditions used in these experiments.
Despite the many potential advantages of stem cell gene therapy for AIDS, there are many obstacles to overcome. The levels of genetically modified hematopoietic cells in most human and nonhuman primate trials of transduced hematopoietic stem cells have been disappointingly low, generally less than 1%.5-7,59,60 This inefficient level of genetic modification is likely to reflect the difficulties in transducing hematopoietic stem cells, which are largely quiescent,61,62 with the current generation of retroviral vectors that are dependent on cell cycling for efficient integration. Additional obstacles to stem cell gene therapy for AIDS are unique to HIV infection. Although CD34+ bone marrow cells appear to be infected only at a very low level (if at all) in vivo63,64 and CD34+ cells from HIV infected subjects can be efficiently transduced with retroviral vectors,17 there are multiple reports of abnormal hematopoiesis in HIV-infected people (reviewed in Harbol et al65 ). Thus, abnormalities in the bone marrow microenvironment of HIV-infected people might interfere with differentiation of genetically modified hematopoietic stem cells. An additional potential barrier to stem cell gene therapy for AIDS is the loss of thymic function, either due to age66 or the effects of HIV infection.67 A final concern is that reservoirs of HIV in relatively long-lived hematopoietic cells (eg, macrophages, dendritic cells, or microglia) or nonhematopoietic cells resident in these microenvironments (eg, thymic epithelial cells68 69 ) may interfere with the ability of genetically modified cells to differentiate or restore immune function.
In light of these challenges, trials in nonhuman primates are likely to play an important role in the development of stem cell gene therapy for AIDS. Carefully designed trials in the SIV/macaque model can address the issues of optimization of transduction of hematopoietic stem cells, engraftment and differentiation of genetically modified cells in an SIV-infected host, the potential for evolution of viruses resistant to a specific therapeutic gene, and the relative efficacy and toxicity of different approaches.
In sum, our results demonstrate potent inhibition of SIV replication in both T cells and macrophages derived from CD34+ progenitor cells transduced with a hairpin ribozyme. These findings provide strong evidence that intracellular immunization of hematopoietic stem cells with a ribozyme can protect progeny from AIDS virus infection and will permit analysis of stem cell gene therapy approaches for AIDS in nonhuman primates.
ACKNOWLEDGMENT
We thank Ron Desrosiers for providing SIVmac239 and SIVmac316, Maurice Gately for recombinant IL-2, Johnson Wong for the rhesus CD3-specific monoclonal antibodies, and Carolyn A. O'Toole for manuscript preparation.
Supported by Public Health Service Grants No. RR-00168, AI-36550, and DK49618.
Address reprint requests to R. Paul Johnson, MD, New England Regional Primate Research Center, Harvard Medical School, PO Box 9102, One Pine Hill Dr, Southborough, MA 01772.