Abstract
Under shear stress, leukocytes use P-selectin glycoprotein ligand-1 (PSGL-1) to tether to and roll on P-selectin expressed on activated platelets or endothelial cells. P-selectin has an NH2-terminal lectin domain, an epidermal growth factor (EGF)-like motif, nine consensus repeats (CRs), a transmembrane domain, and a cytoplasmic tail. To determine whether the CRs are required for P-selectin to bind PSGL-1, we expressed a soluble protein (Lec-EGF) that contained only the lectin and EGF domains, plus a short C-terminal epitope tag. Electron microscopy and hydrodynamic analysis confirmed that Lec-EGF was monomeric, as previously shown for soluble P-selectin (sPS) that contained the lectin and EGF domains plus all nine CRs. Fluid-phase Lec-EGF or sPS inhibited binding of oligomeric125I-labeled membrane-derived P-selectin (mPS) to PSGL-1 on neutrophils and binding of 125I-PSGL-1 to immobilized mPS. The IC50 for inhibiting binding of mPS to neutrophils was fivefold greater for Lec-EGF than for sPS, whereas the IC50 for inhibiting binding of mPS to purified PSGL-1 was indistinguishable for Lec-EGF and sPS. Under static or shear conditions, neutrophils used PSGL-1 to tether to or roll on Lec-EGF that was captured by an immobilized monoclonal antibody to the C-terminal epitope. These data show that P-selectin requires only the lectin and EGF domains to bind to PSGL-1.
P-, E-, AND L-SELECTIN are membrane-anchored lectins that initiate tethering and rolling of flowing leukocytes on the vessel wall through interactions with cell-surface carbohydrate ligands.1,2 P-selectin is redistributed from secretory granules to the surface of thrombin-activated platelets and endothelial cells, and certain cytokines also increase its synthesis.3-5 The major ligand for P-selectin is P-selectin glycoprotein ligand-1 (PSGL-1),1 a disulfide-linked, homodimeric mucin expressed on most leukocytes.6-8 P-selectin binds to an NH2-terminal region of PSGL-1 that includes tyrosine sulfate9-11 and at least one core-2, sialylated, and fucosylated O-glycan.12,13 PL1, a monoclonal antibody (MoAb) that recognizes a protein epitope in this NH2-terminal region,14 blocks binding of purified PSGL-1 to P-selectin as well as tethering and rolling of flowing leukocytes on P-selectin.15-17
Like the other selectins, P-selectin contains an NH2-terminal carbohydrate-recognition domain typical of those in C-type lectins, then an epidermal growth factor (EGF)-like motif, a series of consensus repeats (CRs) like those in complement-regulatory proteins, a transmembrane domain, and a cytoplasmic tail. The functions of these domains in ligand binding and cell adhesion have been a major focus of study. Studies using blocking MoAbs18 and site-directed mutagenesis19-22 suggest that a specific region of the lectin domain of P-selectin binds carbohydate ligands such as those on PSGL-1. Experiments with chimeric selectins suggest that the EGF domain of P-selectin modulates ligand preferences,23,24 although the isolated EGF domain does not detectably bind to leukocytes.25
The role of the CRs of P-selectin in ligand recognition is less clear. Under static conditions, human HL-60 cells adhere equivalently to Chinese hamster ovary cells expressing matched densities of either full-length P-selectin or a chimera in which the lectin and EGF domains of P-selectin are fused to the CRs, transmembrane domain, and cytoplasmic tail of L-selectin.26 This result was interpreted to indicate that the lectin and EGF domains of P-selectin constitute the optimal ligand-recognition unit. However, the data do not exclude the possibility that the L-selectin domains in the chimera substitute adhesive functions normally provided by the corresponding domains of P-selectin. Under shear forces, neutrophils tether and roll much less efficiently on Chinese hamster ovary cells expressing membrane-anchored P-selectin constructs with less than five CRs. But neutrophils roll equivalently on cells expressing either wild-type P-selectin or a chimeric P-selectin construct in which the first two CRs of P-selectin are fused to four heterologous CRs from C4b-binding protein.27 This result suggests that the P-selectin CRs function primarily to extend the lectin and EGF domains of P-selectin sufficiently above the plasma membrane to contact PSGL-1 on flowing leukocytes. However, the data do not exclude a specific function of the first two CRs in ligand recognition. P-selectin Ig chimeras that appear to bind normally to PSGL-1 all contain the first 1 to 2 CRs.19,20,22 To date, no construct with only the lectin and EGF domains of P-selectin has been studied. The potential importance of the CRs in enhancing the binding affinity of P-selectin is underscored by reports that soluble forms of L- and E-selectin containing only the lectin and EGF domains bind to their ligands with much lower affinity than do full-length molecules that contain the CRs.28 29
We previously expressed a soluble form of P-selectin containing the lectin and EGF domains plus all nine CRs. This protein is monomeric in aqueous buffers and binds with relatively high affinity to PSGL-1 on human myeloid cells.30 To address whether the lectin and EGF domains of P-selectin are sufficient to confer specific binding to PSGL-1, we expressed a soluble form of P-selectin containing only these two domains. After confirming that this Lec-EGF construct was also monomeric, we compared its binding properties with those of soluble P-selectin containing the CRs.
MATERIALS AND METHODS
Antibodies and proteins.The anti–P-selectin MoAbs G1 and S12 were prepared as described previously.18,31 G1 blocks binding of P-selectin to PSGL-1, whereas S12 does not. G1 binds to an epitope in the lectin domain,32,33 whereas S12 binds to an epitope that requires the sixth CR.27 The anti–PSGL-1 MoAbs PL1 and PL2 were prepared as described.15 PL1 blocks binding of PSGL-1 to P-selectin, whereas PL2 does not. The anti-protein C MoAb HPC4, which recognizes an 8-residue epitope in a Ca2+-dependent manner, was prepared as described.34 The MoAb 1478 also recognizes the HPC4 epitope; it was raised against the membrane fraction of a cell line transfected with the endothelial protein C receptor carrying an HPC4 epitope tag.35 The MoAb 5A8 was prepared by standard procedures,15 36 using Lec-EGF (see below) as the immunogen. As determined by enzyme-linked immunosorbent assay (ELISA) and Western blotting, 5A8 binds to both Lec-EGF and sPS; therefore, it identifies an epitope in the lectin and/or EGF domains of P-selectin. 5A8 does not block binding of P-selectin to PSGL-1. All MoAbs are of the murine IgG1 subclass.
Native, membrane-derived P-selectin (mPS) was purified from platelet membrane extracts as described.37 Recombinant soluble P-selectin (sPS; formerly called tPS) was expressed in human 293 cells and purified from conditioned medium by MoAb affinity chromatography.30 PSGL-1 was purified from human neutrophil membranes as described.38
Expression and purification of Lec-EGF.A soluble form of P-selectin containing the lectin and EGF domains (Lec-EGF) was made. Lec-EGF was constructed by a two-step polymerase chain reaction (PCR) protocol using the P-selectin cDNA39 in pcDNA1/Neo (Invitrogen, San Diego, CA) as a template. In the first step, the plasmid T7 primer was used as the sense primer. An antisense primer encoded bases 615 to 630 in the EGF domain plus a 12-bp overhang complementary to the 12 bp encoding a 4-residue factor Xa cleavage site. This reaction generated a fragment beginning in the pcDNA1/Neo vector and terminating with the factor Xa cleavage site. The second PCR reaction used the same sense primer and an antisense primer complementary to the sequence encoding the factor Xa cleavage site plus an overhang encoding residues for the HPC4 MoAb epitope, a stop codon, and an Xba I site. This reaction extended the product of the primary PCR reaction to include an 8-residue epitope for the MoAb HPC4, a stop codon, and an Xba I site at its 3′ end. The second PCR product was digested with HindIII and Xba I and cloned into the HindIII and Xba I sites of pRc/RSV (Invitrogen). Assuming cleavage of the signal peptide, the translated Lec-EGF protein contains 156 residues in the lectin and EGF domains (ending with Val156) plus the C-terminal, 12-residue extension.
The Lec-EGF construct in pRc/RSV was transfected into 293 cells by previously described methods.30 Stable clones of cells expressing Lec-EGF in the conditioned medium were selected, and the highest expressing clone was expanded into roller bottle cultures. Cells were first grown to confluence in medium containing 10% serum. Subsequent medium contained only 2% serum.
The concentration of Lec-EGF in the conditioned medium was determined by ELISA, as described previously, for soluble P-selectin,30 except that G1 (5 μg/mL, 100 μL/well) was used as the capture antibody and biotinylated HPC4 (1 μg/mL, 100 μL/well) was used as the detection antibody. Bound HPC4 was quantified using the Immunoselect ELISA Amplification System from GIBCO Labs (Grand Island, NY). The absorbance at 490 nm was measured with a Vmax Kinetic Microtiter Reader interfaced to Softmax software (Molecular Devices, Menlo Park, CA). The concentration of Lec-EGF in the conditioned medium was calculated using a standard curve of purified Lec-EGF.
Conditioned medium from roller bottle cultures expressing Lec-EGF was harvested every 3 to 4 days, centrifuged, and stored at −20°C with 5 mmol/L benzamidine and 0.02% sodium azide as preservative. Lec-EGF from the conditioned medium was purified by affinity chromatography over an HPC4-Affigel column (3 mg/mL of antibody conjugated to Affigel-10 [Bio-Rad Laboratories, Richmond, CA] according to the manufacturer's instructions.) A precolumn of Sephadex G-75 (Sigma, St Louis, MO) was used to reduce the number of contaminants eluted from the HPC4 column. Both columns were equilibrated at 4°C in wash buffer (20 mmol/L Tris, pH 7.4, 0.1 mol/L NaCl, 1 mmol/L CaCl2 , and 0.02% NaN3). The conditioned medium (2L) was thawed, made 1 mmol/L in CaCl2 , and applied to an 18.5 × 1.5-cm G-75 column that was linked in series to a 12.5 × 0.9-cm HPC4 column. The medium was chased with 200 mL of wash buffer, and the precolumn was then disconnected. The HPC4 column was further washed with 20 mmol/L Tris, pH 7.4, 1 mol/L NaCl, 1 mmol/L CaCl2 , and 0.02% NaN3 , followed by 100 mL of wash buffer. The bound protein was eluted with 5 mmol/L EDTA in 20 mmol/L Tris, pH 7.4, 0.1 mol/L NaCl, and 0.02% NaN3 . The protein-containing fractions were pooled, concentrated in a Centriprep-10 device (Amicon, Beverly, MA), and dialyzed against 10 mmol/L MOPS, pH 7.5, 0.1 mol/L NaCl. The purified protein was filtered through a 0.2-μm filter and stored at −20°C. Purified Lec-EGF was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12.5% polyacrylamide gels followed by silver staining.30
Amino acid compositional analysis and assignment of extinction coefficient.Two preparations of purified Lec-EGF were individually subjected to amino acid compositional analysis after hydrolysis in vacuo in 6 N HCl at 107°C for 24 hours in the Molecular Biology Resource Facility of the University of Oklahoma Health Sciences Center. All samples contained an internal standard to determine recoveries. The amino acid compositions of all samples agreed closely with those predicted from their amino acid sequences. Based on the compositional analyses and the molecular weight of Lec-EGF in nondenaturing buffers as determined by sedimentation equilibrium analysis (see Results), the extinction coefficient (E1%280) of Lec-EGF was determined to be 21.0 ± 2.7 (n = 3). Protein concentrations were converted to molar values based on the molecular weight in nondenaturing buffer as determined by sedimentation equilibrium analysis.
Endoglycosidase digestion.Lec-EGF was digested with Peptide N-glycosidase F (PNGase F, Boehringer Mannheim, Indianapolis, IN) as described by the manufacturer. Briefly, the protein (10 μg), diluted in incubation buffer (10 mmol/L phosphate, pH 7.4), was denatured in 10% SDS. It was then made up to a total volume of 50 μL in a final concentration of 2% Triton-X-100, 0.2% SDS, 10 mmol/L EDTA, and 1% 2-mercaptoethanol. PNGase F (20 U/mL) was added to the sample and incubated at 37°C for 16 hours. As a negative control, a parallel sample was handled identically except that no enzyme was added. Both samples were analyzed by SDS-PAGE and silver staining.
Electron microscopy.Lec-EGF was prepared for electron microscopy, as described previously for sPS.30 Briefly, a 200-μL sample of Lec-EGF (∼200 μg/mL) was centrifuged through a glycerol gradient in 0.2 mol/L ammonium bicarbonate. Protein-containing fractions (identified by SDS-PAGE) were sprayed on mica and rotary shadowed.40
Sedimentation equilibrium and sedimentation velocity analysis.Short column sedimentation equilibrium experiments with Lec-EGF were conducted as described previously30,41 in 10 mmol/L MOPS, pH 7.5, 100 mmol/L NaCl at 23.3°C at rotor speeds of 10,000, 20,000, 30,000, and 40,000 rpm in a Beckman model E ultracentrifuge (Beckman Instruments, Palo Alto, CA) equipped with Rayleigh interference optics.42 Global data analysis was performed as described43 using data from all rotor speeds for the protein stock solution (1.9 mg/mL) and serial dilutions of 1:2, 1:4, and 1:8 made using dialysate. A calculated44 partial specific volume of 0.71 mL/g and a solvent density of 1.002 g/mL were used to calculate the solution molecular weight.
Sedimentation velocity analysis of Lec-EGF was conducted at 20°C and 60,000 rpm in a Beckman XLA ultracentrifuge equipped with Rayleigh interference optics.45 Data were acquired at 1-second intervals to produce sedimentation coefficient distributions according to the method described by Stafford.46 The data were interpreted as described previously44 to obtain the axial ratio and hydrodynamic length.
Protein iodination.Carrier-free Na125I (ICN Biomedicals, Costa Mesa, CA) was used to iodinate mPS, PSGL-1, or the MoAb G1. mPS (50 to 75 μg) was radiolabeled with 400 μCi of Na125I using Enzymobeads (Bio-Rad) according to the manufacturer's instructions. G1 (100 to 150 μg) was diluted in 20 mmol/L HEPES, pH 7.4, 0.1 mol/L NaCl, and labeled with 200 μCi of Na125I using Iodogen (Pierce, Rockford, IL).38 After the iodination reaction, free 125I was removed by gel filtration through a Sephadex G-25 column (PD-10; Pharmacia Fine Chemicals, Uppsala, Sweden). The concentration of labeled protein was determined with a micro BCA protein assay kit (Pierce). The proteins were made 0.1% in bovine serum albumin (BSA) and stored at 4°C to 10°C. Labeled proteins were routinely greater than 95% trichloroacetic acid precipitable. The specific activities of mPS and G1 were 2 to 3 μCi/μg protein.
PSGL-1 was diluted in 20 mmol/L HEPES, pH 7.4, 0.1 mol/L NaCl, and labeled with 200 μCi of Na125I using Iodogen. The labeled protein was desalted over PD-10 in buffer containing 0.1% Brij-58, and 125I-PSGL-1 was further fractionated over a Superose 6 PC 3.2/30 column as described (Pharmacia Biotech, Uppsala, Sweden).38
Inhibition of 125I-mPS binding to neutrophils by fluid-phase mPS, sPS, or Lec-EGF.Human neutrophils isolated from healthy volunteer donors were resuspended in Hanks' buffered salt solution (HBSS) containing 1.26 mmol/L Ca2+ and 0.81 mmol/L Mg2+ plus 0.5% human serum albumin (0.5% HSA/HBSS).47 Binding of fluid-phase 125I-mPS to neutrophils was measured as described.30 48 To study the competitive inhibition of 125I-mPS binding to neutrophils by fluid-phase unlabeled P-selectin, 1.5 × 106 neutrophils were incubated with increasing concentrations of unlabeled mPS, sPS, or Lec-EGF. The percentage of inhibition of 125I-mPS binding by each concentration of unlabeled selectin was calculated by taking the specific binding of 125I-mPS alone as 100%.
Inhibition of 125I–PSGL-1 binding to immobilized mPS by unlabeled fluid-phase mPS, sPS, or Lec-EGF.This assay was performed essentially as described.38 Briefly, Immulon-I Removawells (Dynatech, Chantilly, VA) were coated with 5 μg/mL of mPS (50 μL/well) overnight at 4°C. The wells were blocked with 1% HSA in HBSS for 2 hours at room temperature. Dilutions of unlabeled mPS, sPS, or Lec-EGF in 0.1% HSA/HBSS (50 μL) were added to the wells. Then, 125I–PSGL-1 in 0.1% HSA/HBSS (10,000 to 20,000 cpm, 50 μL/well) was added and incubated for 1 hour at room temperature. The wells were extensively washed using a 12-channel plate washer (Nunc-Immuno wash 12; Nunc Inc, Naperville, IL), and individual wells were counted in a γ counter. The assay was performed in triplicate for each concentration of unlabeled selectin. Nonspecific binding of 125I–PSGL-1 to the plate was determined in the presence of 10 mmol/L EDTA. Specific binding was calculated by subtracting this number from the total binding for each concentration of unlabeled selectin. The percentage of inhibition of 125I–PSGL-1 binding to immobilized mPS by each concentration of unlabeled selectin was calculated by taking the specific binding of 125I–PSGL-1 alone as 100%.
Determination of selectin site density in microtiter wells.The site density of antibody-captured sPS or Lec-EGF in microtiter wells was determined as described,15 with the following modifications. 125I-G1 MoAb was used at a concentration of 2 μg/mL, the wells were extensively washed using a 12-channel plate washer, and bound radioactivity was eluted with 1 mol/L NH4OH. The determinations were performed in triplicate for each selectin concentration, and nonspecific binding was determined by adding 125I-G1 to an empty, blocked well. The number of G1 epitopes per square micrometer was calculated from the specific activity of the labeled antibody and the area of the microtiter wells, assuming monovalent binding of antibody at saturating concentrations.
Neutrophil adhesion assay under static conditions.The MoAb 1478 (50 μg/mL) was coated on microtiter wells overnight at 4°C. The wells were blocked with 1% HSA in HBSS for 2 hours and then washed with HBSS. Lec-EGF in 0.1% HSA/HBSS was captured on immobilized 1478 as described.30 Static adhesion of neutrophils to 1478-captured Lec-EGF or to control wells containing only 1478 was measured as described.30 Some of the neutrophils were preincubated with 10 μg/mL of the anti–PSGL-1 MoAbs PL1 or PL2 before their addition to the wells.
Neutrophil adhesion assay under hydrodynamic flow conditions.The MoAbs 1478 or S12 (50 μg/mL) were coated on 35-mm2 tissue culture dishes overnight at 4°C. Lec-EGF or sPS in 1% HSA/HBSS was captured on immobilized 1478 or S12, respectively. The density of G1 epitopes was measured for proteins captured under identical conditions in microtiter wells. Dishes containing the antibody-captured selectins at matched densities were inserted into a parallel-plate flow chamber. The accumulation of neutrophils flowing on the sPS and Lec-EGF surfaces under defined shear stresses was quantified with a video microscopy system as described previously.15,17 The rolling velocities of neutrophils were determined as described.17
RESULTS
Expression and purification of Lec-EGF.We previously characterized a soluble recombinant form of P-selectin, termed sPS (formerly called tPS), that is secreted from 293 cells.30 sPS contains the lectin domain, EGF domain, and all nine CRs, but it lacks the transmembrane and cytoplasmic domains found in mPS (Fig 1). As determined by electron microscopy and hydrodynamic analysis, sPS is monomeric, whereas mPS isolated from human platelets forms heterogeneous oligomers in buffers with low levels of nonionic detergent.30 Both sPS and mPS bind to PSGL-1 on human leukocytes, but mPS binds with significantly higher avidity because of its oligomeric organization.30
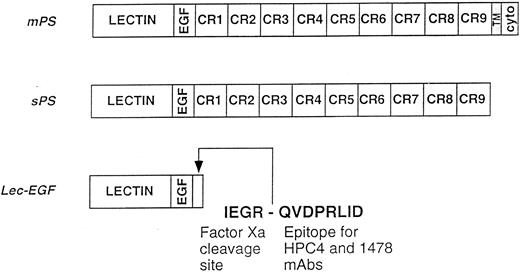
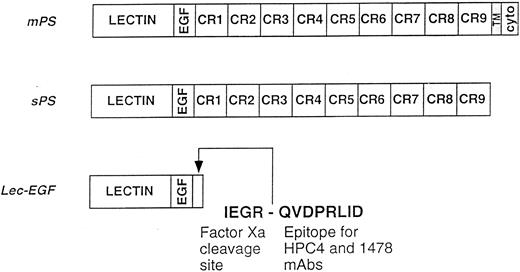
Schematics of the P-selectin constructs. mPS contains an NH2-terminal lectin domain (lectin), followed by an EGF-like domain (EGF), nine consensus repeats (CR1-9) related to those in complement-regulatory proteins, a transmembrane domain (TM), and a cytoplasmic tail (cyto). sPS terminates with Ala730 at the junction of CR9 with the transmembrane domain. Lec-EGF contains the lectin and EGF domains (residues 1-156), plus a C-terminal extension that includes a factor Xa cleavage site and the epitope for the MoAbs HPC4 and 1478. Native mPS was purified from human platelets. Recombinant sPS and Lec-EGF were purified from the conditioned medium of transfected 293 cells. Not shown is the NH2-terminal signal peptide that is cleaved after synthesis of all three proteins.
Schematics of the P-selectin constructs. mPS contains an NH2-terminal lectin domain (lectin), followed by an EGF-like domain (EGF), nine consensus repeats (CR1-9) related to those in complement-regulatory proteins, a transmembrane domain (TM), and a cytoplasmic tail (cyto). sPS terminates with Ala730 at the junction of CR9 with the transmembrane domain. Lec-EGF contains the lectin and EGF domains (residues 1-156), plus a C-terminal extension that includes a factor Xa cleavage site and the epitope for the MoAbs HPC4 and 1478. Native mPS was purified from human platelets. Recombinant sPS and Lec-EGF were purified from the conditioned medium of transfected 293 cells. Not shown is the NH2-terminal signal peptide that is cleaved after synthesis of all three proteins.
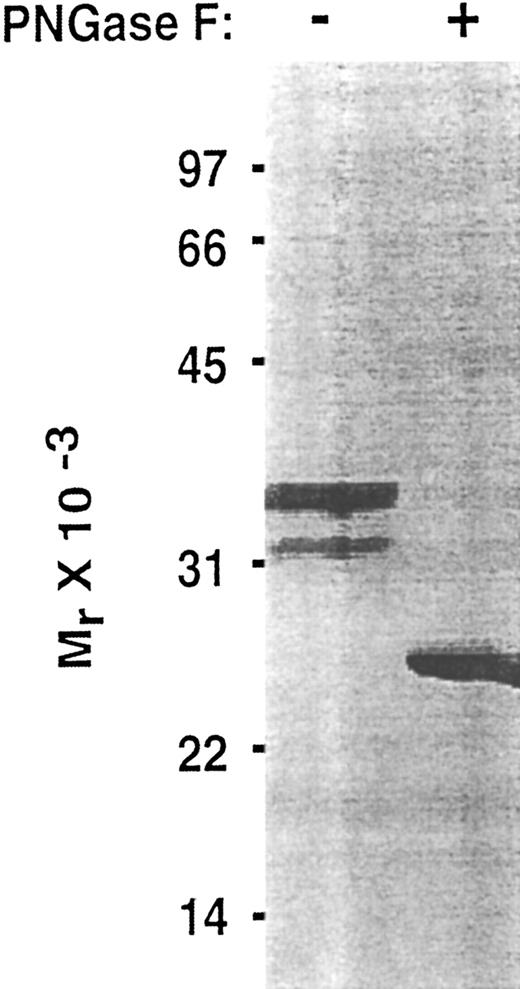
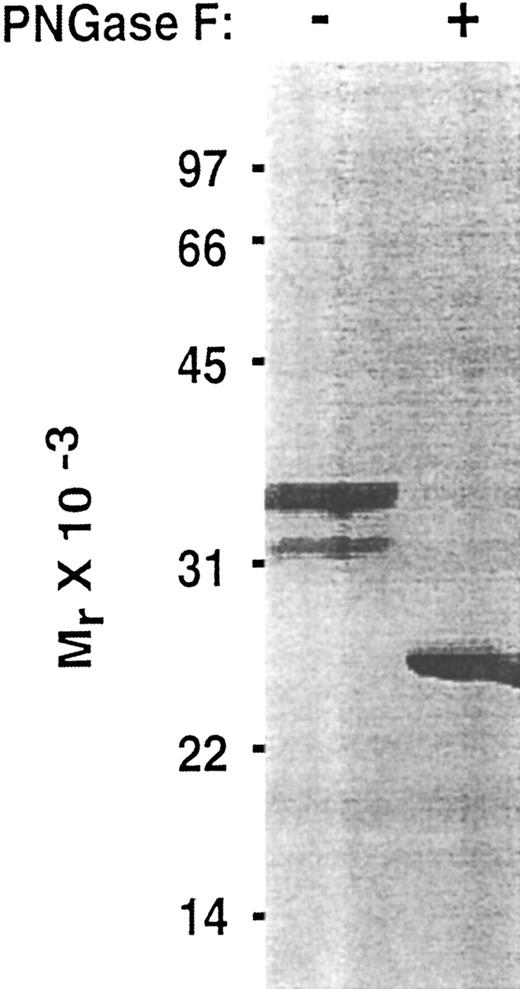
To determine whether the lectin and EGF domains of P-selectin are sufficient to bind PSGL-1, we inserted into an expression vector a cDNA that encoded only the signal peptide, lectin domain, and EGF domain. To facilitate purification and analysis, this Lec-EGF construct also encoded a C-terminal extension consisting of a 4-residue cleavage site for factor Xa and an 8-residue epitope for the Ca2+-dependent MoAbs HPC4 and 1478 (Fig 1). 293 cell clones permanently transfected with this cDNA secreted a protein that was detected with a sandwich ELISA using MoAb HPC4, which binds to the epitope tag, and MoAb G1, which binds to an epitope in the lectin domain. Lec-EGF protein from conditioned medium of the highest-expressing clone was isolated by HPC4 immunoaffinity chromatography. When analyzed by SDS-PAGE and silver staining, Lec-EGF migrated as three distinct species with apparent Mrs of 38,000, 33,000, and 29,000, with the smallest species being much less abundant (Fig 2). In Western blots, the MoAbs HPC4 and G1 recognized all three species, confirming that they were isoforms of Lec-EGF (data not shown). After treatment with PNGase F to remove N-linked glycans, Lec-EGF migrated as a single species with an apparent Mr of 25,000 (Fig 2). There are three potential attachment sites for N-glycans on the lectin and EGF domains of P-selectin.39 The data in Fig 2 suggest that most Lec-EGF molecules have three N-glycans, but there are less common glycoforms with only one or two N-glycans.
Effect of PNGase F treatment of Lec-EGF. Purified Lec-EGF was incubated in the presence or absence of PNGase F. The treated samples were analyzed by SDS-PAGE in a 12.5% polyacrylamide gel under reducing conditions, followed by silver staining.
Effect of PNGase F treatment of Lec-EGF. Purified Lec-EGF was incubated in the presence or absence of PNGase F. The treated samples were analyzed by SDS-PAGE in a 12.5% polyacrylamide gel under reducing conditions, followed by silver staining.
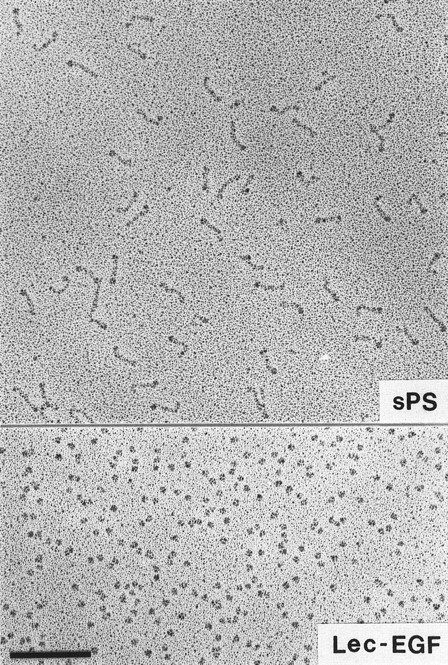
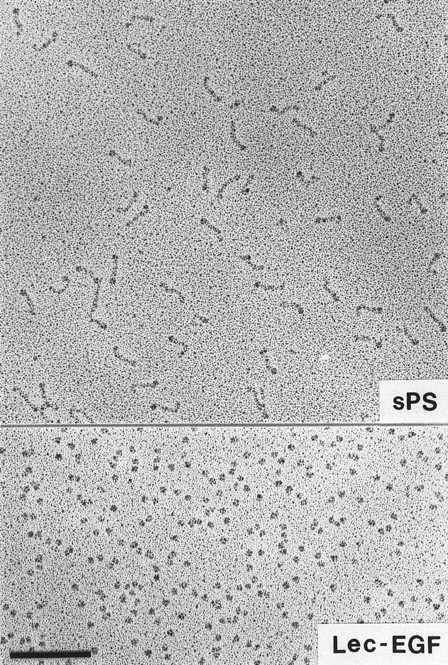
Visualization of Lec-EGF and sPS by electron microscopy.Lec-EGF sedimented as a single peak at 3.5 seconds in a glycerol gradient, relative to a set of standard proteins sedimented in a parallel gradient; this S value is probably less accurate than the S value determined in aqueous buffer (see below). Electron micrographs of rotary-shadowed Lec-EGF showed monomeric globular molecules (Fig 3). These molecules were estimated to be 4 nm in diameter, after subtracting 2 nm contributed by the shell of metal. For comparison, Fig 3 also shows a field of sPS molecules that was previously shown to be highly extended with a globular domain at one end.30 The size and shape of the Lec-EGF molecule is very similar to the globular end of sPS, which presumably corresponds to the lectin and EGF domains.
Electron microscopy of rotary-shadowed P-selectin molecules. sPS molecules are elongated with a globular domain at one end of each molecule as observed previously.30 The globular Lec-EGF molecules are similar in size to the globular ends of the sPS molecules. Original magnification × 150,000. The bar indicates 100 nm.
Electron microscopy of rotary-shadowed P-selectin molecules. sPS molecules are elongated with a globular domain at one end of each molecule as observed previously.30 The globular Lec-EGF molecules are similar in size to the globular ends of the sPS molecules. Original magnification × 150,000. The bar indicates 100 nm.
Sedimentation equilibrium and sedimentation velocity analysis of Lec-EGF.Sedimentation equilibrium analysis showed that Lec-EGF was clearly monomeric, with a solution molecular weight of 28,100. Sedimentation velocity analysis showed that Lec-EGF had a sedimentation coefficient (S20,w) of 2.34. Assuming a reasonable range of hydration (0.3 to 0.4 g H2O/g protein), Lec-EGF sedimented with a calculated equivalent prolate axial ratio under 4. Thus, Lec-EGF, like sPS, is monomeric in aqueous solutions. But sPS is much more asymmetric with a calculated equivalent prolate axial ratio of 19.30
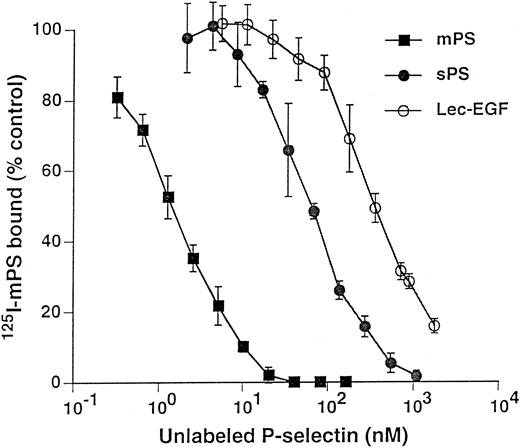
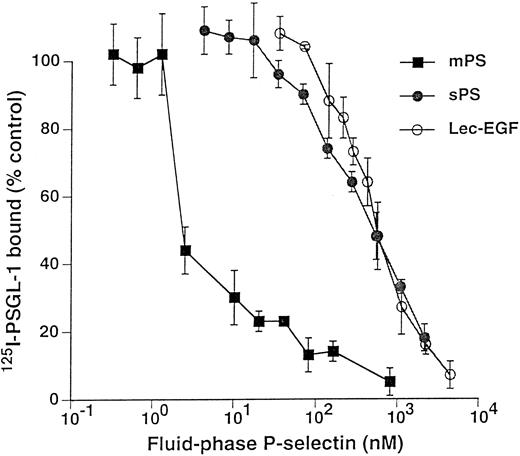
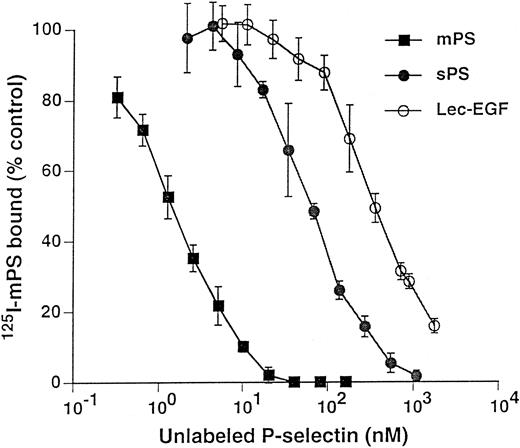
Unlabeled fluid-phase Lec-EGF inhibits binding of 125I-mPS to neutrophils.Fluid-phase 125I-mPS binds saturably to human neutrophils in a Ca2+-dependent manner.30,48 Unlabeled fluid-phase sPS inhibits binding of 125I-mPS, indicating that oligomeric and monomeric forms of P-selectin bind to the same sites.30 These binding sites correspond to PSGL-1, because they are removed or blocked by treatment of the cells with O-sialoglycoprotein endopeptidase or by the anti–PSGL-1 MoAb PL1.7,15,30 Figure 4 shows that increasing concentrations of unlabeled Lec-EGF also inhibited binding of a fixed concentration of 125I-mPS to neutrophils. The IC50 of Lec-EGF was approximately fivefold higher than the IC50 of sPS, which was ∼100 nmol/L, confirming previous results.30 As shown previously,30 unlabeled mPS had an IC50 of only 3 nmol/L, reflecting the enhanced binding avidity contributed by oligomerization. These results indicate that Lec-EGF binds to PSGL-1 on neutrophils. In this assay, the equilibrium binding affinity of Lec-EGF was slightly lower than that of sPS.
Competitive inhibition of fluid-phase 125I-mPS binding to neutrophils by unlabeled mPS, sPS, or Lec-EGF. Neutrophils (1.5 × 106 cells; final volume, 500 μL) were incubated with a near-saturating concentration of 125I-mPS (4.2 nmol/L) and increasing concentrations of unlabeled mPS, sPS, or Lec-EGF for 1 hour at 4°C. Specific binding was determined as described in the Materials and Methods. Each point represents the mean ± standard deviation (SD) of triplicate assays in a representative experiment. Similar results were obtained in three other experiments using different preparations of unlabeled fluid-phase mPS, sPS, and Lec-EGF.
Competitive inhibition of fluid-phase 125I-mPS binding to neutrophils by unlabeled mPS, sPS, or Lec-EGF. Neutrophils (1.5 × 106 cells; final volume, 500 μL) were incubated with a near-saturating concentration of 125I-mPS (4.2 nmol/L) and increasing concentrations of unlabeled mPS, sPS, or Lec-EGF for 1 hour at 4°C. Specific binding was determined as described in the Materials and Methods. Each point represents the mean ± standard deviation (SD) of triplicate assays in a representative experiment. Similar results were obtained in three other experiments using different preparations of unlabeled fluid-phase mPS, sPS, and Lec-EGF.
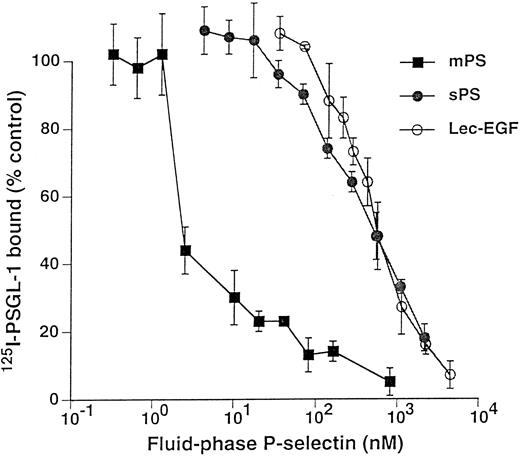
Fluid-phase Lec-EGF inhibits binding of 125I–PSGL-1 to immobilized mPS.125I-PSGL-1 binds specifically to immobilized mPS; the binding is Ca2+-dependent and is blocked by MoAbs to P-selectin and to PSGL-1.15 38 Figure 5 shows that increasing concentrations of fluid-phase mPS, sPS, or Lec-EGF inhibited binding of a fixed concentration of 125I–PSGL-1 to immobilized mPS. The IC50 for mPS was markedly lower than those of sPS or Lec-EGF, again reflecting its higher avidity of binding because of its oligomeric state. In this assay, the IC50 values of sPS and Lec-EGF were indistinguishable. These data show that Lec-EGF binds to purified PSGL-1 with an equilibrium affinity that is similar to that of sPS.
Competitive inhibition of 125I–PSGL-1 binding to immobilized mPS by fluid-phase mPS, sPS, or Lec-EGF. Microtiter wells were coated with mPS (5 μg/mL) and then blocked with HSA. To each well was added a fixed concentration of 125I–PSGL-1 with increasing concentrations of unlabeled mPS, sPS, or Lec-EGF. After incubation for 1 hour at room temperature, specific binding of 125I–PSGL-1 was determined as described in the Materials and Methods. Each point represents the mean ± SD of triplicate assays in a single experiment. Similar results were obtained in three other experiments using different preparations of unlabeled fluid-phase mPS, sPS, and Lec-EGF.
Competitive inhibition of 125I–PSGL-1 binding to immobilized mPS by fluid-phase mPS, sPS, or Lec-EGF. Microtiter wells were coated with mPS (5 μg/mL) and then blocked with HSA. To each well was added a fixed concentration of 125I–PSGL-1 with increasing concentrations of unlabeled mPS, sPS, or Lec-EGF. After incubation for 1 hour at room temperature, specific binding of 125I–PSGL-1 was determined as described in the Materials and Methods. Each point represents the mean ± SD of triplicate assays in a single experiment. Similar results were obtained in three other experiments using different preparations of unlabeled fluid-phase mPS, sPS, and Lec-EGF.
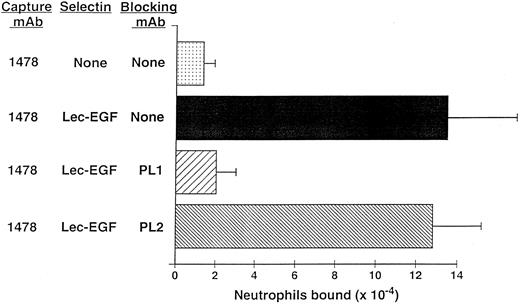
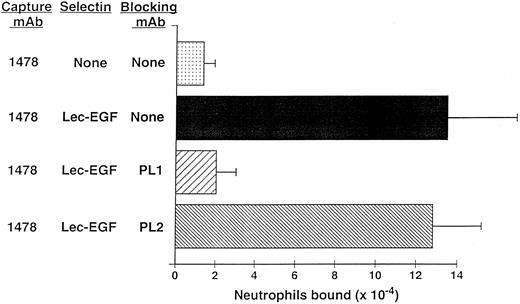
Neutrophils use PSGL-1 to adhere to MoAb-captured Lec-EGF under static conditions.The anti–PSGL-1 MoAb PL1 blocks static neutrophil adherence to P-selectin when it is immobilized over a wide range of densities, indicating that PSGL-1 is the predominant ligand for P-selectin in this multivalent adhesion assay.15 Under static conditions, neutrophils also adhered to Lec-EGF when it was captured at its C-terminus by the immobilized MoAb 1478 (Fig 6). The anti–PSGL-1 MoAb PL1 completely prevented adhesion, whereas the nonblocking anti–PSGL-1 MoAb PL2 had no effect. These results show that PSGL-1 is also the major ligand on neutrophils for Lec-EGF. Thus, the absence of the CRs does not detectably alter the binding specificity of P-selectin for ligands on the neutrophil surface.
Neutrophil adhesion to MoAb-captured Lec-EGF under static conditions. Microtiter wells were coated with the MoAb 1478. Lec-EGF was captured in the indicated wells at a density of 2,780 sites/μm2. Neutrophils preincubated in the presence or absence of the anti–PSGL-1 MoAbs PL1 or PL2 were then added to the wells. Adhesion under static conditions was measured as described in the Materials and Methods. The data represent the mean ± SD of triplicate determinations. Similar results were obtained in two other experiments.
Neutrophil adhesion to MoAb-captured Lec-EGF under static conditions. Microtiter wells were coated with the MoAb 1478. Lec-EGF was captured in the indicated wells at a density of 2,780 sites/μm2. Neutrophils preincubated in the presence or absence of the anti–PSGL-1 MoAbs PL1 or PL2 were then added to the wells. Adhesion under static conditions was measured as described in the Materials and Methods. The data represent the mean ± SD of triplicate determinations. Similar results were obtained in two other experiments.
Neutrophils tether to and roll on Lec-EGF under hydrodynamic flow conditions.Selectins mediate the tethering and rolling of flowing leukocytes on the vessel wall under shear stresses.1,2 This dynamic, reversible adhesion requires that selectin-ligand bonds have fast on and off rates with high tensile strength.49 To determine whether bonds between Lec-EGF and PSGL-1 have similar features, we examined the ability of flowing neutrophils to tether to and roll on Lec-EGF that was captured by immobilized MoAb 1478. For comparison, we measured the rolling of neutrophils on sPS that was captured with S12, an MoAb that recognizes an epitope that requires the sixth CR.
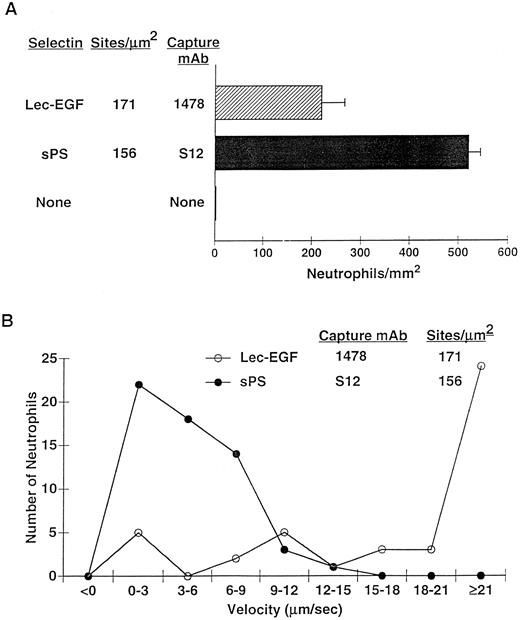
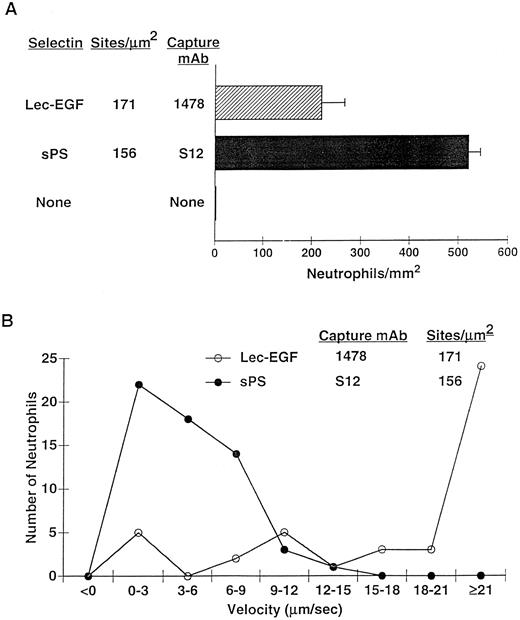
After 4 minutes of perfusion at a shear stress of 1 dyn/cm2, significant numbers of rolling neutrophils accumulated on 1478-captured Lec-EGF, although more neutrophils accumulated on S12-captured sPS at matched densities (Fig 7A). Neutrophils also accumulated on Lec-EGF at shear stresses of 2 and 3 dyn/cm2, although to a lesser degree (data not shown). At 1 dyn/cm2, neutrophils rolled significantly faster on 1478-captured Lec-EGF than on S12-captured sPS (Fig 7B). Rolling of neutrophils on both Lec-EGF and sPS was blocked by the anti–PSGL-1 MoAb PL1 or the anti–P-selectin MoAb G1, confirming that adhesion was mediated through interactions of PSGL-1 with the P-selectin constructs (data not shown). These data show that flowing neutrophils tether to and roll on Lec-EGF when it is captured with an immobilized MoAb to a C-terminal epitope.
Neutrophil adhesion to MoAb-captured Lec-EGF or sPS under hydrodynamic flow conditions. Lec-EGF or sPS was captured at the indicated density on 35-mm dishes with immobilized MoAb 1478 or S12, respectively. As a control, one dish was blocked with HSA but was not incubated with MoAb or selectin. Each dish was inserted into a parallel-plate flow chamber. (A) Neutrophils were perfused through the chamber at a wall shear stress of 1 dyn/cm2. After 4 minutes, the number of adherent cells (all of which rolled on the surface) were quantified. (B) The velocities of 50 individual neutrophils rolling on either Lec-EGF or sPS were measured. Similar results were obtained in two other experiments.
Neutrophil adhesion to MoAb-captured Lec-EGF or sPS under hydrodynamic flow conditions. Lec-EGF or sPS was captured at the indicated density on 35-mm dishes with immobilized MoAb 1478 or S12, respectively. As a control, one dish was blocked with HSA but was not incubated with MoAb or selectin. Each dish was inserted into a parallel-plate flow chamber. (A) Neutrophils were perfused through the chamber at a wall shear stress of 1 dyn/cm2. After 4 minutes, the number of adherent cells (all of which rolled on the surface) were quantified. (B) The velocities of 50 individual neutrophils rolling on either Lec-EGF or sPS were measured. Similar results were obtained in two other experiments.
DISCUSSION
We have expressed a soluble form of P-selectin (Lec-EGF) that contains only the lectin and EGF domains. Electron microscopy and hydrodynamic data indicate that Lec-EGF is monomeric. This enabled direct functional comparison of Lec-EGF with sPS that includes all nine CRs as well as the lectin and EGF domains. Using competitive equilibrium binding assays, we show that Lec-EGF inhibits the binding of oligomeric mPS to PSGL-1 on intact neutrophils and to purified PSGL-1. The IC50 for inhibiting binding to neutrophils is fivefold greater for Lec-EGF than for sPS, whereas the IC50 for inhibiting binding to purified PSGL-1 is indistinguishable for Lec-EGF and sPS. The slight differences observed between the two assays may be due to very small amounts of undetected oligomers in either Lec-EGF or sPS that cause minor shifts in the IC50 values. Regardless, the data show that Lec-EGF and sPS bind to PSGL-1 with similar equilibrium affinities. Thus, the CRs have at most a relatively minor role in enhancing binding of fluid-phase P-selectin to its physiologic ligand. Furthermore, the CRs do not detectably alter the binding specificity of P-selectin, because the anti–PSGL-1 MoAb PL1 blocks static adhesion of neutrophils to either Lec-EGF (this study) or sPS.15 We cannot formally exclude the possibility that the 12-residue C-terminal epitope tag and factor Xa cleavage site modulate binding of Lec-EGF. Even if this were true, there is no requirement for a specific CR.
Assays that measure the kinetics of binding of fluid-phase P-selectin to PSGL-1 are required to determine whether the CRs alter the kon and koff without affecting the overall affinity. However, antibody-captured Lec-EGF does support tethering and rolling of neutrophils under hydrodynamic flow, suggesting that it still binds to PSGL-1 with rapid on and off rates and significant mechanical strength. Neutrophils accumulate in greater numbers and roll at slower velocities on S12-captured sPS than on 1478-captured Lec-EGF. This advantage could reflect the ability of the CRs to increase the kon and/or decrease the koff of binding of P-selectin to PSGL-1. However, the advantage more likely derives from the ability of S12, which binds to the sixth CR of P-selectin, to project the lectin domain of sPS relatively far from the plastic surface. By contrast, 1478, which binds to the C-terminus of the relatively short Lec-EGF protein, probably positions the lectin domain a lesser distance above the surface. Interestingly, neutrophils tether and roll poorly on either sPS or Lec-EGF when they are captured with 5A8, a nonblocking MoAb that binds to an epitope in the lectin and/or EGF domains (Mehta P, Patel KD, and McEver RP, unpublished observations). These data extend previous observations on the ability of flowing neutrophils to attach to and roll on P-selectin constructs of various lengths that are expressed on the surface of transfected Chinese hamster ovary cells.27 Neutrophils roll poorly on membrane-anchored P-selectin containing the lectin and EGF domains plus only the first two CRs. However, neutrophils roll normally on a chimeric protein in which four heterologous CRs from the plasma C4b-binding protein are inserted between the two P-selectin CRs and the transmembrane domain. Collectively, the results suggest that a major function of the CRs of P-selectin is to extend the lectin and EGF domains sufficiently above the cell surface for rapid, efficient contact with PSGL-1 on flowing leukocytes.
Our data can be contrasted with studies suggesting that fluid-phase L- and E-selectin require their CRs for optimal ligand recognition. When expressed as an Ig chimera, an L-selectin construct lacking the two CRs binds much less well to several different glycosylated ligands than does an L-selectin Ig chimera that includes the two CRs.28 It was not determined whether either chimera oligomerizes in solution. Thus, it remains unclear whether the CRs enhance the true affinity of binding of the L-selectin Ig chimera to its ligands or, instead, increase the avidity of binding through oligomerization. Oligomerization of the L-selectin Ig chimera might not represent the self-association potential of the extracellular domains of L-selectin, since Igs themselves can self-associate. Although soluble proteins that contain only the extracellular domains of E-selectin or P-selectin are clearly monomeric,29,30,38,50 a similar analysis of the extracellular domains of L-selectin has not been performed. Fluid-phase, monomeric E-selectin containing the lectin and EGF domains plus all six CRs inhibits adhesion of neutrophils to immobilized E-selectin, whereas fluid-phase proteins containing only the lectin and EGF domains or the lectin and EGF domains plus the first two CRs do not inhibit adhesion even at high concentrations.29 These data imply that more than two CRs are required for optimal binding of E-selectin, because the fluid-phase construct with two CRs functions no better in inhibiting cell adhesion than the construct with no CRs.29 However, the ligand-binding function of an Ig chimera containing the lectin domain of E-selectin fused to the EGF domain and two CRs of L-selectin appears similar to that of an Ig chimera containing all the extracellular domains of E-selectin.51
It remains possible that the CRs are critical for enhancing the binding affinities of L- and E-selectin for their ligands. However, our data show that the lectin and EGF domains are sufficient for most, and perhaps all, of the binding affinity of P-selectin for its physiologic ligand, PSGL-1. The CRs of P-selectin may function primarily to extend the lectin and EGF domains above the surface to enhance encounters with PSGL-1 on flowing leukocytes.
ACKNOWLEDGMENT
The authors thank Drs Greg Bliss and Shigeru Ushiyama for performing pilot experiments during the early phases of this work, Dr Kevin Moore for providing purified PSGL-1, and Cindy Carter, Ginger Hampton, and Chris Titsworth for technical assistance. We are grateful to Drs Richard Cummings and Kevin Moore for critically reading the manuscript. We also thank Drs Charles and Naomi Esmon for providing the MoAbs HPC4 and 1478 and Dr Kenneth Jackson for the amino acid compositional analysis of Lec-EGF.
Supported by National Institutes of Health Grants No. HL 45510 and CA 47056 and by Grant No. DIR 9002027 from the National Science Foundation. K.D.P. is the recipient of a National Research Service Award from the National Institutes of Health.
Address reprint requests to Rodger P. McEver, MD, W. K. Warren Medical Research Institute, University of Oklahoma Health Sciences Center, 825 NE 13th St, Oklahoma City, OK 73104.