Abstract
Wiskott-Aldrich syndrome (WAS) and X-linked thrombocytopenia (XLT), caused by mutations of the WAS protein (WASP) gene, represent different phenotypes of the same disease. To demonstrate a phenotype/genotype correlation, we determined WASP gene mutations in 48 unrelated WAS families. Mutations included missense (20 families) and nonsense (eight) mutations located mostly in exons 1 to 4, and splice-site mutations (seven) and deletions and insertions (13) located preferentially in exons 7 to 11. Both genomic DNA and cDNA were sequenced and WASP expression was measured in cell lysates using peptide-specific rabbit anti-WASP antibodies. WASP was expressed in hematopoietic cell lines including bone marrow–derived CD34+ cells. Missense mutations located in exons 1 to 3 caused mild disease in all but one family and permitted WASP expression, although frequently at decreased concentration. Missense mutations affecting exon 4 were associated with classic WAS and, with one exception, barely detectable WASP. Nonsense mutations caused classic WAS and lack of protein. Insertions, deletions, and splice-site mutations resulted in classic WAS and absent, unstable, truncated, or multiply spliced protein. Using affinity precipitation, WASP was found to bind to Src SH3-containing proteins Fyn, Lck, PLC-γ, and Grb2, and mutated WASP, if expressed, was able to bind to Fyn-glutathione S-transferase (GST) fusion protein. We conclude that missense mutations affecting the PH domain (exons 1 to 3) of WASP inhibit less important functions of the protein and result in a mild phenotype, and that missense mutations affecting exon 4 and complex mutations affecting the 3′ portion of WASP interfere with crucial functions of the protein and cause classic WAS.
WISKOTT-ALDRICH SYNDROME (WAS), an X-linked recessive disorder with variable clinical phenotype,1,2 is caused by mutations of the WAS protein (WASP) gene. Thrombocytopenia and small platelets are characteristic findings present in all WAS patients irrespective of clinical severity. Patients with a classic WAS phenotype present with eczema that may be severe, develop recurrent bacterial and viral infections due to abnormal immune function, and have an increased risk of autoimmune diseases and malignancies.1-5 Immunologic abnormalities typically observed in WAS are complex and involve both B- and T-cell function. Affected male infants have a normal number of circulating lymphocytes but develop lymphopenia by 6 to 8 years of age due to a loss of T lymphocytes.4 Most affected boys present with normal levels of serum IgG, moderately depressed IgM, and elevated IgA and IgE. Antibody responses are normal to some antigens and insufficient to others. A consistent finding is a markedly depressed response to polysaccharides.3,4 Antibody responses to certain T-cell–dependent antigens such as bacteriophage φX174 are quantitatively reduced with lack of amplification and failure to switch from IgM to IgG.4 Abnormal function of WAS T cells is suggested by diminished but not absent lymphocyte responses to mitogens,3 depressed proliferative responses to allogenic cells4 and immobilized anti-CD3 monoclonal antibody,6 and failure to proliferate in response to periodate.7 Distinguishable from the classic WAS phenotype is a milder form designated as hereditary X-linked thrombocytopenia (XLT).7-11 In patients with XLT, eczema is mild, if present, and immune functions may be normal. The genes for both WAS and XLT have been mapped to the pericentromeric short arm of the X chromosome at Xp 11.22,12,13 and sequence analysis of the WASP gene has identified mutations of the same gene in both phenotypes.14 15
The gene responsible for WAS is composed of 12 exons containing 1,823 base pairs and encodes a 502–amino acid protein14 that appears to be of central importance for the function of hematopoietic cells. WASP, expressed in all hematopoietic stem cell–derived lineages,14 is located predominantly in the cytoplasm.16-19 Although several unique binding domains of WASP have been identified, its precise function is unknown. WASP has a proline-rich region with motifs corresponding to the PXXP binding consensus for SH3 domains.20 Interaction of WASP with SH3 domains of selected signaling molecules has been demonstrated, including p47nck, a 47-kD cytosolic adapter protein,16 Fyn,21 cFRG,21,22 c-Src, p47phox,22 Grb2,23 and the Tec family cytoplasmic tyrosine kinases, Btk, Tec, PLC-γ1,22,23 and Itk.23,24 Of these, only p47nck and Fyn were shown to coprecipitate in vivo with WASP. In addition, WASP may play a role in the regulation of the actin/cytoskeleton system by interacting directly with the Rho-like GTPase, cdc42.25-27
To investigate a possible correlation between genotype and phenotype, we identified the mutations of the WASP gene in a large cohort of patients with WAS or XLT, determined the effect of these defects on gene transcription, protein expression, and SH3 binding, and correlated the molecular findings with the clinical phenotype.
MATERIALS AND METHODS
Patients and clinical phenotypes.Affected members of 48 unrelated WAS families were included in this analysis. Using previously published criteria,19 28 the severity of WAS-associated symptoms was estimated and expressed as a score of 1 to 5. A score of 1 was assigned to a patient with thrombocytopenia and small platelets without any other symptoms, clinical findings, or laboratory abnormalities. Patients with platelet abnormalities and mild, transient eczema with or without minor infections received a score of 2. Patients with persistent but manageable eczema or recurrent infections or both received a score of 3. Patients with persistent and difficult-to-control eczema and frequent life-threatening infections were scored as a 4. A score of 5 was assigned if patients presenting with eczema and/or frequent infections developed autoimmune diseases or malignancies.
Cell lines.B-lymphoblastoid cell lines (B-LCLs) were established by inoculating peripheral blood mononuclear cells (PBMCs) from normal controls and WAS patients with Epstein-Barr virus (EBV)-containing supernatant, as previously described.29 The human kidney tumor cell line 293 and the hepatoma cell line SK HEP1 (both provided by Dr Mark Kay, University of Washington, Seattle, WA), the human megakaryoblastic cell line MEG-01 (a gift from Dr Thalia Papayannopoulou, University of Washington, Seattle), and HEL, HE-60, Jurkat T-cell line, and K562 cells (obtained from the American Type Culture Collection, Rockville, MD) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum. CD34+ bone marrow cells from a normal human donor were prepared by Dr Robert Andrews (University of Washington, Seattle).
RNA isolation and reverse transcriptase–polymerase chain reaction.RNA isolation, using a single-step method and Trizol (GIBCO-BRL, Gaithersburg, MD), and reverse transcriptase–polymerase chain reaction (RT-PCR), using the SuperScript Preamplification System kit (GIBCO-BRL), were performed as previously described.30 WASP cDNA was amplified by PCR in two overlapping fragments using the previously reported primers28 and the Expand High Fidelity PCR System (GIBCO-BRL). PCR products were purified by agarose gel electrophoresis.
Dideoxynucleotide fingerprinting and direct sequencing of cDNA.To screen the amplified WASP cDNA for mutations, we modified the dideoxynucleotide fingerprinting (ddF ) method originally designed by Sarkar et al31 as previously described,28 using the fmol cycle-sequencing kit (Promega, Madison, WI). The resulting PCR products were electrophoresed in 5% polyacrylamide nondenaturing gels at 4°C. The mutations suggested by ddF were confirmed by direct sequencing using selected primers and the fmol cycle-sequencing kit.
DNA purification and sequencing of genomic DNA.DNA was extracted from B-LCLs as previously described.30 Purified genomic DNA samples were amplified with primer pairs designed to span the suspected mutation sites, including exon/intron junctions, using previously reported conditions.28 The amplified DNA fragments were separated by agarose gel electrophoresis and directly sequenced using the fmol cycle-sequencing kit.
Northern blot analysis.PolyA(+) mRNA samples isolated from 30 μg total RNA with the polyATtract mRNA isolation system (Promega) were electrophoresed through agarose/formaldehyde gels and transferred to nylon membranes (Magna NT; MSI, Westboro, MA). The WASP cDNA clone M5.5, a 750–base pair cDNA fragment of the WAS gene, was radiolabeled by random primer extension and used as a probe for hybridization. A human actin cDNA probe was used to monitor mRNA loading.
Anti-WASP antisera.Three hydrophobic peptide sequences were selected from the reported WASP sequence.14 Peptide 1050 (ADEDEAQAFRALVQEK) represents part of exon 4, peptide 1048 (SSRYRGLPAPGPSPADKK) part of exon 7, and peptide 1049 (KRSRAIHSSDEGRDQ) part of exons 11 and 12. Following synthesis, the peptides were coupled to ovalbumin, suspended in Freund adjuvant, and used to immunize rabbits to raise polyclonal anti-WASP antisera. The antisera were designated Ab953 (peptide 1050), Ab503 (peptide 1048), and Ab1468 (peptide 1049). Specificity was determined by Western blot analysis of in vitro translated WASP gene product and lysates from normal B-LCLs.
Western blot analysis.B-LCLs from normal control subjects and WAS patients were suspended at 2.5 × 107/mL in lysis buffer containing 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mmol/L PMSF, 0.5% aprotinin, and 10 μg/mL leupeptin at pH 7.5 and kept on ice for 10 minutes. The protein concentration was determined in each lysate by a protein assay kit (Bio-Rad Laboratories, Hercules, CA). From each sample, 40 μg total protein was loaded onto a sodium dodecyl sulfate (SDS)-polyacrylamide gel, electrophoresed, and transferred to a PVDF Immobilon-P membrane (Millipore, Bedford, MA). After blocking with 10% nonfat milk, the membranes were incubated with anti-WASP antibody 503 (and, if indicated, with antibodies 953 or 1468) at 1:5,000 dilution for 1 hour at room temperature. After washing with 0.1% Tween/phosphate-buffered saline four times, the membranes were incubated with horseradish peroxidase–conjugated goat anti-rabbit Igs (Biosource International, Camarillo, CA) at a concentration of 1:10,000 for 1 hour at room temperature. The results were visualized by an enhanced chemiluminescence method (Amersham, Arlington Heights, IL).
Affinity precipitation of WASP binding proteins.B-LCL lysates containing 80 μg total protein were incubated with 1 μg SH3 (Fyn, Lck, GAP, PLC-γ, and Grb2) or SH2 (Fyn, GAP, PI-3, and PLC-γ) glutathione S-transferase (GST) fusion protein agarose beads (UBI, Lake Placid, NY) and 25 μL Sepharose in 500 μL lysis buffer at 4°C for 2 hours. After extensive washing, the pelleted beads were incubated with 20 μL 2× loading buffer at 95°C for 3 minutes, and the supernatants were loaded onto SDS-polyacrylamide gel for electrophoresis. The membranes were incubated with anti-WASP antibody 503.
In vitro translation of WASP.The entire coding sequence of WASP cDNA was amplified using the Expand High Fidelity PCR System (GIBCO-BRL) and primers with restriction enzyme sites (5′-AAG ACA GGA TCC AGA AAG CAC CAT GAG TGG, forward [BamHI, cDNA position 25] and 5′-GGC CFA TCT AGA CTC AGC CAC TCA GTC ATC, reverse [Xba I, cDNA position 1554]). The digested PCR products and the pd16 vector were ligated using T4 DNA ligase (GIBCO-BRL) and competent cells (DH5α) transformed using standard electroporation. Using the TNT in vitro Translation system (Promega) and a nonradiolabeled amino acid mixture, plasmid DNA was translated and the product was used for Western blot analysis.
RESULTS
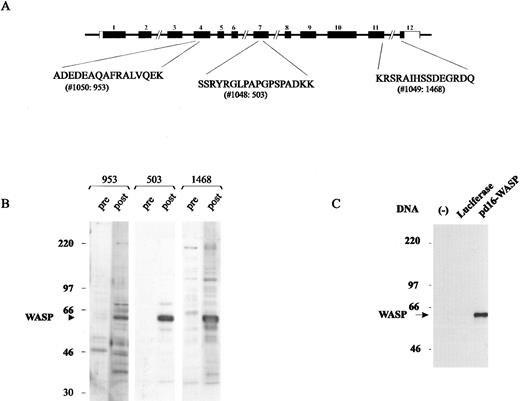
WASP-specific antisera.Expression of WASP was determined by rabbit anti-WASP antibodies raised against peptides encoded by exon 4 (Ab953), exon 7 (Ab503), and exon 11 (Ab1468) (Fig 1A). Postimmunization sera but not preimmunization sera recognized a band of 62 kD in Western blots generated from normal B-LCL lysates (Fig 1B). Ab503 had the highest titer and the least background, and was used in most experiments. Figure 1C shows that the protein translated from the pd16-WASP plasmid DNA, but not that expressed by a control plasmid containing the luciferase gene, was recognized by the anti-WASP Ab503, demonstrating that the antibody was specific for WASP.
Anti-WASP antisera. (A) WASP-derived peptides used in the production of rabbit anti-WASP antibody (Ab). (B) Assessment of Ab953, Ab503, and Ab1468 using Western blot analysis of normal B-LCL lysate and preimmunization and postimmunization antisera. (C) Only protein translated from WASP gene mRNA, not from luciferase gene mRNA, was recognized by Ab503 as a single band of 62 kD, demonstrating that the antibody was specific for WASP.
Anti-WASP antisera. (A) WASP-derived peptides used in the production of rabbit anti-WASP antibody (Ab). (B) Assessment of Ab953, Ab503, and Ab1468 using Western blot analysis of normal B-LCL lysate and preimmunization and postimmunization antisera. (C) Only protein translated from WASP gene mRNA, not from luciferase gene mRNA, was recognized by Ab503 as a single band of 62 kD, demonstrating that the antibody was specific for WASP.
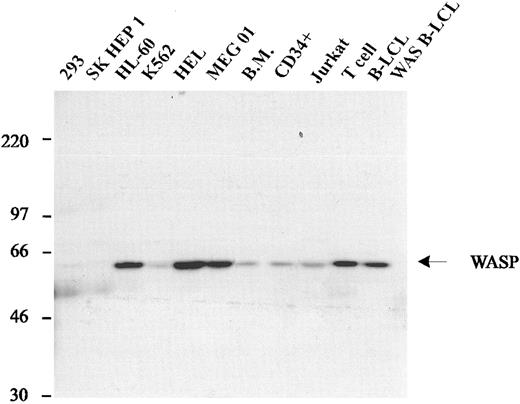
WASP is expressed by hematopoietic cells.Previous studies demonstrated that WASP mRNA was expressed in all hematopoietic cells.14 To examine WASP expression at the protein level, we studied cell lysates from unseparated and CD34+-enriched human bone marrow cells and several human cell lines by Western blot analysis. WASP was abundant in lysates of MEG-01, HEL, HL-60, and cultured normal T- and B-cell lines, and was detectable in lysates of bone marrow, CD34+ cells, and Jurkat and K562 cells (Fig 2). WASP was not detected in the human kidney tumor cell line 293, the hepatoma cell line SK Hep 1, or a B-LCL derived from a WAS patient with a nonsense mutation in exon 1 (DM, Table 1).
Expression of WASP in human cell lines. Western blot analysis of lysates from various hematopoietic cell lines (HL-60, K562, HEL, MEG01, Jurkat, normal IL-2–dependent T-cell line, normal B-LCL, and a B-LCL from a WAS patient with the classic phenotype) and 2 nonhematopoietic cell lines (293 and SK HEP1). All hematopoietic cell lines express WASP, with the exception of the B-LCL derived from a WAS patient. Nonhematopoietic cell lines do not express WASP.
Expression of WASP in human cell lines. Western blot analysis of lysates from various hematopoietic cell lines (HL-60, K562, HEL, MEG01, Jurkat, normal IL-2–dependent T-cell line, normal B-LCL, and a B-LCL from a WAS patient with the classic phenotype) and 2 nonhematopoietic cell lines (293 and SK HEP1). All hematopoietic cell lines express WASP, with the exception of the B-LCL derived from a WAS patient. Nonhematopoietic cell lines do not express WASP.
Mutation analysis.RT-PCR products obtained from PBMCs or B-LCLs derived from patients with WAS or XLT were screened by the ddF method. Mutations were confirmed by direct sequencing of the RT-PCR products and of amplified genomic DNA. Patients with multiple RT-PCR products and/or splice-site mutations were further studied by subcloning the cDNA and sequencing up to 20 individual clones. Mutations of the WASP gene were identified in 59 affected members of 48 unrelated families. Table 1 summarizes the results, dividing patients into groups with mild (scores 1 to 2) and classic (scores 3 to 5) WAS phenotypes. Mutations in 17 of these families were previously reported by us14,28,34 or by others (Table 1).32,33 Of 48 mutations identified, 40 are unique within this patient population, and compared with the mutations reported to date,14,15,28 32-40 20 are novel and found only in our patient population. The mutations identified are distributed throughout the WASP gene and affect all exons except 5, 8, and 12. All but one of 20 missense mutations identified are located in exons 1 to 4. Eight unrelated families had five unique nonsense mutations located in exons 1, 3, 7, 9, and 10, including patient RM, in whom the nonsense mutation affects codon 99 in exon 3, resulting in two transcripts, one containing the stop codon and the other deleting exon 3. Five of the six unique point mutations (from seven families) resulting in abnormal splicing were located in exons/introns 6 to 11. Deletions and insertions of genomic DNA were identified in 13 unrelated patients, representing 12 unique mutations, and were located either in exons 1 to 3 (six patients) or exons 10 and 11 (seven patients). Mutation hotspots observed include the known hotspots C290T (Arg86Cys), G291A (Arg86His), and C665T (Arg211 → stop) and the two splice-site mutations, t → c + 2 on intron 9 and t → c + 2 on intron 11 (X2). In addition, a series of five guanines at positions 1301 to 1305 were involved in three unrelated families: a G insertion resulting in a frameshift and stop at amino acid 494 was observed in two families, and a G deletion resulting in a frameshift and stop at amino acid 444 was observed in the third family.
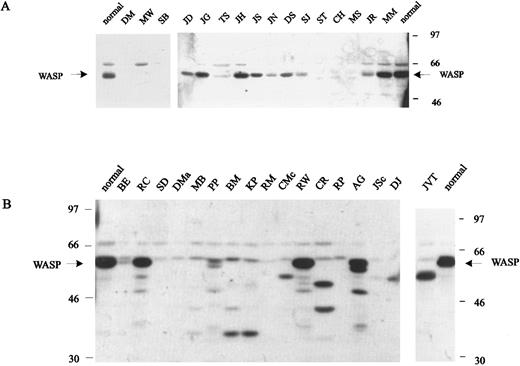
WASP expression by cells from WAS/XLT patients.B-LCLs from affected males of 41 unrelated families and frozen PBMCs from two additional unrelated males were available for Western blot analysis of WASP (Table 1). Selected blots are shown in Fig 3. As expected, all eight patients with nonsense mutations, including three patients (TH, Kpe, and AT) not shown in the figure, had a classic phenotype with scores of 3 to 5 and failed to express WASP (Table 1 and Fig 3A). Five of 20 unrelated patients with missense mutations had transitional mutations in exon 4. All five presented with a severe phenotype and had either WASP that was normal in amount and size (CC) or WASP that was barely detectable by Western blot and appeared slightly shorter (MS, CH, and ST) (Table 1 and Fig 3A), although gene expression measured by Northern blot analysis was normal in the three patients studied (Table 1). Of the remaining 15 unrelated patients with missense mutations, 14 had mutations located in exons 1 to 3 and one in exon 9; all but one had a mild phenotype, and where determined, all had normal mRNA expression and detectable WASP by Western blot analysis (Table 1 and Fig 3A). WASP expression was either markedly decreased as in patient TS (9% of normal by densitometry) or near normal as in patients MM (75%) or JH (60%).
Western blot analysis of cell lysates (B-LCLs) from WAS patients (see Table 1 for detailed description of the mutations). (A) Blots from patients with nonsense mutations or missense mutations affecting exons 1 to 4. (B) Blots from patients with splice-site mutations, insertions, or deletions located in exons 7 to 11 (except patient SD, who has a splice-site mutation affecting intron 3).
Western blot analysis of cell lysates (B-LCLs) from WAS patients (see Table 1 for detailed description of the mutations). (A) Blots from patients with nonsense mutations or missense mutations affecting exons 1 to 4. (B) Blots from patients with splice-site mutations, insertions, or deletions located in exons 7 to 11 (except patient SD, who has a splice-site mutation affecting intron 3).
Seven unrelated patients had point mutations affecting either a regular or cryptic splice site or a nonsense mutation resulting in exon-skipping (RM). In patient BE with XLT and a score of 1, we found two splicing products: 70% of 20 RT-PCR clones generated from a B-LCL showed an insertion of 38 nucleotides derived from intron 6 resulting in frameshift and premature stop; however, 30% of the clones had a normal cDNA sequence and, on Northern blot, a normal quantity of mRNA. The presence of normal-size WASP in a cell extract, although considerably decreased in quantity, confirms this explanation (Table 1 and Fig 3B). The other six patients with splice-site mutations had classic WAS phenotypes with scores of 3 to 5 and either a complete absence of WASP (SD, DMa, MB, and RM) or the presence of truncated WASP (BM and KP). The latter two patients, who are unrelated, have identical mutations that result in deletion of exon 11, frameshift, and loss of the termination codon (TGA). The pattern of multiple bands observed in Western blots suggests that the use of normally nontranslated sequences of exon 12 render WASP unstable.
The remaining 13 patients had deletions or insertions of one or more nucleotides affecting genomic DNA. All but one (JVT) had classic WAS and a score of 3 to 5. WASP, evaluated in 11 patients, was either truncated or unstable or, as in patient RP, not demonstrable (Fig 3B). Patients RW and AG, who are unrelated but have the same mutation, showed multiple bands of WASP, suggesting unstable protein or, less likely, multiple splicing products.
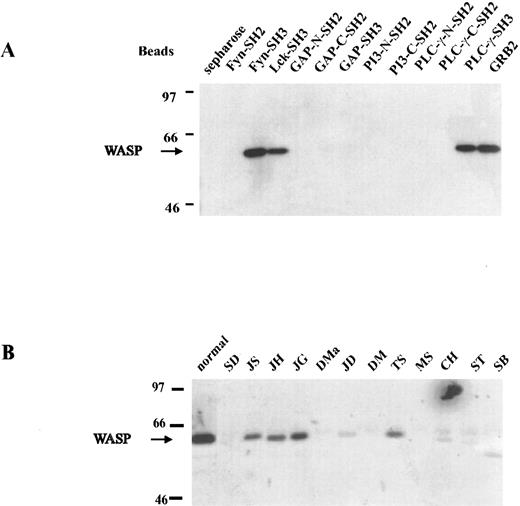
In vitro interaction of WASP and SH3 domains.To determine whether wild-type WASP interacts in vitro with proteins containing SH3 domains, we used affinity precipitation and Western blot analysis to assess five SH3-GST fusion proteins (Fyn, Lck, GAP, PLC-γ, and Grb2) and, as controls to rule out nonspecific binding, seven SH2-GST fusion proteins bound to agarose beads. If the beads were incubated with normal B-LCL lysate and the eluted proteins subjected to Western blot analysis, SH3 domains of Fyn, Lck, PLC-γ, and Grb2 were found to bind to WASP in vitro (Fig 4A). However, the SH3 domain of GAP and all SH2 domains studied (Fyn, GAP, PI-3, and PLC-γ) failed to bind to WASP, suggesting that WASP selectively binds to many SH3 proteins and that the binding is not due to a nonspecific association. Attempts to confirm the interactions between WASP and Fyn, Lck, PLC-γ, and Grb2 by in vivo coprecipitation experiments have been unsuccessful. To determine if mutated WASP from selected patients, known to express protein, can bind SH3-containing protein in vivo, we used a Fyn-GST fusion protein and affinity precipitations. Mutated WASP was precipitated in vitro by Fyn SH3-GST fusion protein agarose beads from lysates of B-LCLs established from 12 WAS patients. Only patients with demonstrable WASP in the cell lysates showed a band, as demonstrated by direct Western blot analysis (Fig 4B). The amount of WASP precipitated by Fyn SH3-GST agarose beads seemed to correlate with the amount of WASP in the cell lysate (Fig 3).
In vitro interaction of WASP and SH3-containing proteins. (A) Proteins containing SH2 and SH3 domains were used to affinity-precipitate WASP. Of 5 SH3-GST fusion proteins, 4 (Fyn, Lck, PLC-γ, and Grb2) were able to bind WASP. None of the SH2-GST fusion proteins were able to bind WASP. (B) Lysates of B-LCLs established from 12 WAS patients, some expressing WASP, were incubated in vitro by Fyn SH3-GST fusion protein agarose beads. Only patients with demonstrable WASP showed a band (JS, JH, JG, JD, TS, and possibly CH).
In vitro interaction of WASP and SH3-containing proteins. (A) Proteins containing SH2 and SH3 domains were used to affinity-precipitate WASP. Of 5 SH3-GST fusion proteins, 4 (Fyn, Lck, PLC-γ, and Grb2) were able to bind WASP. None of the SH2-GST fusion proteins were able to bind WASP. (B) Lysates of B-LCLs established from 12 WAS patients, some expressing WASP, were incubated in vitro by Fyn SH3-GST fusion protein agarose beads. Only patients with demonstrable WASP showed a band (JS, JH, JG, JD, TS, and possibly CH).
DISCUSSION
The recent cloning of the WASP gene has made possible the genetic definition, carrier detection, and prenatal diagnosis of WAS. Mutation analysis has demonstrated that a milder WAS phenotype described variably as XLT, “isolated” thrombocytopenia, and “atypical” or “attenuated” WAS is caused by mutations of the same gene responsible for “classic,” “full-blown” WAS.14 15
Attempts to correlate these strikingly different WAS phenotypes with specific genotypes have resulted in controversial interpretations. One group of investigators reported families whose affected males presented with the same clinical phenotypes,2,32,37 whereas others have described families whose affected members had different phenotypes.5 38 To assess the clinical phenotype objectively, we designed a simple scoring system based on characteristic clinical symptoms. This allowed us to differentiate patients with symptoms limited to platelet abnormalities and bleeding (XLT) from those with a classic WAS phenotype. To define in more detail the nature of each mutation, we not only sequenced genomic DNA but also examined the effect of the mutations on RNA transcription, expression of WASP in patient lymphocytes, and binding of mutated WASP to the SH3-containing protein-tyrosine kinase Fyn.
To quantify WASP, we generated rabbit polyclonal antibodies to three different peptides derived from WASP. The antibody preparation (Ab503) we selected for most experiments was highly specific and suitable for immunostaining and Western blot analysis. This antibody detected WASP in all hematopoietic cell lines studied, including T- and B-cell lines, erythroleukemia and myelogenous leukemia cell lines, and a megakaryocytic cell line. WASP was also present in human bone marrow–derived CD34+ cells, but not in nonhematopoietic cell lines. A protein, N-WASP, resembling the structure of WASP and having many of the same functional domains has been recently identified in nonhematopoietic cells.41
To examine the effect of individual WASP mutations on the clinical phenotype, we identified mutations of the WASP gene in 48 unrelated families. Twenty (50%) of 40 unique mutations observed in this group of patients have not been reported by others.40 This heterogeneity has been observed in other X-linked immunodeficiency disorders, and may reflect the high rate of new mutations characteristic for X-linked disorders. The most frequent mutations identified (20 of 48) were missense mutations, all but one within exons 1 to 4. The remaining 28 mutations consisted of nonsense mutations (eight families), deletions and insertions (13 families), and splice-site mutations (seven families) and were located preferentially in exons 7 to 11. Deletions and/or insertions affecting genomic DNA resulted in direct stop codons, frameshift and early termination, or, as in patient JVT, multiple gene products. Mutations affecting splice sites resulted frequently in multiple splicing products.
To determine the effects of the mutations on WASP expression, we established B-LCLs from affected members of the majority of our WAS families. Patients with missense mutations located in exons 1 to 3, with one exception, had mild disease (a score of 1 to 2), and cell extracts contained WASP at concentrations that varied between 3.6% and 75% of normal control values. The exception was JG, with a score of 5, who had two consecutive point mutations resulting in substitution of leucine by histidine at codon 35 and whose PBMCs had WASP of normal size and moderately reduced amount (30% of normal). Five patients with missense mutations affecting exon 4 presented with a classic WAS phenotype and a score of 3 to 5 and, with one exception, had barely detectable WASP of slightly smaller size. As expected, patients with nonsense mutations lacked WASP and were found to have a classic WAS phenotype with a score of 4 or 5. The 5′ region of WASP (exons 1 to 3) where most missense mutations are located has been designated WH127 and has the characteristics of a PH domain.41 We hypothesize that the amino acid substitutions (except Leu35His) located in exons 1 to 3 of the WASP gene only partially inhibit the function of the protein, and affect platelet number and platelet size, possibly by interfering with platelet release from megakaryocytes.4 In contrast, exon 4 appears to be critical for the stability and function of WASP, since most of the missense mutations we observed in exon 4 resulted in lack of detectable WASP expression or in truncated/unstable protein although normal amounts of transcript were present. The importance of exon 4 is further underlined by the observation that all of our patients with missense mutations in exon 4 have developed a classic severe WAS phenotype with a score of 4 to 5. Insertions, deletions, and splice-site mutations led to the absence of WASP, unstable or truncated WASP, or multiple splicing products and, as a rule, were associated with severe, classic WAS. The exceptions were two patients with complex mutations. Further analysis provided an explanation for their mild XLT phenotype. Patient BE, with a score of 1, has a g → a transition at nucleotide +5 of intron 6, which does not completely inactivate the splice donor site. As a consequence, a portion of the gene transcript is normally spliced and translated into a product of normal size by Western blot analysis. However, the dominant splicing product of this patient uses a cryptic splice site, and contains the 5′ portion of intron 9, leading to frameshift and early termination. The other exception is patient JVT who, by genomic DNA analysis, has a T insertion at position 1,029. This mutation is expected to result in frameshift and premature termination three codons downstream of the T insertion (Asn335stop). However, cloning of the RT-PCR product revealed that only approximately 25% (five clones) of WASP cDNA derived from B-LCLs in patient JVT shows the T insertion. Two additional cDNA species were identified, one with a deletion of the entire 407 nucleotides of exon 10, resulting in truncation, frameshift, and loss of the last nine amino acids of exon 12. The third and most frequent (10 of 20 clones) cDNA species identified in patient JVT had an inframe deletion of the first 52 amino acids of exon 10. Whereas the former deletion translates into a severely truncated protein with loss of the proline-rich regions and the WH2 domain,27 the inframe deletion of 52 amino acids results in a less truncated, stable protein of 450 amino acids, demonstrable by Western blot, and contains all known functionally important domains of WASP, possibly explaining the mild phenotype in JVT. A similar situation may account for the mild WAS phenotype observed in a recently reported family whose affected members had both Fanconi syndrome and XLT. Sequence analysis of genomic DNA of these patients revealed a G insertion just one nucleotide downstream of mutation in patient JVT.37
We conclude from this analysis that mutations affecting the downstream portion of the WASP gene more effectively interfere with the function of WASP, causing a severe phenotype. In contrast, missense mutations located within the PH domain (exons 1 to 3) were consistently, with one exception, associated with a mild phenotype, presumably by leaving intact the functionally important cdc42 binding site, the SH3 binding motifs, and the WH2 domain, which includes a verprolin and cofilin homology domain.41 On the other hand, genetic determinants not related to WASP and environmental factors undoubtedly influence the clinical phenotype, possibly explaining the discordant WAS phenotypes observed in members of some families.5 38
The proline-rich region of exon 10 contains multiple minimal PXXP motifs and two PPPPXXRG SH3 binding motifs and is considered important for the interaction of WASP with SH3-containing proteins.16,20-24 Using affinity precipitation and Western blot analysis, four of five SH3-GST fusion proteins were found to bind to WASP in vitro, including Fyn, Lck, PLC-γ, and Grb2, suggesting that WASP may selectively bind to many SH3-containing proteins. To explore whether mutated WASP loses its capability to bind to SH3 domains, we analyzed WASP in cell extracts from B-LCLs derived from selected patients with missense and nonsense mutations or splice-site mutations and deletions/insertions (not shown) by affinity precipitation of the Fyn-GST fusion protein. Whenever the mutation allowed protein expression, the Fyn-GST fusion protein was able to bind the mutated WASP, and the amount precipitated by Fyn SH3-GST agarose beads correlated, in most instances, with the amount of mutated WASP present in the cell lysate. Since neither we nor others40 have yet found missense mutations that selectively affect SH3 binding motifs, the importance of these motifs for the function of WASP and the direct interaction with SH3-containing proteins is unknown.
Although the function of WASP has not been clearly defined, recent observations support the hypothesis that WASP plays a prominent role in the regulation of the actin/cytoskeleton system. Interaction of WASP with the small GTPase cdc42, a key element in the dynamic organization of the actin/cytoskeleton, has been suggested.25-27 However, missense mutations that would result in substitution of amino acids 238 to 257, the suggested cdc42 binding domain, have not yet been identified. It is presently unknown whether WASP interacts directly with actin or through actin binding proteins, and if the underlying problem results in a transmembrane signaling defect. The observations that actin bundling is necessary for T-cell activation by anti-CD3 antibody42 and that cdc42 is required for the polarization of T cells toward antigen-presenting cells43,44 suggest that the abnormal antibody responses characteristic of most WAS patients are a direct consequence of defective T-/B-cell interaction. In the absence of functional WASP, T cells fail to provide adequate help to B cells, resulting in impaired B-cell function. Such a mechanism would explain the characteristic defect in antibody responses to bacteriophage φX174 by WAS patients,4,19 a pattern similar to that observed in patients with X-linked hyper-IgM syndrome.45
ACKNOWLEDGMENT
The following individuals allowed us to study their patients and provided material that was invaluable for this investigation: N. Day, A. Filipovitch, C. Frantz, E. Gillian, R. Good, H. Hill, R. Insel, A. Junker, A. Laszlo, J. Oleske, L. Pachman, R. Roberts, R. Schiff, A. Shigeoka, C. Stotts, K. Sullivan, L. Vogler, and J. Winkelstein.
Supported by grants from the National Institutes of Health (HD17427), the March of Dimes Birth Defects Foundation (FY96-0330), and the DeJoria Wiskott-Aldrich Research Fund; conducted in part at the Clinical Research Center at the University of Washington (RR-37).
Address reprint requests to Hans D. Ochs, MD, Division of Infectious Diseases, Immunology and Rheumatology, Department of Pediatrics, University of Washington School of Medicine, Box 356320, Seattle, WA 98195-6320.