Abstract
We analyzed a genetic polymorphism of Fcγ receptor IIIa (CD16) that is present on position 158 (Phe or Val) in the membrane-proximal, IgG-binding domain. With a polymerase chain reaction–based allele-specific restriction analysis assay we genotyped 87 donors and found gene frequencies of 0.57 and 0.43 for FcγRIIIA-158F and −158V, respectively. A clear linkage was observed between the FcγRIIIA-158F and −48L genotypes on the one hand and the FcγRIIIA-158V and −48H or −48R genotypes on the other hand (χ2 test; P < .001). To determine the functional consequences of this FcγRIIIa-158V/F polymorphism, we performed IgG binding experiments with natural killer (NK) cells from genotyped donors. All donors were also typed for the recently described triallelic FcγRIIIa-48L/R/H polymorphism. NK cells were treated with lactic acid to remove cell-associated IgG. FcγRIIIaNK158F bound significantly less IgG1, IgG3, and IgG4 than did FcγRIIIaNK-158V, irrespective of the FcγRIIIa-48 phenotype. Moreover, freshly isolated NK cells from FcγRIIIa-158VV individuals carried significantly more cytophilic IgG than did NK cells from FcγRIIIa-158FF individuals. In addition, CD16 monoclonal antibody (MoAb) MEM154 bound more strongly to FcγRIIIa-158V, compared with -158F, again independently of the FcγRIIIa-48 phenotype. The binding of MoAb B73.1 was not influenced by the FcγRIIIa-158V/F polymorphism, but proved to depend solely on the amino acid present at position 48 of FcγRIIIa. In conclusion, the previously reported differences in IgG binding among the three FcγRIIIa-48L/R/H isoforms are a consequence of the linked, biallelic FcγRIIIa-158V/F polymorphism at amino-acid position 158.
SEVERAL IgG Fc receptor (FcγR) polymorphisms that influence the binding of IgG have been described.1 On neutrophils, the FcγRIIIb-NA1 and -NA2 isoforms, which differ by four amino acids in the membrane-distal Ig-like loop of the receptor, interact differently with IgG-opsonized particles,2-4 and influence the interaction of the receptor with FcγRIIa.5 On FcγRIIa, the extensively investigated high responder/low responder polymorphism on amino-acid position 131 in the membrane-proximal, IgG-binding domain is critical for the interaction with human IgG2.6 Moreover, the FcγRIIa-131R/H polymorphism was found to be associated with several diseases, such as bacterial infections in children, heparin-induced thrombocytopenia, juvenile periodontitis, and systemic lupus erythematosus.7-12
Previously, Ravetch and Perussia described a polymorphism in the membrane-proximal domain of FcγRIIIa.13 A nucleotide substitution at position 559 of FcγRIIIA predicts either a valine or a phenylalanine at amino-acid position 158 of FcγRIIIa. Because the IgG binding site is most probably located in this part of FcγRIIIa,14 15 we determined the gene frequency and functional consequences of this polymorphism in the context of the recently described FcγRIIIa-48L/R/H polymorphism. As expected, a clear although incomplete linkage was observed between the two polymorphisms. Moreover, we found that the previously described differences in binding between the FcγRIIIa-48 isoforms of IgG and of some of the CD16 monoclonal antibodies (MoAbs) are attributable to the FcγRIIIa-158 phenotype.
MATERIALS AND METHODS
MoAbs.Anti-pan FcγRIII (CD16) MoAbs used were CLBFcRgran1 (mIgG2a) and MEM154 (mIgG1). B73.1 (mIgG1) reacts with NA1-FcγRIIIb and with FcγRIIIa,16 and was kindly provided by Dr B. Perussia (Thomas Jefferson University, Philadelphia, PA). MEM154 was obtained through the 5th Leukocyte Typing Workshop. Phycoerythrin (PE)-labeled Leu19 (CD56; mIgG1) was purchased from Becton Dickinson (San Jose, CA). Fluorescein isothiocyanate (FITC)-conjugated goat–antimouse Ig and irrelevant control MoAbs of the IgG1 and IgG2a subclasses were from the Central Laboratory of the Netherlands Red Cross Blood Transfusion Service (CLB), Amsterdam, The Netherlands.
Isolation of cells.Fresh, EDTA-anticoagulated blood from healthy volunteers was diluted with two volumes of phosphate-buffered saline (PBS) and was centrifuged over a Ficoll gradient (Pharmacia Fine Chemicals AB, Uppsala, Sweden) with a specific gravity of 1.076 g/mL. Peripheral blood mononuclear cells (PBMC) were obtained from the interphase and were washed twice with PBS, containing 0.2% (wt/vol) bovine serum albumin (BSA).
Flow cytometry.PBMC were incubated with CD16 MoAbs for 25 minutes at room temperature. The cells were washed with PBS/BSA, and were incubated with FITC-labeled goat–antimouse-Ig for 25 minutes at room temperature. Free F(ab′ )2 regions of the conjugate were blocked with a mixture of irrelevant mIgG1 and mIgG2a. Thereafter, the cells were incubated with PE-labeled CD56. Only CD56+ lymphocytes were analyzed in a FACScan flowcytometer (Becton Dickinson).
FcγRIIIA-48L/R/H genotyping.Genotyping for the FcγRIIIA-48L/R/H polymorphism was performed as previously described,17 with a polymerase chain reaction (PCR)-based allele-specific restriction analysis assay. Briefly, a 91-bp FcγRIIIA-specific fragment containing the polymorphic site was amplified from genomic DNA and digested with Mnl I. Digested fragments were electrophoresed in 10% acryl amide gels, stained with ethidium bromide, and visualized with UV light. Homozygous FcγRIIIA-48LL individuals showed 40-bp, 34-bp, and 17-bp bands, whereas PCR fragments of individuals carrying no or only one FcγRIIIA-48L allele showed a 51-bp band. The genotype of these latter individuals was determined by direct sequencing of the amplified fragments, using one of the PCR primers, end-labeled with 32P (Amersham International, Buckinghamshire, UK), with the Life Technologies cycle sequencing kit, following the instructions of the manufacturer (Life Technologies, Gaithersburg, MD).
FcγRIIIA-158V/F genotyping.Genotyping of the FcγRIIIA-158V/F polymorphism was performed by means of a (nested) PCR-based allele-specific restriction analysis assay. Two FcγRIIIA gene-specific primers (sense A013: 5′-ATA TTT ACA GAA TGG CAC AGG-3′; antisense A012: 5′-GAC TTG GTA CCC AGG TTG AA-3′; italic characters denote mismatches that were introduced to increase specificity) were used to amplify a 1.2-kb fragment containing the polymorphic site. This PCR assay was performed with 5 ng of genomic DNA, 150 ng of each primer, 200 μmol/L of each dNTP, and 2 U of Taq DNA polymerase (Promega, Madison, WI), diluted in a buffer recommended by the manufacturer in a total volume of 50 μL in a Perkin Elmer Cetus cycler (Norwalk, CT). The first PCR cycle consisted of 10 minutes denaturation at 95°C, 11/2 minute primer annealing at 56°C, and 11/2 minute extension at 72°C. This was followed by 35 cycles in which the denaturing time was decreased to 1 minute. The last cycle was followed by 8 minutes at 72°C to complete extension. The sense primer in the second PCR contained a mismatch that created an NlaIII restriction site only in FcγRIIIA-158V-encoding DNA (A014: 5′-atc aga ttc gAT CCT ACT TCT GCA GGG GGC AT-3′; uppercase characters denote annealing nucleotides, lowercase characters denote nonannealing nucleotides), the antisense primer was chosen just 5′ of the fourth intron (A016: 5′-acg tgc tga gCT TGA GTG ATG GTG ATG TTC AC-3′ ). This “nested” PCR was performed with 1 μL of the amplified fragment, 150 ng of each primer, 200 μmol/L of each dNTP, and 2 U of Taq DNA polymerase, diluted in the recommended buffer. The first cycle consisted of 5 minutes' denaturing at 95°C, 1 minute primer annealing at 64°C, and 1 minute extension at 72°C. This was followed by 35 cycles in which the denaturing time was 1 minute. The last cycle was followed by 91/2 minutes at 72°C to complete extension. The 94-bp fragment was digested with NlaIII, and digested fragments were electrophoresed in 10% polyacrylamide gels, stained with ethidium bromide, and visualized with UV light. Cycle sequencing of first-round fragments to check for specificity was performed with 32P end-labeled primer A016 with the Life Technologies cycle sequencing kit. Genomic DNA of individuals whose FcγRIIIa encoding cDNA sequence was known were used to optimize the PCR assay.
IgG-binding experiments.Quantification of IgG binding to natural killer (NK) cells of genotyped donors was performed as previously described.17 In short, mononuclear cells were pretreated with 0.1% (wt/vol) lactic acid (pH 3.9) to remove NK-cell–associated IgG. This treatment did not alter FcγRIIIa expression as measured with CD16 MoAb CLBFcRgran1 (not shown). The cells were incubated with saturating amounts of human IgG subclasses purified from sera of patients suffering from multiple myeloma. The antibodies were at least partially aggregated, because binding to PMN was observed with the IgG1 and IgG3 preparations (not shown). After washing, bound IgG was detected by FITC-labeled F(ab′ )2 fragments of goat–antihuman-IgG (Kallestad, South Austin, TX), and NK cells were identified with PE-labeled CD56. Only CD56+ lymphocytes were analyzed in a FACScan flowcytometer.
Statistical analysis.The data were analyzed with the Student's t-test and the χ2-test. Data with different standard deviations (SDs) were compared with the Welch's approximate t-test. P values below .05 were considered significant.
RESULTS
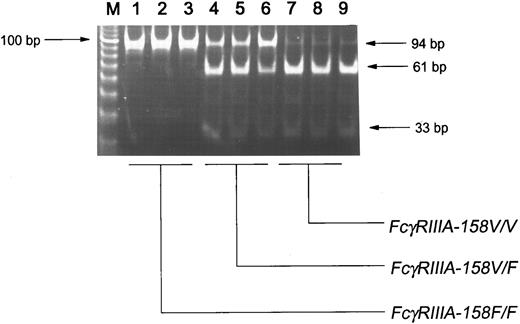
FcγRIIIA-158V/F genotyping.An FcγRIIIA-derived fragment containing the polymorphic site was amplified from genomic DNA with two FcγRIIIA gene-specific primers. Subsequently, a nested PCR was performed in which an NlaIII restriction site was created only in the FcγRIIIA-158V allele. Figure 1 shows digestion of the 94-bp PCR fragment with NlaIII. In lanes 1 to 3, PCR fragments from homozygous FcγRIIIA-158FF donors are not digested by NlaIII. In lanes 4 to 6, three bands of 94 bp, 61 bp, and 33 bp are visible, indicating the FcγRIIIA-158VF heterozygosity of these individuals. Although the fragments in lanes 7 to 9 were from homozygous FcγRIIIA-158VV donors, a 94-bp band of low intensity remained, which was not removed by longer digestion, higher enzyme concentrations, or use of isoschizomer Nsp I. The 94-bp band in these donors was not a result of amplification of FcγRIIIB fragments due to unspecificity of the first-round PCR, because the FcγRIIIB gene carries a G at nucleotide position 559, which results in digestion by NlaIII. Direct sequencing of several first-round PCR products confirmed the FcγRIIIA gene specificity of the assay and the FcγRIIIA-158 genotype of the donors. Genotyping of 87 healthy white individuals yielded gene frequencies of 0.57 and 0.43 for FcγRIIIA-158F and −158V, respectively.
NlaIII restriction analysis of the 94-bp FcγRIIIA-specific fragment, containing the polymorphic nucleotide 559. A 1.2-kb FcγRIIIA-specific fragment was amplified from genomic DNA, followed by a nested PCR. The sense primer of this nested PCR contained a mismatch that introduced a NlaIII restriction site only in the FcγRIIIA-158V (559G) allele. Homozygous FcγRIIIA-158FF fragments were not digested (lanes 1 through 3). Three bands (94 bp, 61 bp, and 33 bp) were visible in heterozygous individuals, whereas homozygous FcγRIIIA-158VV fragments were maximally digested (lanes 7 through 9). A 92-bp fragment of low intensity remained in homozygous 559G fragments (lanes 7 through 9).
NlaIII restriction analysis of the 94-bp FcγRIIIA-specific fragment, containing the polymorphic nucleotide 559. A 1.2-kb FcγRIIIA-specific fragment was amplified from genomic DNA, followed by a nested PCR. The sense primer of this nested PCR contained a mismatch that introduced a NlaIII restriction site only in the FcγRIIIA-158V (559G) allele. Homozygous FcγRIIIA-158FF fragments were not digested (lanes 1 through 3). Three bands (94 bp, 61 bp, and 33 bp) were visible in heterozygous individuals, whereas homozygous FcγRIIIA-158VV fragments were maximally digested (lanes 7 through 9). A 92-bp fragment of low intensity remained in homozygous 559G fragments (lanes 7 through 9).
Table 1 displays the correlation between the FcγRIIIa-48 and the FcγRIIIa-158 phenotypes. A clear linkage was observed between the FcγRIIIA-158V/F and the FcγRIIIA-48L/R/H polymorphisms (χ2-test, P < .001). All FcγRIIIA-158FF individuals were homozygous FcγRIIIA-48LL, whereas all donors with heterozygous FcγRIIIA-48LR or −48LH genotypes carried at least one FcγRIII-158V allele.
Binding of CD16 MoAbs to NK cells of genotyped donors.To determine the influence of the FcγRIIIa-158V/F and -48L/R/H polymorphisms on the binding of a panel of CD16 MoAbs, we investigated binding patterns by NK cells of genotyped donors (Table 2). The binding of MoAb CLBFcRgran1 was not significantly different among NK cells from all donors. In contrast, FcγRIIIa-158VF-positive NK cells from donors homozygous positive for FcγRIIIa-48L bound more MEM154 than did NK cells of FcγRIIIa-158FF-positive donors (Welch's approximate t-test, P =.01). In turn, FcγRIIIa-158VF NK cells from donors who had only one FcγRIIIA-48L allele bound less MEM154 than did FcγRIIIa-158VV NK cells (P = .04).
Comparison of NK cells of FcγRIIIa-158VF phenotyped donors who were either homozygous FcγRIIIa-48LL, or heterozygous -48LR or -48LH showed different binding patterns with MoAb B73.1. NK cells positive for the FcγRIIIa-48R or -48H isoforms showed reduced binding of B73.1 compared with FcγRIII-48LL homozygous NK cells (P = .05). Binding of MoAb MEM154 was equal among these donors. Three FcγRIIIa-158VV individuals who were either homozygous FcγRIIIA-48RR or −48HH were tested. Binding of MEM154 to cells of these donors was equal compared to the binding to NK cells from FcγRIIIa-158VV individuals with a heterozygous FcγRIIIa-48 phenotype, whereas the binding of B73.1 was diminished (Table 2).
Binding of IgG subclasses by FcγRIIIaNK-158 isoforms in the context of FcγRIIIaNK-48 phenotype.To determine the functional consequences of the FcγRIIIa-158V/F polymorphism, we compared the IgG binding capacity of NK cells from genotyped donors. We studied the differences in IgG binding between individuals who had different FcγRIIIA-158V/F genotypes, but were identical regarding the FcγRIIIA-48L/R/H genotype. Preincubation of PBMC with F(ab′ ) fragments of CD16 MoAb CLBFcRgran1 reduced the binding of IgG to less than 10% of control values, indicating that the interaction was FcγRIIIa-mediated (data not shown). As shown in Table 3, lactic acid-treated homozygous FcγRIIIa-48LL NK cells from individuals heterozygous at position 158 of FcγRIIIa bound more IgG3 than did NK cells from FcγRIIIa-158FF phenotyped donors (P = .02). Although a trend was observed, the difference in binding of IgG1 was not statistically significant. In one single experiment, NK cells from the one available donor who was homozygous FcγRIIIA-48LL as well as −158VV bound more IgG3 than did FcγRIIIA-8LL-matched homozygous FcγRIIIA-158FF or heterozygous donors, without clear differences in binding of IgG1 (data not shown). In individuals who were heterozygous for the FcγRIIIa-48L/R/H polymorphism, FcγRIIIa-158VV-positive NK cells bound more IgG1 compared with FcγRIIIa-158VF-positive NK cells (Table 3; Welch's approximate t-test, P = .03). For these donors, the differences in binding of IgG3 were not statistically significant, although a trend was observed. When we determined the levels of NK-cell–associated IgG of freshly isolated NK cells, we observed that FcγRIIIa-158FF-positive cells carried less IgG than did FcγRIII-158VV-positive cells. Comparing NK cells from donors of the same FcγRIIIa-48L/R/H phenotype, we observed a trend toward higher levels of cytophilic IgG bound by FcγRIIIa-158VF and -158VV-positive cells, as compared with FcγRIIIa-158FF-positive cells (Table 3). The levels of NK-cell–associated IgG of FcγRIIIa-48LL and -158VF-positive NK cells did not significantly differ from that of FcγRIIIa-48LR or -48LH NK cells of the same -158VF phenotype.
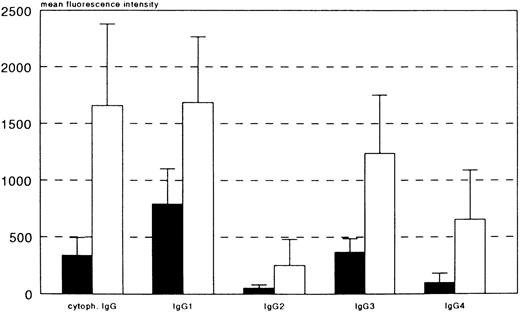
To exclude any effect of the triallelic FcγRIIIa-48L/R/H polymorphism on IgG binding, we compared FcγRIIIa-48LLpositive NK cells with FcγRIIIa-48LR-or -48LH-positive NK cells from donors who were all heterozygous FcγRIIIa-158VF. As shown in Table 3, no statistically significant differences were observed in either the amount of NK-cell–associated IgG or in the binding of IgG subclasses to lactic acid–treated NK cells. Therefore, we depicted the results of IgG-binding experiments, comparing FcγRIIIa-158FF NK-cells with FcγRIIIa-158VV NK cells, disregarding the FcγRIIIa-48 phenotype (Fig 2). Significant differences in levels of IgG1 (P = .007), IgG3 (.005), IgG4 (.02), and cytophilic IgG (.004) were observed.
FcγRIIIaNK-158V binds more IgG than does FcγRIIIaNK-158F. IgG binding by NK cells from individuals either homozygous FcγRIIIA-158FF (▪) or homozygous FcγRIIIA-158VV (□) was compared, irrespective of the FcγRIIIa-48L/R/H genotype. At least three different donors of each genotype were tested. The level of cytophilic IgG and the binding of IgG1, IgG3, and IgG4 was significantly higher in NK cell from FcγRIIIA-158VV individuals (P < .05 in all cases).
FcγRIIIaNK-158V binds more IgG than does FcγRIIIaNK-158F. IgG binding by NK cells from individuals either homozygous FcγRIIIA-158FF (▪) or homozygous FcγRIIIA-158VV (□) was compared, irrespective of the FcγRIIIa-48L/R/H genotype. At least three different donors of each genotype were tested. The level of cytophilic IgG and the binding of IgG1, IgG3, and IgG4 was significantly higher in NK cell from FcγRIIIA-158VV individuals (P < .05 in all cases).
DISCUSSION
Recently we identified a triallelic polymorphism in the membrane-distal Ig-like domain of FcγRIIIa, in which a leucine, an arginine, or a histidine can be present at amino acid position 48.17 In the present report, we functionally characterized a previously described genetic polymorphism of FcγRIIIA, encoding either a phenylalanine (F ) or a valine (V) at amino acid position 158 in the membrane-proximal Ig-like loop of FcγRIIIa.13 We determined that the gene frequencies for FcγRIIIA-158F and FcγRIIIA-158V were 0.57 and 0.43, respectively. The FcγRIIIA-158F genotype was shown to be clearly linked to the FcγRIIIA-48L genotype, whereas the FcγRIIIA-158V was found to be linked to FcγRIIIA-48R and −48H. A linkage between allotypes of the FcγRIIIA-158V/F and the FcγRIIIA-48L/R/H polymorphisms was expected because the two polymorphisms are located within the same gene.
In our previous work, we attributed differences in binding of several CD16 MoAbs to the amino acid present on position 48 of FcγRIIIa. We now show that the binding of MEM154 depends on the presence of a valine at amino acid position 158 of FcγRIIIa and is not influenced by the amino acid polymorphism at position 48. These findings are in conformity with data from Tamm and Schmidt,19 who found that the epitope recognized by MEM154 is located in the membrane-proximal domain. We did not observe any effect of the FcγRIIIa-158V/F polymorphism on B73.1 binding but confirmed our initial observations that only the amino acid at position 48 influenced the binding of MoAb B73.1.17 18
The membrane-proximal domain, carrying the FcγRIIIa-158V/F polymorphism, is generally accepted to contain the IgG-binding site in FcγRs, and experiments with mutants of FcγRIIIb showed that the membrane-proximal loop is essential for IgG binding in FcγRIIIb.14,15 To study the functional consequences of the FcγRIIIa-158V/F polymorphism in the context of the FcγRIIIa-48L/R/H polymorphism, we performed IgG binding experiments with NK cells from genotyped donors. NK cells from homozygous FcγRIIIa-158VV-positive individuals bound more IgG1 and IgG3 than did FcγRIIIa-158FF-positive NK cells, irrespective of the amino acid present on position 48 of the receptor. NK cells of FcγRIIIa-158VF heterozygous donors showed intermediate levels of IgG binding, indicating a gene-dosis effect. These data indicate that the donor-dependent differences in IgG binding of FcγRIIIa isoforms are attributable to the FcγRIIIa-158 polymorphism and not to the FcγRIIIa-48L/R/H polymorphism, as we previously suggested.17 Due to the linkage between the two FcγRIIIA polymorphisms, the FcγRIIIa-48LL-positive donors described in our previous work were all homozygous FcγRIIIa-158FF and therefore their NK cells showed low IgG binding. In contrast, the tested FcγRIIIa-48LR or -48LH-positive donors all carried one or two FcγRIIIa-158V alleles and therefore bound high levels of IgG.
Vance et al20 described a donor-dependent difference in binding of CD16 MoAb 3G8 and monomeric IgG to NK cells. Although we did not observe clear differences in 3G8 binding among our donors (not shown), our IgG binding experiments suggest that the polymorphism described by these investigators might be the result of different FcγRIIIa-158V/F phenotypes.
Hulett and Hogarth21 extrapolated FcγRII data to FcγRIII and suggested that the phenylalanine on position 158 of FcγRIIIa might be partly responsible for the medium affinity of the receptor for IgG, thus attributing the low-affinity receptor of FcγRIIIb to the valine at position 158. Our results are contradicting with this theory because FcγRIIIa-158V bound more IgG than did FcγRIIIa-158F. Recent studies suggest that association of FcγRIIIa with the FcεRI-γ chain might be responsible for the medium affinity, because association of FcγRI with the γ-chain increases the affinity of the receptor for IgG, and results from experiments with FcγRIIIb mutants that could associate with the γ-chain pointed in the same direction.22
The clinical consequences of our findings remain to be established. A higher affinity for IgG could result in higher clearance of immune complexes in patients suffering from (auto)immune disease. On the other hand, a higher amount of cytophilic IgG might imply a decreased availability of FcγRIIIa and, thus, obstruction of receptor-immune complex interaction. Recently we23 and others18 separately described children with a homozygous FcγRIIIA-48HH genotype suffering from recurrent viral infections. We genotyped the two unrelated patients from the former study and found that they were both homozygous FcγRIIIA-158VV. Previous work has suggested that cytophilic IgG on NK cells has inhibitory effects on natural cytotoxicity.24 25 It is tempting to speculate that in the described pediatric patients, NK cells are loaded with a high amount of FcγRIIIa-158V-bound IgG, thereby hampering a proper antiviral response.
ACKNOWLEDGMENT
We thank Dr E. De Vries (Department of Pediatrics of the University Hospital Leiden, Leiden, The Netherlands) for providing the patient samples.
Supported by Grant No. 900-512-092 from the Netherlands Organization for Scientific Research (NWO).
Address reprint requests to Masja de Haas, PhD, Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands.