Abstract
The platelet glycoprotein Ib (GpIb) complex is composed of four polypeptides: the disulfide-linked GpIbα and GpIbβ and the noncovalently associated GpIX and GpV. GpIbα contains binding sites for von Willebrand factor and for thrombin and mediates platelet adhesion to the subendothelium under conditions of high shear stress. We have previously shown the presence of GpIbα and GpIbβ mRNA and protein in cultured human umbilical vein endothelial cells (HUVECs) as well as the presence of GpIbα mRNA and protein in tonsillar endothelium. We, therefore, probed ECs for the presence of the other components of the GpIb/IX/V complex. We have identified the presence of GpIX and GpV mRNA in cultured HUVEC monolayers. The sequence of HUVEC GpIX cDNA was identical to the previously published human erythroleukemia (HEL) cell GpIX cDNA sequence. Two species of GpV mRNA, one of 3 kb and one of 4.4 kb, were found in HUVECs, whereas HEL cells displayed only the 4.4-kb species and the megakaryocytic cell line CHRF-288 contained only the 3-kb species. We previously showed that EC GpIbα protein is identical in molecular weight to platelet GpIbα. HUVEC GpIbβ, in contrast to its platelet counterpart, has a molecular weight of 50 kD and forms a correspondingly larger disulfide-bonded complex with EC GpIbα. The molecular weights of GpIX and GpV were 22 and 88 kD, respectively, identical to the corresponding platelet polypeptides. Furthermore, we have identified all four components of the complex in tonsillar vessels. Using flow cytometry, we have established that all four polypeptides of the GpIb/IX/V complex are expressed on the surface membranes of cultured HUVECs and adult aortic ECs. Furthermore, using two-color fluorescence, we have shown that all ECs expressing GpIbα also express GpIX and GpV on their surface. The ratio of GpIbα:GpIX:GpV is 1:1:0.5, which is identical to the ratio present in platelets. None of the polypeptides of the GpIb complex could be identified on the surface of human smooth muscle cells or lymphocytes. The presence of all members of the GpIb complex in the EC membrane suggests that this complex may play a role in endothelial function in vivo.
THE PLATELET glycoprotein (Gp) Ib complex is composed of four membrane-spanning polypeptide chains: GpIbα, GpIbβ, GpIX, and GpV.1,2 GpIbα is a 143-kD protein disulfide-linked to the 22-kD GpIbβ chain. The 20-kD GpIX is tightly but noncovalently bound to GpIbα-β in a 1:1 complex and is coprecipitated by antibodies to GpIbα. Platelet GpV, an 83-kD protein, is more loosely associated with GpIb and GpIX in a molar ratio of 0.5:1.3 GpIbα contains specific binding sites for von Willebrand factor (vWF ) and for thrombin.1,2 The congenital bleeding disorder, Bernard-Soulier syndrome, is caused by mutations in genes of this complex, primarily GpIbα or GpIX, usually resulting in absence or decreased expression of all four components of the complex on the platelet membrane.4
We5 and others6 have published evidence that human umbilical vein endothelial cells (HUVECs) synthesize and express in their membrane a protein closely related to or identical with GpIbα. Furthermore, we have shown tumor necrosis factor-α–enhanced expression of GpIbα mRNA and protein in human endothelium.7,8 In addition, we recently reported the cDNA sequence of a novel HUVEC GpIbβ species and the presence of the corresponding protein in cultured HUVECs.9 We now report the presence and membrane expression of all four components of the GpIb/IX/V complex in HUVECs and adult aortic ECs (HAECs).
MATERIALS AND METHODS
Materials
Collagenase was purchased from Boehringer Manheim (Indianapolis, IN). Tissue culture flasks were obtained from Corning Glassware, Inc (Corning, NY). Fungizone, penicillin–streptomycin sulfate, medium 199, RPMI-1640, L-glutamine, and 0.05% trypsin-0.53 mmol/L EDTA were purchased from GIBCO/BRL (Grand Island, NY). Endothelial cell growth factor was kindly provided by Dr Elliot Levine (The Wistar Institute, Philadelphia, PA). Heparin was purchased from Sigma Chemical Co (St Louis, MO). Fetal bovine serum was obtained from Hyclone Labs (Logan, UT). Oligo (dT)-cellulose was purchased from Collaborative Biomedical Products (Bedford, MA). Hybond-N was obtained from Amersham (Arlington Heights, IL). Avian myeloblastosis virus reverse transcriptase was purchased from Seikagaku America (Rockville, MD). Mouse antirabbit IgG, rabbit antimouse IgG, and alkaline phosphatase-antialkaline phosphatase (APAAP) complex were obtained from the DAKO Corp (Carpenteria, CA).
Antibodies
AP-1,10 6D1,11 and Ib1,12 all murine monoclonal antibodies (MoAbs) specific for human platelet GpIbα, were generous gifts of Dr Robert Montgomery (The Blood Center of Southeastern Wisconsin, Milwaukee, WI), Dr Barry Coller (Mount Sinai Medical Center, New York, NY), and Dr Zaverio Ruggeri (The Scripps Research Institute, La Jolla, CA), respectively. The GpIX-specific MoAbs FMC-2513 and KMP-914 were gifts of Dr Michael Berndt (University of Sidney, Sidney, Australia) and Dr Shosaku Nomura (Kansai Medical University, Osaka, Japan), respectively. The GpV-specific MoAb 1D9 was prepared as previously described.15 Polyclonal rabbit antibody to human glycocalicin (the majority of the extracellular portion of GpIbα) was a generous gift of Dr Graham Jamieson (The American Red Cross Jerome H. Holland Laboratories, Rockville, MD). Affinity-purified rabbit antibodies to human platelet GpIbβ, GpIX, and GpV16,17 were generous gifts of Dr Gerald Roth (University of Washington, Seattle, WA). A platelet glycocalicin-specific rabbit polyclonal antibody and a platelet GpIbβ-specific rabbit polyclonal antibody raised against two extracellular sequences of the GpIbβ chain were produced in this laboratory, as described.18 An MoAb specific for LFA-3, used as a positive control in flow cytometry of lymphocytes (see below), was a gift of Dr Bice Perussia (Kimmel Cancer Institute, Thomas Jefferson University, Philadelphia, PA). An MoAb to the human integrin β1 chain (clone P4C10; GIBCO/BRL) was used as a positive control in the flow cytometry of smooth muscle cells. MOPC 21 (a mouse IgG1κ), normal mouse, and rabbit IgG and phycoerythrin (PE)-labeled goat antimouse IgG were obtained from Sigma Chemical Co and used as negative controls for the appropriate primary antibodies. Fluorescein isothiocyanate (FITC)-labeled goat antimouse IgG (Fab′)2 and goat antirabbit IgG (Fab′)2 , biotin-labeled goat antimouse IgG, and alkaline phosphatase-labeled streptavidin were purchased from Kirkegaard & Perry Laboratories (Gaithersburg, MD).
Cell Culture
HUVECs were harvested from human umbilical cords and cultured at 37°C in a 5% CO2 incubator essentially as described.5 For immunostaining, confluent HUVECs were detached with 0.05% trypsin-0.53 mmol/L EDTA, seeded at half-confluent concentrations onto four-chamber Lab-Tek microscope slides (Nunc, Inc, Naperville, IL) that had been coated with 0.2% gelatin, and grown to confluence (in 2 to 3 days). For flow cytometry and Western blotting, HUVECs were detached in a different buffer (0.01 mol/L phosphate, 0.15 mol/L NaCl, 5 mmol/L NaHCO3 , 10 mmol/L EDTA, 5 mmol/L EGTA, 0.1% bovine serum albumin, 1 mmol/L phenylmethyl sulfonyl fluoride [PMSF ], pH 7.2). HAECs were obtained from Dr Elliot Levine (The Wistar Institute) at a cumulative population doubling of 16. Human erythroleukemia (HEL) cells were obtained from the American Type Culture Collection (ATCC; Rockville, MD; ATCC TIB 180, HEL 92.1.7) and propagated in RPMI 1640 with 10% fetal bovine serum. CHRF-288 cells (kindly provided by Dr M. Lieberman, Children's Hospital Medical Center, Cincinnati, OH) were propagated in Fischer's medium with 20% horse serum (GIBCO/BRL). Human aortic smooth muscle cells were obtained from Dr Elaine Tan (Department of Anatomy, Pathology and Cell Biology, Thomas Jefferson University) and passaged in RPMI/medium 199 containing 20% fetal calf serum, fungizone, penicillin, and streptomycin. We thank Dr James San Antonio (Cardeza Foundation, Thomas Jefferson University) for providing the passaged cells. Smooth muscle cells were used at passage four.
RNA Isolation and Evaluation
Total cellular RNA was isolated by solubilization of confluent cells in guanidine hydrochloride, as previously described.7 19 Poly (A+) RNA was isolated directly from cells using oligo (dT)-cellulose and analyzed by electrophoresis in a denaturing formaldehyde gel followed by Northern blotting onto a nylon membrane. The RNA was fixed to the membrane with UV irradiation; prehybridized in a solution of 1 mol/L NaCl, 0.1% sodium dodecyl sulfate (SDS), 1.5 mg/mL sonicated herring sperm DNA, and 10% dextran for 3 hours at 65°C, hybridized at 65°C for 12 to 24 hours in the prehybridization solution with additional herring sperm DNA (1.5 mg/mL) and radiolabeled GpV cDNA probe (kindly provided by Dr Gerald Roth, University of Washington, Seattle, WA). The blot was washed to high stringency in 0.1× SSC (2.25 mol/L NaCl, 0.225 mol/L sodium citrate), 0.1 % SDS, 1 mmol/L Na2 EDTA, 10 mmol/L sodium phosphate, pH 6.8, at 65°C and analyzed by autoradiography.
Nuclear Run-On Transcription Assay
Fourth passage HUVECs were washed with Dulbecco's Ca2+- and Mg2+-free phosphate-buffered saline (D-PBS), exposed to trypsin-EDTA for 10 seconds, and harvested by rinsing with D-PBS. Nuclei were harvested by gently vortexing the cells in Nonidet 40 lysis buffer (10 mmol/L Tris-HCl, pH 7.4, 100 mmol/L NaCl, 3 mmol/L MgCl2 , 0.5% Nonidet 40), followed by centrifugation at 800g. Newly transcribed RNA was isolated as described by Lindsten et al.20 The radiolabeled RNA was hybridized to pUC-based plasmids (5 μg) with the gene-specific inserts indicated, denatured by alkali treatment, and transferred to nitrocellulose filters in a slot blot apparatus. Plasmids contained cDNA for a 3.8-kb vWF gene fragment (pVWH33)21 or full-length cDNAs for GpIbα,22 GpIbβ,23 or GpIX.24 The plasmid pGEM without a cDNA insert was used as a control.
Polymerase Chain Reaction (PCR) Amplification and Sequence Analysis
GpIX was amplified from reverse-transcribed HUVEC, HEL cell, and CHRF-288 RNA using PCR, as previously described.25 Briefly, 1 μg of total cellular RNA was used as template with 10 U of avian myeloblastosis virus reverse transcriptase for 1 hour at 41°C in 50 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 8 mmol/L MgCl2 , 10 mmol/L dithiothreitol, and dNTPs at 500 mmol/L each in a total volume of 50 μL with 0.02 A260 units of oligo(dT)16 as primer. Ten microliters of the reverse transcriptase reaction was adjusted to PCR buffer conditions in a total volume of 50 μL, with 0.002 A260 units of each PCR primer and 5 U of Taq polymerase (Thermus aquaticus DNA polymerase; Perkin-Elmer Cetus, Norwalk, CT). PCR was performed for 5 cycles in an automated thermocycler (Perkin-Elmer Cetus) with cycle times of 1 minute at 94°C, 1 minute at 55°C, and 2 minutes at 72°C. After this, 0.02 A260 units of each primer was added to the reactions and 30 additional cycles were performed. The GpIX primers (99-118, 500-51924) span the 108-bp intron within the GpIX 5′ untranslated region.26 Hepatoma G2 cell RNA was a gift of Dr Jaime Caro (Cardeza Foundation, Thomas Jefferson University). Genomic DNA was prepared from peripheral blood mononuclear cells and amplified under the conditions described above. For GpIX cDNA sequence analysis, HUVEC RT-RNA was amplifed by PCR as described above using primers spanning nucleotides 99-519 and 421-727 of the cDNA sequence.24 The PCR products were ligated into the vector PCR 1000 (Invitrogen Corp, San Diego, CA), propagated, and sequenced using a modification of the dideoxy-chain termination method to sequence double-stranded DNA.27
Immunohistochemical Studies
For the immunostaining of tonsillar endothelium, fresh tonsils obtained at surgery from two patients were snap-frozen and processed for immunohistochemistry as described.7 Briefly, 5-μm sections were made on a cryostat, mounted on gelatinized glass slides, and fixed with acetone at 4°C for 30 minutes. After the slides were air-dried, they were incubated for 1 hour with affinity-purified rabbit antibodies to platelet glycocalicin or to intact platelet GpIbβ, GpIX, or GpV (the latter 3 supplied by Dr Gerald Roth) or were incubated for 1 hour with normal rabbit IgG. For studies using HUVEC monolayers, confluent cells grown on chamber slides were washed twice with medium 199, air-dried for 2 hours, and fixed with acetone at 4°C for 30 minutes. The same rabbit antibodies were used for HUVECs and for tonsillar sections. For both tonsillar tissue and cultured ECs, a “mousifying” step was introduced by incubation with mouse antirabbit IgG (1:40 dilution) for 30 minutes. The remainder of the immunostaining procedure was performed as previously described.8
Characterization of EC GpIb Polypeptides by Western Blotting
GpIbα and GpIbβ.Affinity-purified rabbit polyclonal anti-glycocalicin (Gc) IgG, prepared in our laboratory,18 was conjugated to CNBr-activated Sepharose CL-4B. Approximately 108 HUVECs were pelleted and incubated for 30 minutes in lysis buffer (0.5% Triton X-100, 0.15 mol/L NaCl, 10 mmol/L Tris-HCl, 1 mmol/L PMSF, 25 mg/mL leupeptin, 10 mmol/L benzamidine, 200 KIU/mL aprotinin, and 2 mg/mL DNase I, pH 7.5). Larger debris was removed by centrifugation and the supernatant was precleared by incubation for 2 hours with Sepharose CL-4B followed by a second centrifugation. The precleared HUVEC lysate was loaded onto a 1.5 × 8 cm anti-Gc–Sepharose CL-4B column and incubated at 4°C for 2 hours. The column was thoroughly washed with 0.01 mol/L phosphate-0.15 mol/L NaCl, pH 7.4, and the bound protein was eluted with 0.05 mol/L diethylamine, pH 11.5. Western blotting was performed after SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of nonreduced and reduced samples in 10% polyacrylamide gels, as described.18 Affinity-purified polyclonal anti-Gc IgG, affinity-purified polyclonal anti-GpIbβ peptide IgG (both prepared in our laboratory), or preimmune rabbit IgG (as a negative control) was incubated with the PVDF membranes for 1 hour, followed by three washes with 20 mmol/L Tris-HCl, 500 mmol/L NaCl, pH 7.4. Membranes were incubated for 1 hour with alkaline phosphatase-labeled goat antirabbit IgG, washed three times, and developed by incubation with phosphatase substrate for 10 minutes.
GpIX and GpV.Approximately 5 × 107 ECs were lysed as described above in lysis buffer modified to contain 1% Triton X-100. The freshly prepared lysates were immediately centrifuged, precleared as described above, loaded onto 1.5 × 3 cm MoAb-Sepharose columns (FMC-25 for isolation of GpIX and 1D9 for isolation of GpV), and incubated overnight at 4°C. The columns were washed extensively with a buffer containing 0.5% Triton, 20 mmol/L Tris, 0.5 mol/L NaCl and PMSF, leupeptin, benzamidine, and aprotinin (at the concentrations used in the lysis buffer) before elution with 0.05 mol/L diethylamine, pH 11.5; for GpV preparation, the elution buffer also contained all four inhibitors. Column eluates were concentrated using Centricon concentrators (No. 4224; Amicon, Beverly, MA) and run on SDS-PAGE (12.5% and 10% polyacrylamide for GpIX and GpV, respectively). Western blotting was performed as described above, using MoAb KMP-9 for GpIX and MoAb 1D9 for GpV, except that, after incubation with the MoAbs and before application of the biotin-labeled goat antimouse IgG and streptavidin-phosphatase, the blots were incubated for 2 hours with normal goat serum (Sigma; 10% for GpIX and 2% for GpV).
Binding of MoAb Ib1 to Platelets and HUVECs
MoAb Ib1 was labeled by the Iodogen method to a specific activity of 5,000 to 6,000 counts per minute (cpm)/ng. Gel-filtered platelets were isolated from platelet-rich plasma on Sepharose CL-2B using modified Tyrode's buffer containing 0.1 μg/mL prostaglandin E1 . HUVECs (105/incubation) and platelets (106/incubation) were incubated in suspension at room temperature for 1 hour with MoAb Ib1 (0 to 20 μg/mL final concentration). The cells were washed three times with PBS containing 3% bovine serum albumin and bound radioactivity was measured in a gamma counter. Nonspecific binding was measured in the presence of a 100-fold excess of unlabeled MoAb and subtracted from the total bound cpm to determine specific bound radioactivity. The number of molecules of bound Ib1 was calculated by Scatchard analysis.
Flow Cytometry
For single antibody studies, a suspension of 106 ECs, human lymphocytes (kindly provided by Dr Bice Perussia, Kimmel Cancer Institute, Thomas Jefferson University), or smooth muscle cells in 100 μL was incubated for 1 hour at 22°C with a saturating concentration (previously determined) of mouse MoAbs Ib1 (anti-GpIbα), FMC-25 (anti-GpIX) or 1D9 (anti-GpV), or rabbit polyclonal antibodies to GpIbα or to platelet GpIbβ peptide. Aliquots of the same cells were incubated with normal mouse or rabbit IgG to assess background binding. As positive controls, lymphocytes and smooth muscle cells were incubated with MoAbs against LFA-3 and the human integrin β1 chain, respectively, as positive controls. All samples were then washed three times by resuspension with PBS and centrifugation at 2,000g for 10 minutes and incubated for 1 hour with a 1:10 dilution of FITC-conjugated goat antimouse IgG or goat antirabbit IgG, after which the cells were washed three times with PBS, resuspended in 200 μL PBS, and analyzed in a Coulter EPICS Profile II fluorescence-activated cell sorter (Coulter Corp, Hialeah, FL). For two-color studies, HUVECs were first incubated for 1 hour with MoAbs FMC-25 or 1D9 or with normal mouse IgG, washed three times with PBS, and then incubated for 1 hour with PE-labeled goat antimouse IgG. The cells were then incubated for 2 hours with normal mouse IgG (2 mg/mL) and washed twice with PBS. Finally, FITC-labeled MoAb Ib1 or FITC-labeled normal mouse IgG, both prepared in our laboratory, was added and incubation was allowed to proceed for 1 hour. The cells were washed three times with PBS and examined in the flow cytometer, as described above.
RESULTS
GpIbα, GpIbβ, GpIX, and GpV Gene Expression in HUVECs
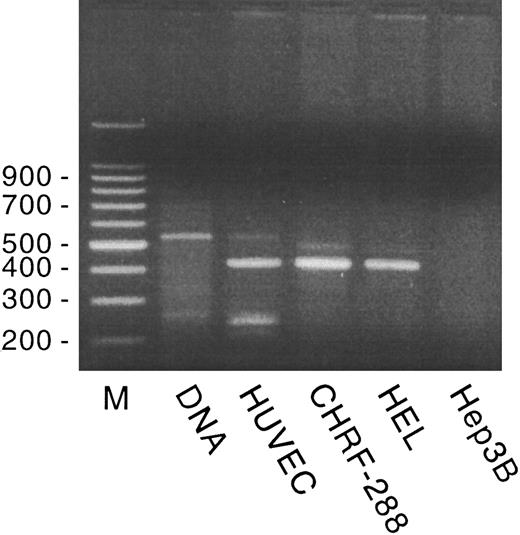
In previously published studies, we showed GpIbα and GpIbβ mRNA expression in HUVECs. Using size comparison and using restriction digest and partial sequence analysis, GpIbα mRNA appears to be identical in HUVECs, platelets, and HEL cells.7,22 However, HUVECs contain only a 3.5-kb GpIbβ mRNA species and express a correspondingly larger protein; megakaryocytes and HEL cells contain only a 1.1-kb species. All of the platelet GpIbβ cDNA sequence is contained within the HUVEC GpIbβ cDNA.9 In the present study, we found that GpIX mRNA is expressed in cultured HUVECs at a very low level. GpIX mRNA could not be detected in Northern blot analysis of poly A+ HUVEC RNA, but could be detected by RT-PCR, as shown in Fig 1. The GpIX PCR primers span the 108-bp intron located within the 5′ untranslated region and amplified a 420-bp fragment from HUVEC, CHRF-288, and HEL cell RT-RNA, a 528-bp fragment from genomic DNA, and no product from hepatoma G2 cell RT-RNA (used as a negative control). PCR fragments spanning the entire GpIX coding region (nt 99-727)24 were subcloned, propagated, and sequenced. The sequence was identical to that published for HEL cell GpIX cDNA by Hickey et al.24 In addition, GpIX, as well as GpIbα and GpIbβ, gene transcription was detected in cultured HUVECs using a nuclear run-on transcription assay (data not shown). vWF cDNA was used as a positive control and the plasmid pGEM was used as a negative control.
GpIX expression in HUVECs, CHRF-288, and HEL cells. Genomic DNA or RT-RNA from HUVECs, CHRF-288, and HEL cells and from hepatoma 3B cells was amplified by PCR, as described in Materials and Methods, using GpIX primers that span the 108-bp intron in the GpIX 5′ untranslated region. A 100-bp marker ladder (M) is shown on the left.
GpIX expression in HUVECs, CHRF-288, and HEL cells. Genomic DNA or RT-RNA from HUVECs, CHRF-288, and HEL cells and from hepatoma 3B cells was amplified by PCR, as described in Materials and Methods, using GpIX primers that span the 108-bp intron in the GpIX 5′ untranslated region. A 100-bp marker ladder (M) is shown on the left.
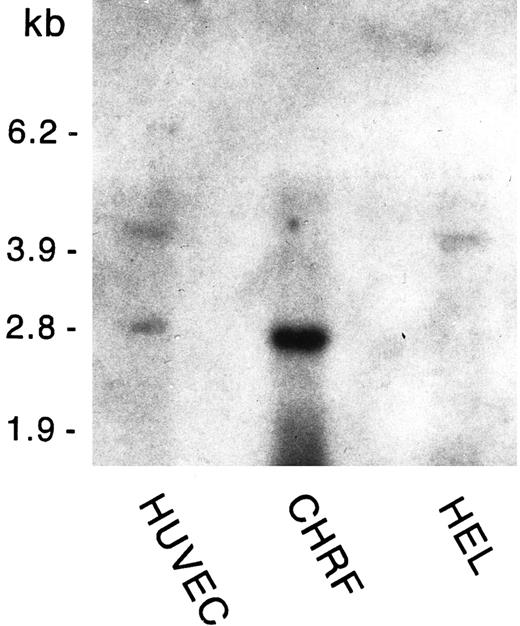
Poly A+-selected RNA from HUVECs, HEL cells, and the megakaryoblastic cell line CHRF-288 was analyzed for GpV mRNA expression. As shown in Fig 2, we detected 4.4- and 3-kb species of GpV mRNA in ECs; HEL cells displayed only the 4.4-kb message, whereas the megakaryoblastic cell line CHRF-288 contained only the 3-kb species. The 4.4-kb message is similar in size to that reported by Lanza et al28 in HEL cells; the 3-kb message is what would be predicted from the GpV genomic sequence if the earlier polyadenylation signal28 is used.
GpV mRNA expression in HUVECs, CHRF, and HEL cells. Poly A+-selected RNA was extracted from confluent HUVECs (third passage), CHRF-288 cells, and HEL cells. Four micrograms of RNA was analyzed by Northern blotting using radiolabeled GpV cDNA as the probe. An RNA size ladder is shown on the left.
GpV mRNA expression in HUVECs, CHRF, and HEL cells. Poly A+-selected RNA was extracted from confluent HUVECs (third passage), CHRF-288 cells, and HEL cells. Four micrograms of RNA was analyzed by Northern blotting using radiolabeled GpV cDNA as the probe. An RNA size ladder is shown on the left.
Immunostaining of HUVECs and Tonsillar Endothelium
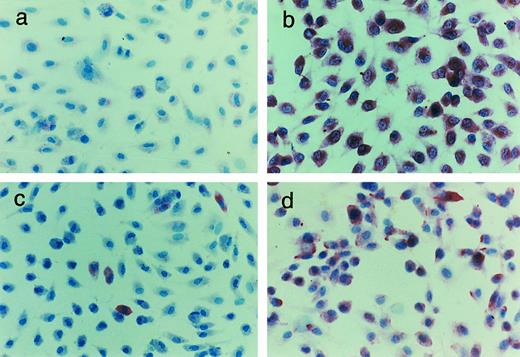
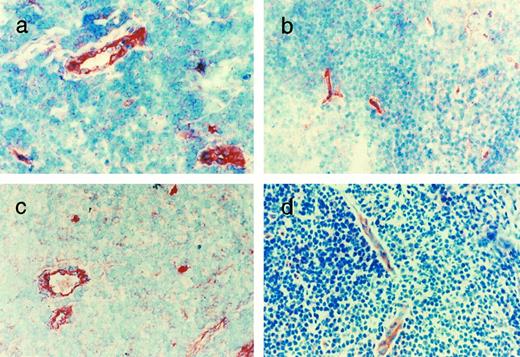
As shown in Fig 3, using polyclonal, affinity-purified antibodies to the entire polypeptide chains, GpIbα, GpIbβ, GpIX, and GpV protein staining was present in confluent HUVEC monolayers. GpIbβ was easily detectable in most cells. GpIX and GpV were detectable in 10% to 15% and 40% to 50% of HUVECs, respectively. As shown in Fig 4, GpIbα, GpIbβ, GpIX, and GpV could be identified in the vascular endothelium of human tonsils in frozen sections. The endothelium of each vessel stained uniformly for each of the four proteins, although the intensity of staining varied somewhat in different areas. The vessels also showed positive staining for GpIbα using the MoAbs AP-1 and 6D1 (data not shown). No other cells or structures in the frozen sections reacted uniformly with these antibodies. Furthermore, in the absence of the primary antibodies, no significant reaction was seen (data not shown).
Immunohistochemical identification of the GpIb complex in HUVECs. Cultured HUVECs were stained with affinity-purified rabbit polyclonal antibodies to the whole glycoprotein chains: (a) GpIbα, (b) GpIbβ, (c) GpIX, and (d) GpV. For experimental details, see Materials and Methods.
Immunohistochemical identification of the GpIb complex in HUVECs. Cultured HUVECs were stained with affinity-purified rabbit polyclonal antibodies to the whole glycoprotein chains: (a) GpIbα, (b) GpIbβ, (c) GpIX, and (d) GpV. For experimental details, see Materials and Methods.
Immunohistochemical identification of the GpIb complex in tonsillar endothelium. Sections of tonsil were stained with affinity-purified rabbit polyclonal antibodies to the whole glycoprotein chains: (a) GpIbα, (b) GpIbβ, (c) GpIX, and (d) GpV. For experimental details, see Materials and Methods.
Immunohistochemical identification of the GpIb complex in tonsillar endothelium. Sections of tonsil were stained with affinity-purified rabbit polyclonal antibodies to the whole glycoprotein chains: (a) GpIbα, (b) GpIbβ, (c) GpIX, and (d) GpV. For experimental details, see Materials and Methods.
Characterization of EC GpIb Complex Polypeptides by Western Blotting
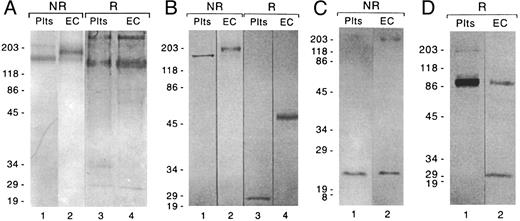
As shown in Fig 5A and B, under nonreducing conditions, bands of approximate molecular weight 190 and 165 kD were seen, respectively, when HUVEC and platelet anti-Gc–Sepharose column eluates were stained with antibodies to GpIbα (Fig 5A) or platelet GpIbβ peptide (Fig 5B). When reduced samples were stained with antibody to GpIbα, a band of approximately 145 kD was seen in both platelet and EC eluates (Fig 5A); the high molecular weight band seen in the reduced samples is nonspecific and was present in the absence of the primary antibody. When reduced samples were developed with the platelet GpIbβ peptide antibody (Fig 5B), a band of molecular weight 50 kD was seen in EC eluates, whereas a band of 24 kD was seen in platelet eluates; no 24-kD band was visualized in EC eluates. As shown in Fig 5C and D, the molecular weights of GpIX and GpV that we purified from ECs were 22 and 88 kD, respectively, which is identical to the sizes we observed in platelets. The 200- and 30-kD bands seen in the EC lanes in Fig 5C and D, respectively, are nonspecific, because they were present when the respective preparations were incubated with the goat antimouse IgG second antibody in the absence of the specific MoAbs (data not shown). The blocking step with normal goat serum decreased these bands considerably, but did not eliminate them completely.
Characterization of the EC GpIb complex by Western blotting. Eluates from an anti-GpIbα affinity column were analyzed for (A) GpIbα and (B) GpIbβ, using affinity-purified rabbit polyclonal antibodies to glycocalicin and to a peptide sequence in platelet GpIbβ, respectively. Eluates from anti-GpIX and anti-GpV affinity columns were analyzed for (C) GpIX and (D) GpV, respectively, using MoAbs. Eluate from 2 to 3 × 106 cells was loaded onto each lane. The numbers to the left of each set of gels are the molecular weights (in kilodaltons) of prestained standards. For experimental details, see Materials and Methods.
Characterization of the EC GpIb complex by Western blotting. Eluates from an anti-GpIbα affinity column were analyzed for (A) GpIbα and (B) GpIbβ, using affinity-purified rabbit polyclonal antibodies to glycocalicin and to a peptide sequence in platelet GpIbβ, respectively. Eluates from anti-GpIX and anti-GpV affinity columns were analyzed for (C) GpIX and (D) GpV, respectively, using MoAbs. Eluate from 2 to 3 × 106 cells was loaded onto each lane. The numbers to the left of each set of gels are the molecular weights (in kilodaltons) of prestained standards. For experimental details, see Materials and Methods.
Surface Expression of GpIbα, GpIbβ, GpIX, and GpV
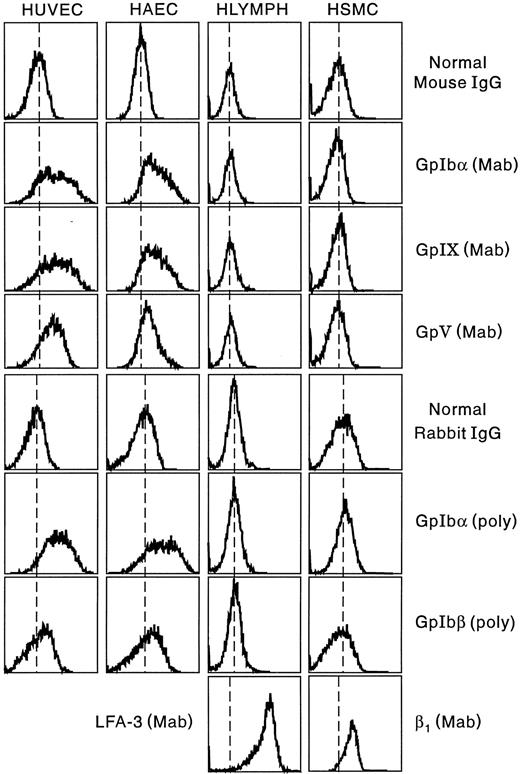
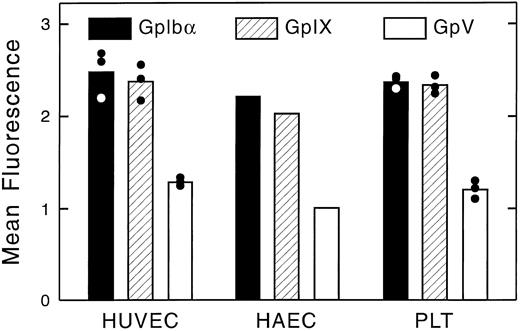
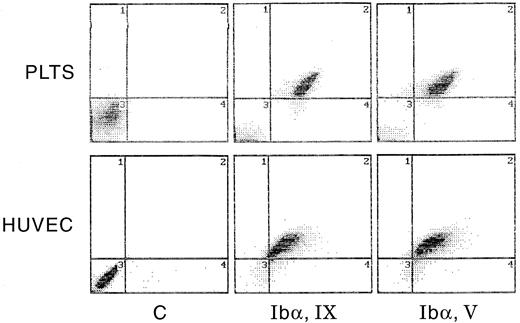
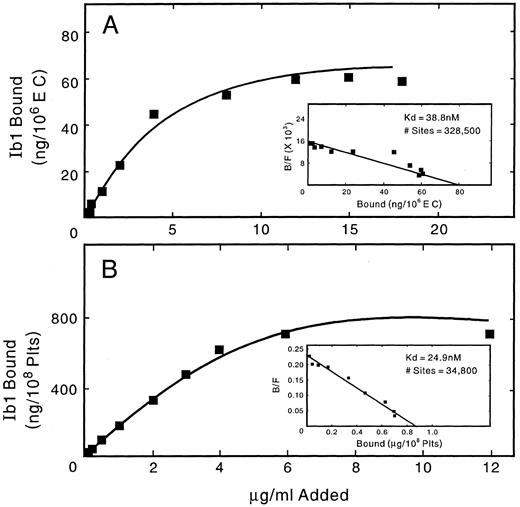
Flow cytometric analysis of the expression of the GpIb complex polypeptides on the surface of HUVECs and HAECs is shown in Fig 6. Both types of ECs show positive staining for each of the four polypeptides, although results are more striking using monoclonal, rather than polyclonal, antibodies. Human lymphocytes and smooth muscle cells did not react with any of the antibodies, whereas these cells were strongly positive with MoAbs to LFA-3 and the human integrin β1 chain, respectively. As shown in Fig 7, surface-bound fluorescence of the mouse MoAbs against GpIbα, GpIX, and GpV gave a ratio of 1:1:0.5 on HUVECs and HAECs. Although possible differences in affinity of the three antibodies makes such an analysis only approximate, the ratios are identical to those we determined for human platelets, as also shown in Fig 7. As shown in Fig 8, two-color fluorescence showed that all cells positive for surface GpIbα were also positive for surface GpIX and GpV. Surface GpIbα was quantified directly by the binding of radiolabeled MoAb Ib1. As shown in Fig 9A, HUVECs bound 328,500 molecules of Ib1 per cell; for comparison, platelets bound 34,800 molecules of Ib1 per platelet (Fig 9B), a result in the expected range. Association constants for the binding were similar, ie, 24.9 nmol/L for platelets and 38.8 nmol/L for HUVECs.
Surface expression of the GpIb complex on ECs, lymphocytes, and smooth muscle cells. For experimental details, see Materials and Methods. Results are shown for MoAbs to GpIbα, GpIX, and GpV and for affinity-purified rabbit polyclonal antibodies to GpIbα (glycocalicin) and a peptide sequence in platelet GpIbβ.
Surface expression of the GpIb complex on ECs, lymphocytes, and smooth muscle cells. For experimental details, see Materials and Methods. Results are shown for MoAbs to GpIbα, GpIX, and GpV and for affinity-purified rabbit polyclonal antibodies to GpIbα (glycocalicin) and a peptide sequence in platelet GpIbβ.
Quantitation of surface fluorescence of GpIbα, GpIX, and GpV on HUVECs, HAECs, and platelets. Surface immunofluorescence was analyzed by flow cytometry, as shown in Fig 6. HUVECs and platelets were analyzed three times; HAECs were studied only once.
Quantitation of surface fluorescence of GpIbα, GpIX, and GpV on HUVECs, HAECs, and platelets. Surface immunofluorescence was analyzed by flow cytometry, as shown in Fig 6. HUVECs and platelets were analyzed three times; HAECs were studied only once.
Paired measurement of GpIbα, GpIX, and GpV by flow cytometry. Human platelets and HUVECs were analyzed by two-color fluorescence for GpIbα and GpIX or GpIbα and GpV. PE-labeled goat antimouse IgG was used to detect GpIX and GpV (abscissa); FITC-labeled MoAb Ib1 was used to detect GpIbα (ordinate). See Materials and Methods for experimental details.
Paired measurement of GpIbα, GpIX, and GpV by flow cytometry. Human platelets and HUVECs were analyzed by two-color fluorescence for GpIbα and GpIX or GpIbα and GpV. PE-labeled goat antimouse IgG was used to detect GpIX and GpV (abscissa); FITC-labeled MoAb Ib1 was used to detect GpIbα (ordinate). See Materials and Methods for experimental details.
Quantitation of surface GpIbα on HUVECs and platelets. HUVEC and platelet suspensions were incubated with 125I-MoAb Ib1, as described in Materials and Methods, and specific binding was analyzed using the method of Scatchard.
Quantitation of surface GpIbα on HUVECs and platelets. HUVEC and platelet suspensions were incubated with 125I-MoAb Ib1, as described in Materials and Methods, and specific binding was analyzed using the method of Scatchard.
DISCUSSION
Using MoAbs and affinity-purified polyclonal antibodies, we have shown the presence of all four constituents of the platelet GpIb/IX/V complex in HUVECs, HAECs, and tonsillar endothelium. The nonuniform immunohistochemical distribution of GpIX and GpV in HUVECs, compared with the uniform distribution of these two proteins in ECs in situ, is puzzling and may represent a culture artifact; similar differences have been noted in other systems.25 However, flow cytometry showed the presence of both proteins on most HUVECs, suggesting that the discrepancy observed is more likely due to sensitivity differences between the two methods. We have further shown that HUVEC GpIX mRNA has a sequence identical to that of platelet/HEL cell mRNA and gives rise to a protein identical in size to platelet GpIX. Our partial sequence and restriction digest analyses of EC GpIbα cDNA are identical to results reported for platelet/HEL cell GpIbα cDNA,22 and EC GpIbα is the same size as the corresponding platelet protein. Thus, almost certainly, EC and platelet GpIbα are identical. The detection of GpV mRNA in ECs and other cells has been somewhat problematic, especially in view of the relatively weak signal observed by us and by others.28,29 We found 3- and 4.4-kb mRNA species in ECs, but only the smaller species in CHRF-288 and only the larger species in HEL cells. Hickey et al29 did not detect GpV mRNA in HEL cells, but found 3.8-, 4.2-, and 5.2-kb species in platelet RNA. Lanza et al28 detected GpV in HEL cells, but not in ECs, using RT-PCR, and only a 4.5-kb mRNA species in human platelets. The differences in GpV detection by different investigators may reflect variable expression of GpV in cell lines, a conclusion in keeping with our own observation of variable expression of GpV protein in HUVEC cultures. The genomic clone of Lanza et al28 exhibited four potential polyadenylation signals. Use of the two most proximal signals would produce mRNA species of 3.2 and 4.4 kb, the sizes we observed in the RNA we evaluated.
The disparity between EC and platelet GpIbβ, on the other hand, is striking. We have previously reported the cloning of endothelial GpIbβ cDNA.9 Sequence analysis of this cDNA predicts a 411 amino acid protein whose C-terminal half corresponds to the platelet GpIbβ sequence, but whose N-terminal half represents a unique protein sequence not previously reported. Consistent with these findings, we detected in ECs a 45- to 48-kD protein that reacted on Western blots with an affinity-purified polyclonal antiserum prepared against intact platelet GpIbβ. Recently, Zieger et al30 cloned and sequenced a 3.4-kb cDNA species from the megakaryoblastic cell line CHRF-288 containing two open reading frames, the 3′ of which contains the platelet GpIbβ sequence. These investigators concluded that the 3.4-kb transcript was the product of the promoter for a separate gene residing 5′ to the platelet GpIbβ gene and was produced because of the presence of an aberrant polyadenylation signal sequence within the more 5′ gene; such a transcript, if present in ECs, could not give rise to a translation product consistent with our findings. However, the results we have presented in Fig 5A and B, using an affinity-purified antibody to peptide sequence from platelet GpIbβ, seem unequivocal and confirm our previous results9 obtained using a slightly different approach and a different antibody to GpIbβ. Using an MoAb to GpIbα, we have isolated a complex of molecular weight 190 kD that can be visualized on Western blotting with an affinity-purified antibody to either platelet GpIbα or to a peptide sequence from platelet GpIbβ; this complex is roughly 25 kD larger than the corresponding platelet complex. On reduction, platelet and EC GpIbα both have a molecular weight of approximately 145 kD; however, as detected with the platelet GpIbβ peptide antibody, EC GpIbβ has a molecular weight of approximately 50 kD, which is more than twofold greater than platelet GpIbβ. In this regard, it is interesting that we have not been able to identify either a 24- or a 50-kD GpIbβ protein in CHRF-288 cells, although both the 1.1- and 3.5-kb GpIbβ mRNA species are easily demonstrable.
Using flow cytometry and MoAbs and polyclonal antibodies to the individual polypeptides, we have shown the presence of all four components of the GpIb complex on the surface of HUVECs and HAECs. Binding studies with iodinated MoAb Ib1 identified approximately 325,000 copies of GpIbα per EC, a figure 2.5 times greater than our initial estimate using MoAb AP-1.5 However, we have observed that many MoAbs to platelet GpIbα interact poorly with ECs and with isolated EC GpIb complex (Wu et al, unpublished observations), possibly related to the larger GpIbβ chain in the EC complex, and we consider the present results to be the more reliable. All cells bearing GpIbα also display GpIX and GpV on their surface. Furthermore, the ratio of GpIbα:GpIX:GpV on ECs is identical to the ratio we observed on platelets, ie, 1:1:0.5, suggesting, but not proving, that a fully formed GpIb complex is present on ECs. The stoichiometry we have observed on platelets is identical to that previously reported by other methods.3,31 However, neither our studies nor the previous studies have shown incontrovertibly that all copies of the four component polypeptides participate in complexes. It has been stated that low levels of GpIbα synthesis have been observed in other cells, particularly smooth muscle cells,28 although the only suggestion of such a finding is the modest immunohistochemical staining of smooth muscle cells seen by Asch et al.6 We have seen no evidence of such staining and, furthermore, human cultured smooth muscle cells, as we have shown, are entirely negative for surface expression of the GpIb complex. Thus, our data are consistent with the view that the GpIb complex is normally expressed exclusively on megakaryocytes, platelets, and ECs. However, we cannot rule out trace levels of expression in other cell types or in other cell lines, such as has been reported by Oleksowicz et al32 in some malignant cell lines, and we also cannot rule out the possibility that individual polypeptides of the complex, particularly EC GpIbβ, may be expressed in other cell types.
The role of the EC GpIb complex is still enigmatic. Asch et al6 showed the capacity of ECs to bind vWF in a GpIb-dependent manner and to aggregate in response to ristocetin. We have shown that ECs can adhere to a vWF substratum in a GpIb-dependent manner and that, in the presence of cytokines, the GpIb complex can become the sole mediator of this adhesion.33 It seems likely that the quantitative interaction of vWF with EC GpIbα and platelet GpIbα will differ because of the much larger extracellular domain of EC GpIbβ, and preliminary data are consistent with this possibility.
Such observations lead us to hypothesize that, under certain conditions, EC GpIb-vWF interactions may play a role in hemostasis, in platelet-EC interactions, and in wound healing. Although other ligands, in addition to vWF, are certainly involved, the role of the endothelial GpIb complex in these processes clearly is deserving of further study.
ACKNOWLEDGMENT
The authors thank Marjorie Aldrich and Rosemarie Silvano for preparation of the manuscript, Laural Ludlow for expert technical assistance, and Andrew Likens for preparation of the photographs and illustrations.
Supported in part by Grants No. HL09163 (S.S.S.) and T-32 HL07371 (D.W.E.) from the National Institutes of Health, Grant-in-Aid 93009050 (B.A.K.) from the American Heart Association, and grants from the Delaware Valley Chapter, National Hemophilia Foundation (T.T.), and the Brandywine Valley Hemophilia Foundation (D.W.E.).
Address reprint requests to Sandor S. Shapiro, MD, Cardeza Foundation for Hematologic Research, Jefferson Medical College of Thomas Jefferson University, 1015 Walnut St, Philadelphia, PA 19107-5099.