Abstract
We describe two new monoclonal antibodies specific for the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) that are suitable for the immunohistochemical analysis of routinely processed paraffin sections. These antibodies were applied to the immunohistochemical detection of LMP2A in Hodgkin's disease (HD). LMP2A-specific membrane staining was seen in the Hodgkin and Reed-Sternberg (HRS) cells of 22 of 42 (52%) EBV-positive HD cases, but not in 39 EBV-negative HD cases. In lymphoid tissues from patients with acute infectious mononucleosis (IM), interfollicular immunoblasts were shown to express LMP2A. This is the first demonstration of LMP2A protein expression at the single-cell level in EBV-associated lymphoproliferations in vivo. The detection of LMP2A protein expression in HD and IM is of importance in view of the proposed role of this protein for maintaining latent EBV infection and its possible contribution for EBV-associated transformation. Because LMP2A provides target epitopes for EBV-specific cytotoxic T cells, the expression of this protein in HRS cells has implications for the immunotherapeutic approaches to the treatment of HD.
THE EPSTEIN-BARR virus (EBV) causes infectious mononucleosis (IM) and is associated with several malignant tumors.1 In vitro, EBV can transform human B cells into immortalized lymphoblastoid cell lines (LCL).1 In virus-associated tumors, EBV latent gene expression is restricted facilitating the escape of virus-carrying tumor cells from the EBV-specific host immunity.1 Three major forms of EBV latency have been defined.2 In Burkitt's lymphoma (BL), the EBV-encoded nuclear RNAs (EBER1 and 2) and the nuclear antigen, EBNA1, are expressed (latency I).3,4 In nasopharyngeal carcinoma (NPC), the latent membrane proteins, LMP1, LMP2A, and LMP2B, are expressed in addition to the EBERs and EBNA1 (latency II).5-8 In LCLs, all six nuclear antigens (EBNA1, 2, 3A, 3B, 3C, and -LP), all three LMPs, and the EBERs are expressed (latency III).1
Hodgkin's disease (HD) is characterized by the presence of a small number of Hodgkin and Reed-Sternberg (HRS) cells embedded in a background of abundant reactive cells.9 The nature and clonality of HRS cells are a matter of controversy, with some recent studies suggesting a polyclonal origin.10-13 It is now established that up to 50% of HD cases in Western countries and a far higher proportion in some developing regions are associated with EBV infection.14-18 Southern blot analysis of the terminal repeat (TR) regions of the EBV genomes present in HD have usually identified monoclonal viral episomes.14,15,18,19 Together with the detection of EBV DNA in HRS cells by in situ hybridization,14-16 this has pointed to the HRS cells as the source of the monoclonal EBV genomes and has been taken as evidence supporting the monoclonal nature of HRS cells. However, in situ hybridization studies targeting the abundantly expressed EBERs have found variable numbers of EBV-carrying nonneoplastic lymphoid cells in HD tissues in addition to the EBV-positive HRS cells, raising doubts as to the origin of the viral episomes detected by Southern blotting.20-22 The detection of LMP1 and EBNA1 in the absence of EBNA2 in HRS cells is consistent with a type II latency of EBV.23-25 Transcriptional studies have suggested that LMP2A and LMP2B may be expressed in HRS cells, but direct demonstration of the proteins in the HRS cells has not been possible due to the lack of suitable monoclonal antibodies (MoAbs).26 Localization of the LMP2s is of interest in this context because their expression is possible only from the circularized viral genome.27 LMP2A and LMP2B are transcribed in rightward direction from separate promoters located at the right end of the EBV genome across the fused TR region and into open reading frames at the left end of the viral genome.27,28 The LMP2A protein displays a 119 amino acid N-terminal cytoplasmic tail that is absent from LMP2B, followed by 12 hydrophobic transmembrane domains and a short C-terminal cytoplasmic domain that are common to both proteins.29 Neither LMP2A nor LMP2B is essential for virus-induced B-cell transformation,30,31 but expression of LMP2A in B cells blocks the surface Ig-mediated activation and entry into the lytic viral cycle. This function locates to the N-terminal cytoplasmic tail absent from LMP2B and may be important for the maintenance of latent EBV infection in vivo.32-34 We describe here the generation of LMP2A-specific MoAbs and their application to the detection of LMP2A in formalin-fixed paraffin-embedded tissues with emphasis on HD.
MATERIALS AND METHODS
Cell cultures.Cos cells were transiently transfected using the plasmids pSG5-LMP2A and pSG5-LMP2B containing the complete open reading frames of the LMP2A and LMP2B genes, respectively (kindly provided by Dr R. Longnecker, Chicago, IL).35 For control, Cos cells were transfected with the pSG5 vector alone. The cells were harvested by trypsinization and spun onto glass slides using a cytocentrifuge. Alternatively, cells were fixed in formalin and processed into paraffin wax blocks using the Shandon cytoblock kit (Shandon, Pittsburgh, PA). The EBV-immortalized LCLs X50/7 and AG876 and the EBV-negative B-cell lymphoma cell line BJAB were used for control purposes.
Generation and biochemical characterization of MoAbs.The production of MoAbs directed against LMP2A was described already.36 These MoAbs and additional hybridomas not used previously were tested for their ability to recognize LMP2A in paraffin sections of transiently transfected Cos cells. Two clones, designated 4E11 and 15F9, were selected on the basis of these results. Western blotting analysis was performed as described previously.25
Tissues.Paraffin blocks from 81 cases of HD were available. Of these, 42 were from the West Midlands and the Southwest of England and 39 were from Hamburg, Germany. Most of the British HD cases have been included in previous studies.37,38 Cases were selected to give groups of EBV-positive and EBV-negative cases of roughly equal size. From the United Kingdom, 23 EBV-positive HD cases, including 14 of nodular sclerosis (ns) and 9 of mixed cellularity (mc) subtype, were studied. Nineteen EBV-negative HD cases consisted of 15 ns, 2 mc, and 2 lymphocyte predominant (lp) types. The cases from Hamburg comprised 19 EBV-positive and 20 EBV-negative cases, consisting of 21 mc, 12 ns, and 2 ld types (Table 1). Four cases could not be histotyped. Three lymph nodes and one tonsil from patients with acute IM have been characterized previously.39
In situ hybridization.The EBV status of the cases was determined by in situ hybridization of paraffin sections using EBER-specific RNA probes as described.37,39 Digoxigenin- or 35S-labeled RNA probes were generated from plasmids harboring EBER-specific inserts by in vitro transcription.40 After hybridization and washing, bound probes were detected using a MoAb specific for digoxin (Sigma, Poole, UK) and the alkaline phosphatase antialkaline phosphatase (APAAP) method or autoradiography as appropriate.
Immunohistology and double-labeling studies.Cytospin preparations of transfected Cos cells were fixed for 10 minutes in acetone, air-dried, and then incubated with appropriately diluted primary MoAbs. Paraffin sections of cell blocks and of biopsy material were dewaxed and rehydrated. The sections were subjected to irradiation in a domestic microwave oven (750 W) for 30 minutes at maximum power in 1 L of 0.1 mol/L citrate buffer followed by incubation with the primary MoAbs. Bound primary MoAbs were then detected using a Tyramide Signal Amplification system according to the manufacturer's instructions (Renaissance; DuPont NEN, Boston, MA).41 42 The deposited biotin was detected using a streptavidin-biotinylated alkaline phosphatase complex (Dako, High Wycombe, UK). Alternatively, a polyclonal rabbit antiserum against rat Igs followed by incubation with a rat APAAP complex (Dako) was used for the detection of primary MoAbs. Alkaline phosphatase activity was visualized using Fast Red TR salt (Sigma). For controls, the primary MoAb was omitted from the staining procedure and sections were stained using a rat anti-CD3 MoAb (E. Kremmer, unpublished data).
RESULTS
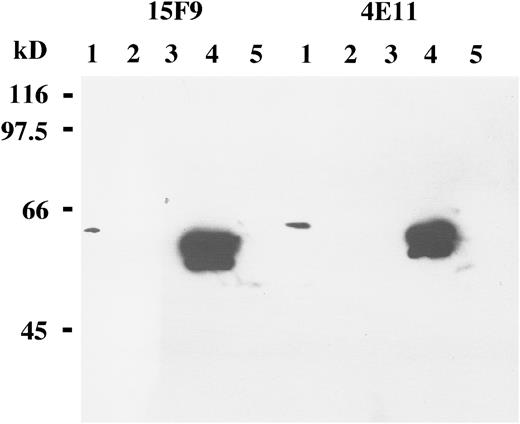
Reactivity of the 4E11 and 15F9 MoAbs with transiently transfected Cos cells.Western blot analysis of Cos cells transiently transfected with the LMP2A gene yielded a strong band of the appropriate size with both MoAbs (Fig 1). No corresponding bands were seen with LMP2B- or vector-transfected cells using these antibodies. This is in agreement with a previous study showing that the 4E11 MoAb recognizes the N-terminal cytoplasmic tail of the LMP2A protein that is not present in the LMP2B protein36 and demonstrates that the MoAb 15F9 is also LMP2A-specific. Protein extracts from the X50/7 LCL did not yield a specific band in Western blot analysis with these antibodies, presumably reflecting low levels of LMP2A protein expression (Fig 1).
Western blot analysis of B-cell lines and transiently transfected Cos cells with the MoAbs 15F9 and 4E11 shows LMP2A expression in protein extracts from Cos cells transiently transfected with the LMP2A open reading frame (lanes 4). By contrast, no specific bands are detected in an EBV-negative B-cell lymphoma line, BJAB (lanes 1); an EBV-positive lymphoblastoid cell line, X50/7 (lanes 2); Cos cells transfected with the vector only (lanes 3); or Cos cells transfected with the LMP2B gene (lanes 5). A weak band is seen in BJAB cells (lane 1), probably representing cross-reactivity with a nonviral cellular protein.
Western blot analysis of B-cell lines and transiently transfected Cos cells with the MoAbs 15F9 and 4E11 shows LMP2A expression in protein extracts from Cos cells transiently transfected with the LMP2A open reading frame (lanes 4). By contrast, no specific bands are detected in an EBV-negative B-cell lymphoma line, BJAB (lanes 1); an EBV-positive lymphoblastoid cell line, X50/7 (lanes 2); Cos cells transfected with the vector only (lanes 3); or Cos cells transfected with the LMP2B gene (lanes 5). A weak band is seen in BJAB cells (lane 1), probably representing cross-reactivity with a nonviral cellular protein.
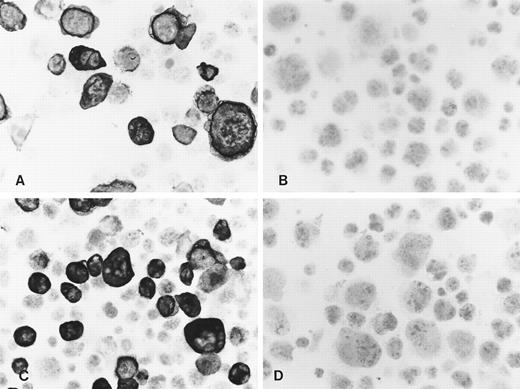
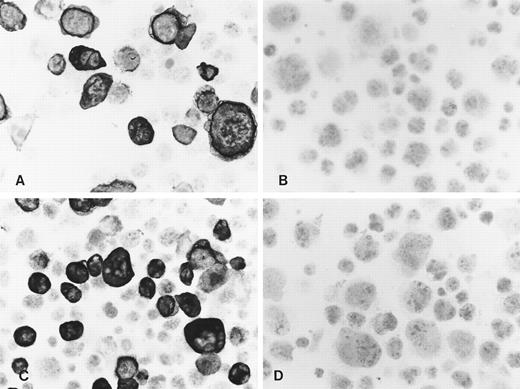
In accordance with the Western blot results, immunocytochemistry of acetone-fixed LMP2A-transfected Cos cells resulted in a strong membrane staining of a proportion of cells using both MoAbs (Fig 2A and B). Only weak background staining was observed in LMP2B-transfected cells (not shown) and in Cos cells transfected with the vector alone (Fig 2C and D). No specific staining was seen in any of the cell lines with the CD3-specific rat MoAb or when the primary antibody was omitted (not shown). Similar results were obtained using sections of formalin-fixed paraffin-embedded cell blocks from these transfectants, confirming that these MoAbs recognize LMP2A but not LMP2B in routinely processed material. In agreement with the Western blot results, no specific staining was observed with these MoAbs in two LCLs, X50/7 and AG876, using both immunohistochemical detection methods, suggesting low levels of expression.
Immunocytochemistry shows reactivity of the MoAbs 4E11 (A) and 15F9 (C) with a proportion of Cos cells transiently transfected with the LMP2A open reading frame. No specific staining was seen with 4E11 (B) and 15F9 (D) in vector control transfectants.
Immunocytochemistry shows reactivity of the MoAbs 4E11 (A) and 15F9 (C) with a proportion of Cos cells transiently transfected with the LMP2A open reading frame. No specific staining was seen with 4E11 (B) and 15F9 (D) in vector control transfectants.
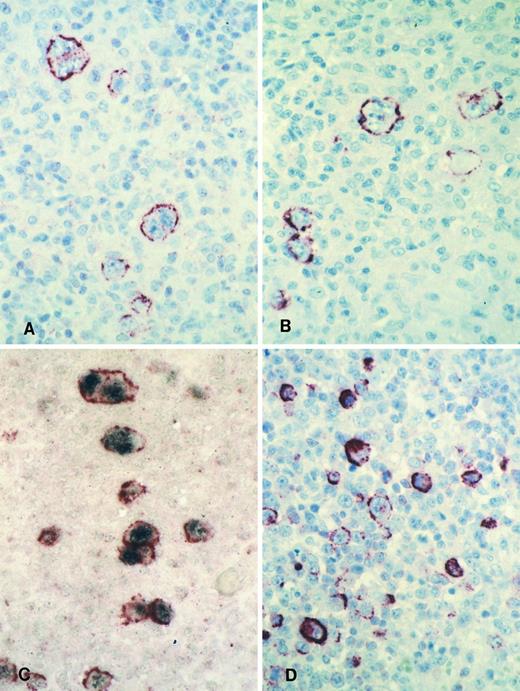
LMP2A expression in HD and IM.The HD cases from the UK consisted of 23 EBV-positive cases and 19 EBV-negative cases as defined by EBER in situ hybridization. Twenty-two of the EBV-positive cases were shown to express the LMP1 protein of EBV. The staining results obtained with the two LMP2A-specific MoAbs, 4E11 and 15F9, were in agreement in 21 of the 23 EBV-positive cases. Thirteen cases showed LMP2A expression in the HRS cells with both MoAbs (Figs 3A and B and 4). HRS cells in 2 cases displayed LMP2A expression only with the MoAb, 15F9. Thus, in 15 of 23 (65%) EBV-positive HD cases from the United Kingdom, LMP2A expression was detectable in HRS cells by at least one of the antibodies (Table 1). In situ hybridization and immunohistochemical analysis of the HD cases from Hamburg, Germany, identified 19 EBER/LMP1-positive cases and 20 EBER/LMP1-negative cases. Seven of 19 EBER-positive cases (37%) were found to express LMP2A (Table 1). In all cases, the results obtained with the two MoAbs were identical. The lower proportion of HD cases with detectable LMP2A expression in the series from Hamburg in comparison to the cases from the UK is probably due to differences in tissue preservation.
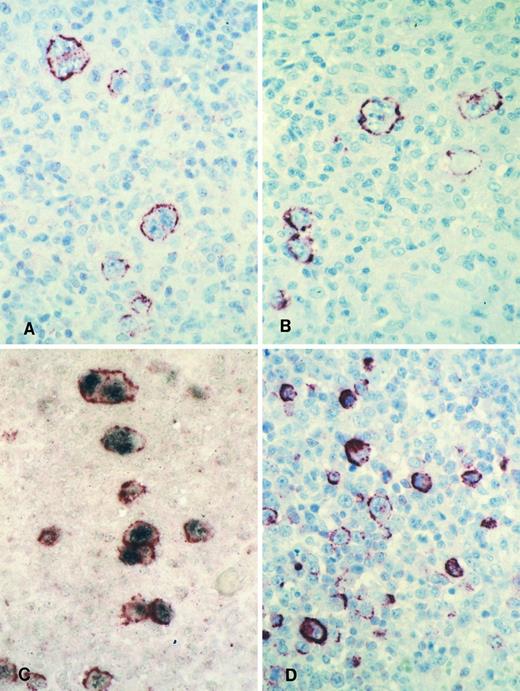
Immunohistology shows LMP2A expression in HRS cells of a case of mixed cellularity HD using the MoAbs 4E11 (A) and 15F9 (B). Double-labeling shows colocalization of the LMP2A-specific signal (red membrane staining) and the EBER in situ hybridization product (blue/black nuclear staining) in HRS cells of the same case (C). Immunohistology with the MoAb 15F9 shows expression of LMP2A in numerous extrafollicular lymphoid blasts in a tonsil from a patient with acute IM (D).
Immunohistology shows LMP2A expression in HRS cells of a case of mixed cellularity HD using the MoAbs 4E11 (A) and 15F9 (B). Double-labeling shows colocalization of the LMP2A-specific signal (red membrane staining) and the EBER in situ hybridization product (blue/black nuclear staining) in HRS cells of the same case (C). Immunohistology with the MoAb 15F9 shows expression of LMP2A in numerous extrafollicular lymphoid blasts in a tonsil from a patient with acute IM (D).
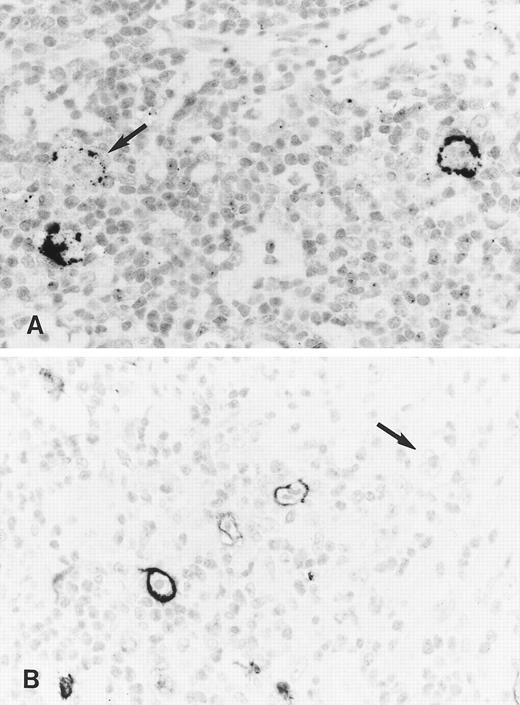
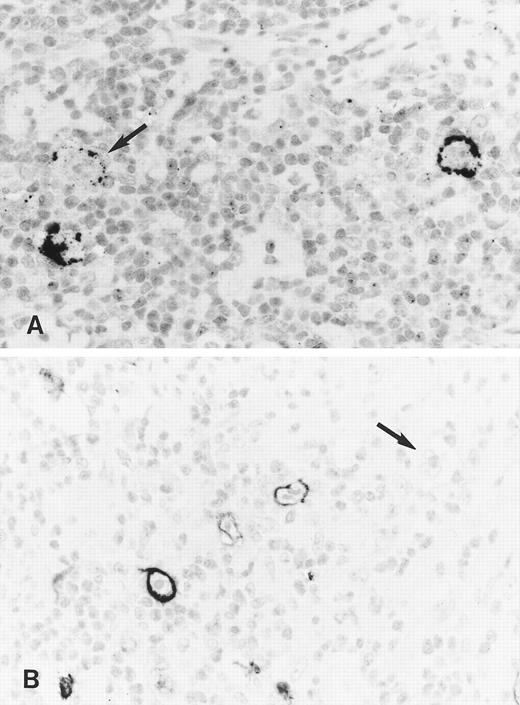
Immunohistology shows expression of LMP2A in HRS cells of two further cases of ns HD using the MoAbs 4E11 (A) and 15F9 (B). Note patchy staining of some HRS cells (arrow in A) in the first case and the absence of detectable LMP2A in some tumor cells of the second case (arrow in B).
Immunohistology shows expression of LMP2A in HRS cells of two further cases of ns HD using the MoAbs 4E11 (A) and 15F9 (B). Note patchy staining of some HRS cells (arrow in A) in the first case and the absence of detectable LMP2A in some tumor cells of the second case (arrow in B).
Staining with both MoAbs resulted in a crisp membrane staining of HRS cells. This was often patchy with parts of the membrane remaining unstained and appeared granular in some cases (Figs 3A and B and 4). Focal dot-like staining of the Golgi region was also occasionally observed, similar to the pattern seen with LMP1-specific MoAbs. The staining intensity varied between cases and between HRS cells within individual cases and some HRS cells remained unlabeled in all cases (Fig 4B). The proportion of HRS cells expressing LMP2A was estimated by comparison with sections stained with the LMP1-specific and the CD30-specific MoAbs. This showed that between less than 5% and up to 60% of the neoplastic cells showed detectable LMP2A expression. LMP2A immunostaining followed by EBER in situ hybridization in one case confirmed that LMP2A expression was confined to the EBER-positive tumor cells (Fig 3C). Nineteen cases from the United Kingdom and 20 cases from Hamburg were EBV-negative. Thirty-six of these showed no labeling of the HRS cells. In 2 cases from the United Kingdom and in one from Hamburg, strong diffuse cytoplasmic staining of most HRS cells was observed (Table 1). This staining pattern differed from the membrane staining observed in the EBV-positive cases and was considered nonspecific.
Numerous LMP2A-expressing cells were seen in 1 lymph node and in a tonsil from patients with acute IM. The other 2 specimens could not be evaluated due to excessive background staining (see below). In the 2 positive cases, LMP2A-expressing cells were mainly interfollicular immunoblasts including HRS-like cells (Fig 3D). Double-labeling in 1 case confirmed colocalization of the EBER in situ hybridization signal and the LMP2A-specific immunostaining (not shown). However, the number of EBER-positive cells greatly exceeded that of LMP2A-expressing cells.
In all cases, staining of sections using the Tyramide Signal Amplification system resulted in a stronger signal than that observed in consecutive APAAP-stained sections. In a few cases, nonspecific labeling of small lymphocytes, predominantly of the follicle mantle, and of plasma cells was noticed with both methods. This did not interfere with the interpretation of HRS cells, but precluded the confident evaluation of two IM biopsies.
DISCUSSION
We describe the production and characterization of two LMP2A-specific MoAbs, 4E11 and 15F9, that specifically stain acetone- or formalin-fixed transiently LMP2A-transfected Cos cells, but not LMP2B or vector control transfectants. The specificity of these MoAbs for LMP2A was confirmed by Western blot analysis of transiently transfected Cos cells. These results are in agreement with a previous study showing reactivity of the 4E11 MoAb with an epitope within amino acids 21 to 36 of the LMP2A protein.36 The MoAb 15F9 has not been tested previously, but the similar reactivity compared with 4E11 in our experiments suggests detection of a neighboring or identical epitope. However, both MoAbs failed to detect LMP2A in 2 LCLs with a type III EBV latency by Western blot and immunocytochemistry. This is presumably due to low levels of expression, because LCLs usually express LMP2A and these cell lines have been shown to express LMP2A mRNA by reverse transcription-polymerase chain reaction (not shown).45 46
In acute IM, LMP2A was expressed predominantly in EBER-positive immunoblasts including HRS-like cells. This finding is in line with previous studies demonstrating type II or type III forms of EBV latent gene expression in IM.32,39 In IM tissues, expression of the BZLF1 protein, which triggers the switch from virus latency to replication, is usually found in small cells, often with plasma cell differentiation.39 Thus, the detection of LMP2A in large immunoblasts is in good agreement with the proposed role for LMP2A in maintaining virus latency.29
It is well established that HRS cells of a proportion of HD cases harbor EBV, and transcriptional studies have suggested a type II of EBV latency with expression of the EBERs, EBNA1, LMP1, LMP2A, and LMP2B.9,14,15,17,18,26 This has been confirmed previously at the protein level for EBNA1 and LMP1.23-25 Using the 4E11 and 15F9 MoAbs, we have now localized LMP2A expression to the EBER-expressing HRS cells in a proportion of EBV-positive HD cases, confirming previous transcriptional studies and showing that the LMP2A mRNA is translated into protein in HRS cells. The detection of LMP2A protein in HRS cells is of relevance because it proves the presence of circularized viral genomes and suggests therefore that the monoclonal viral episomes previously detected by Southern blotting were indeed derived from the HRS cell population.14,15,18,27 Both antibodies produced identical staining results in virtually all cases. A crisp membrane staining of HRS cells was seen often accentuated in patches, reminiscent of the staining seen with LMP1-specific reagents. This is in agreement with the known colocalization of LMP1 and LMP2 in plasma membranes.46 The staining seen with both LMP2A MoAbs was particularly intense using the Tyramide Signal Amplification system (DuPont NEN), confirming the suitability of this method for enhancing relatively weak conventional immunoenzymatic staining results.41 42 However, because of the low expression of the protein and the possibility of nonspecific staining of small lymphocytes and plasma cells, LMP2A staining results should be evaluated together with EBER in situ hybridization to avoid false-positive results.
Two previous studies from Europe have failed to detect LMP2A-specific antibodies in sera from a total of 175 HD patients.47,48 Although the EBV status of these cases was not known, between 30% and 50% of these cases would have been EBV-positive, based on previous studies from European populations.16,23,49 Thus, the consistent absence of LMP2A-specific antibodies from HD patients' sera is unexpected and cannot be explained by a lack of protein expression in the neoplastic cells. Frisan et al50 have shown the absence of EBV-specific CTLs from lymph nodes affected by EBV-associated HD, suggesting a disturbed EBV-specific immunity in the microenvironment of HRS cells, and it has been proposed that interleukin-10 expression in HRS cells may be partly responsible for this.51 Although this might also explain the failure to mount an LMP2A-specific serologic response, the detection of antibodies against LMP1, another viral protein consistently expressed in EBV-positive HRS cells, in HD patients argues against this notion.52 Thus, the absence of LMP2A-specific antibodies in sera of HD patients remains unexplained. By contrast, LMP2A-specific antibodies are frequently detected in sera of patients with undifferentiated NPC.47,48 A preliminary analysis of EBV-associated NPCs and gastric carcinomas did not show convincing staining of the epithelial tumor cells using the MoAbs described here (unpublished observation). Together with previous serologic and transcriptional studies, this suggests that the protein is expressed at low levels in the tumor cells of these neoplasms.5 6
Immunotherapy targeted against viral antigens has been proposed as a possible alternative strategy for the management of HD and other EBV-associated malignancies.53 This approach should have limited side effects because the expression of viral antigens is usually restricted to the tumor cells and a small number of persistently infected lymphocytes. Of the viral proteins expressed in HRS cells, EBNA1 is known not to elicit a CTL response.54-56 LMP1 expression induces cellular phenotypic changes facilitating T-cell recognition, but the detection of LMP1-specific CTLs is at least difficult.57 LMP2A epitopes are recognized by virus-specific CTLs58 and HRS cells in EBV-positive HD express MHC class I proteins and TAP-1.59 Thus, HRS cells should be able to present viral CTL target peptides. Moreover, at least some LMP2A peptides are processed in a TAP-independent fashion.60 Thus, the demonstration of LMP2A expression at the protein level in HRS cells is of pivotal importance for the implementation of immunotherapeutic strategies.53
LMP2A and LMP2B are not essential for EBV-induced B-cell transformation.30,31 It has been suggested that a major function of LMP2A is to maintain EBV latency by blocking the transition into the lytic cycle after signaling through the B-cell antigen receptor (BCR) complex.29,34 LMP2A is a dominant negative inhibitor of Src and Syk protein tyrosine kinases (PTK) and blocks tyrosine phosphorylation and calcium mobilization in response to BCR cross-linking,33 whereas LMP2B inhibits LMP2A by increasing the spaces between the functionally important N-terminal cytoplasmic tails of LMP2A molecules.29 In line with this model and with the frequent detection of LMP2A in HRS cells, EBV lytic cycle proteins are only infrequently expressed in HRS cells.61,62 However, using mini-EBV plasmids containing all viral genes required for B-cell immortalization, Brielmeier et al63 have shown that mini-EBVs lacking the LMP2 gene display a greatly reduced ability to induce B-cell transformation. This implies an important role for LMP2A in enhancing the efficiency of transformation that is not dependent on preventing entry into the lytic cycle.63 This raises the possibility that LMP2A expression may contribute to the malignant phenotype of HRS cells in EBV-associated HD.
In summary, we describe two MoAbs directed against the EBV LMP2A protein suitable for the immunohistologic detection of this protein in routinely processed tissue sections. These MoAbs represent useful additions to the panel of antibodies against EBV-encoded proteins.
ACKNOWLEDGMENT
The authors are grateful to Lindsey Taylor for excellent photographic assistance.
Supported by grants from the Wellcome Trust (to G.N.) and from the Werner-Otto-Foundation (to H.H.).
Address reprint requests to G. Niedobitek, MD, Pathologisch-anatomisches Institut, Friedrich-Alexander Universität, Krankenhausstr. 8-10, 91054 Erlangen, Germany.