To the Editor:
The t(2; 5)(p23; q35) is an established cytogenetic abnormality associated with 40% of cases of anaplastic large-cell lymphoma (ALCL). Cloning of the translocation breakpoint has shown the fusion of a previously unidentified protein tyrosine kinase, anaplastic lymphoma kinase (ALK) gene with nucleophosmin (NPM),1 a housekeeping gene. The hybrid gene contains the portion of the ALK gene encoding the entire intracytoplasmic region, including the catalytic domain fused to the 5′ end of the NPM gene containing the active promoter. Through the chromosome translocation, the NPM gene contributes an active promoter to drive the synthesis of the fusion protein in lymphoma cells leading to transformation.
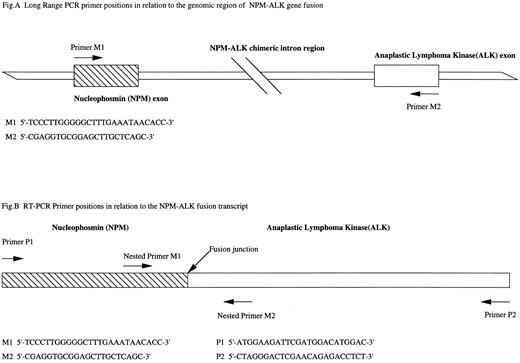
(A) Long-range PCR primer sequences and their positions on the exons of nucleophosmin gene and ALK gene exons involved in the NPM-ALK gene fusion. (B) RT-PCR primer sequences in relation to the fusion transcript. Primers P1 and P2 designed by Orscheschek et al is nested by primers M1 and M2 designed by Morris et al yielding a 177-bp product.
(A) Long-range PCR primer sequences and their positions on the exons of nucleophosmin gene and ALK gene exons involved in the NPM-ALK gene fusion. (B) RT-PCR primer sequences in relation to the fusion transcript. Primers P1 and P2 designed by Orscheschek et al is nested by primers M1 and M2 designed by Morris et al yielding a 177-bp product.
The question of whether the abnormality is associated with Hodgkin's disease (HD) has been addressed. Bullrich et al2 initially observed rearrangement of the NPM locus by Southern blotting in 2 of 9 cases of HD. Orscheschek et al3 detected NPM-ALK transcripts in 11 of 13 cases of HD using reverse transcriptase-polymerase chain reaction (RT-PCR). Their subsequent Northern blot study on a single case of HD showed a transcript of the correct molecular weight. Provocatively, this case showed a positive signal in HD and Reed-Sternberg cells using polyclonal antisera against the ALK protein.4NPM-ALK transcripts have also been observed in HD by other RT-PCR studies but at a considerably lower incidence. Ngan5 reported an incidence of 5 of 24 cases of HD and Yee et al6 reported an incidence in 3 out of 72 cases. Other RT-PCR–based studies have found no evidence at all for the NPM-ALK gene fusion in HD (summarized by Sarris et al7 ).
Several technical issues have been raised regarding the use of RT-PCR to account for these contrasting RT-PCR findings. The design of RT-PCR primers may be critical to achieve the high sensitivity required in amplification of NPM-ALK mRNA8 from HD tissues. RNA is prone to degradation and cross-linking in tissues that are fixed in formalin,9 reducing the efficiency of PCR.4 In addition, control primers may inadvertently amplify genomic pseudo-genes from traces of genomic DNA in RNA preparations, yielding RT-PCR products of the same molecular weight as that expected from cDNA and thereby giving a false impression of the integrity and presence of RNA.10
In view of the limitations of RT-PCR methodology, DNA may be a preferable assay substrate because it is relatively stable and its integrity may be assessed reliably. We thus read with interest the article of Sarris et al7 reporting the absence of NPM-ALK gene fusion among 31 cases of HD as well as among other non-Hodgkin's diseases using the long-range PCR methodology on genomic DNA,7 a methodology that we developed to detect the t(2; 5)(p23; q35) genomic translocation breakpoint in ALCL.11 Long-range PCR amplifies the genomic region of the translocation breakpoint that may extend beyond the maximum amplifyable length of conventional PCR. Individual ALCL cases yielded a PCR product with a unique size reflecting the variation in the precise position of the translocation breakpoint between cases. Contamination, an established problem in PCR-based assays, may thus be controlled for by the detection of PCR product of a unique size, in addition to the use of a negative control.
We report here our investigations into the presence of NPM-ALK gene fusion in HD at the genomic level using long-range PCR methodology. Expand PCR System (Boehringer Mannheim, Mannheim, Germany) was used to amplify the genomic region of the NPM-ALK fusion gene (Fig 1A). NPM-ALK gene fusion was detected in all positive control cases (DNA from cell line materials SU-HL-1 and Karpas299 and also DNA from 4 ALCL cases). However, we found no evidence of NPM-ALK gene fusion in 46 cases of HD (31 mixed cellularity, 12 nodular sclerosis, 1 lymphocyte predominant, and 2 uncharacterized). The integrity of DNA was checked by concurrent amplification of a 1,000-bp region at the NPM locus. To determine the sensitivity of our DNA assay, a dilution series of DNA from the ALCL cell line (SU-DHL-1) was prepared in normal DNA. Successful amplification was achieved down to a level equivalent to 1 tumor cell in a background of 1,500 normal cells.
In addition, using RT-PCR with the primers designed by Orscheschek et al, we investigated samples by attempting to avoid the technical pitfalls in RT-PCR methodology. RT-PCR (Fig 1B) primers designed by Morris et al1 (primers M1 and M2) were used to nest these primers to confirm the product identities. We detected no amplification of NPM-ALK fusion transcripts in a different set of 25 HD cases (5 mixed cellularity, 14 nodular sclerosis, 2 lymphocyte predominant, and 4 unclassified). RNA integrity was checked by a concurrent RT-PCR designed from retinoic acid receptor α cDNA. To show that amplification of the control primers represented the presence of mRNA, a mock RT-PCR reaction was performed on genomic DNA, omitting the reverse transcription step. Amplification was not achieved, demonstrating that amplification of the control primers solely represent the presence of an mRNA species. RT-PCR sensitivity was assessed on RNA extracted from a dilution series of cultured SU-DHL-1 cells in a background of cultured K562 cells. We could detect 1 tumor cell in a background of 100,000 K562 cells.
We have therefore failed to observe NPM-ALK gene fusion in the genomic DNA in 46 cases of HD. It is conceivable that cells carrying this translocation may have been present below the 1:1,500 cell level. However, using a highly sensitive RT-PCR assay, we also failed to detect any NPM-ALK transcripts, supporting the findings of the present DNA study. We therefore conclude that ALCL bearing the t(2; 5)(p23; q35) abnormality and HD do not share a common mode of pathogenesis with respect to the NPM-ALK gene fusion.
ACKNOWLEDGMENT
This work was supported by the Leukaemia Research Fund.