Abstract
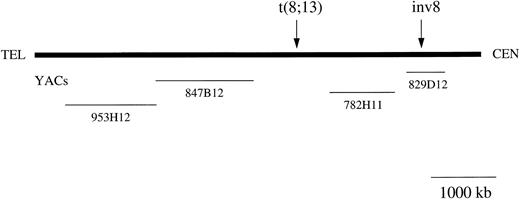
Two distinct leukemia syndromes are associated with abnormalities of chromosome band 8p11. First, a myeloproliferative disorder with features characteristic of both chronic myeloid leukemia and non-Hodgkin's lymphoma and second, an acute myeloid leukemia (AML) with French-American-British (FAB) M4/5 morphology and prominent erythrophagocytosis. The two syndromes are exemplified by a t(8; 13)(p11; q12) and a t(8; 16)(p11; p13), respectively, but cytogenetic variants of both have been described. Recently, the t(8; 16) has been cloned and shown to fuse the MOZ gene at 8p11 to the CBP gene at 16p13. We have used fluorescence in situ hybridization (FISH), Southern blotting, and reverse transcriptase-polymerase chain reaction (RT-PCR) to refine the 8p11 breakpoint in three cases with t(8; 13)(p11; q12) and in a single case of AML-M5 with a clinical picture apparently identical to that found in patients with a t(8; 16), but characterized by an inv(8)(p11q13). FISH analysis was performed with several 8p11 CEPH yeast artificial chromosome (YAC) clones. YAC 782H11 was centromeric to the one case with t(8; 13) tested, but was telomeric to the inv(8). YAC 847B12 was telomeric to both the t(8; 13) and the inv(8), whereas YAC 829D12 was centromeric to the t(8; 13), but split by the inv(8). Southern blotting and PCR of YAC 829D12 showed that it contained the MOZ gene. A 900-bp MOZ fragment encompassing the published t(8; 16) breakpoint was amplified by PCR from normal peripheral blood leukocyte cDNA and used to probe Southern blots of patient DNA. A rearrangement was detected in the case with inv(8), but not in any of the three cases with t(8; 13). Southern blotting with a CBP probe and RT-PCR with MOZ and CBP primers suggested that the inv(8) does not result in a cryptic MOZ-CBP fusion. It is likely, therefore, that MOZ is fused to a novel gene at 8q13 in this case. We conclude that the t(8; 13) breakpoint is flanked by YACs 782H11 and 847B12 and is at least 1 Mb telomeric to MOZ. MOZ is involved, however, in a new variant of the t(8; 16).
THE DISRUPTION OF oncogenes or inactivation of tumor suppressor genes plays a central role in the pathogenesis of malignant diseases; such anomalies may be associated with visible chromosomal abnormalities. In leukemia, detailed characterization of nonrandom chromosome changes has provided a basis for disease classification and has helped to define prognosis in individual patients. Furthermore, the molecular cloning of the genes involved in the chromosomal changes has advanced our understanding of the mechanisms of leukemogenesis and has been valuable in the design of molecular approaches to diagnosis and to monitoring response to treatment.
The recurrent translocation t(8; 16)(p11; p13) associated with acute myeloid leukemia (AML) defines a particular subtype of this disease. AML with the t(8; 16) is predominantly classified as French-American-British (FAB) M4 (myelomonocytic) or M5 (monocytic) and is characterized by pronounced erythrophagocytosis by the blast cells, young age at diagnosis, and poor outcome.1-3 This translocation is found in approximately 2% of cases of AML with FAB M4 or M5 phenotype.4 Chromosome band 8p11, however, has been shown to be involved in other cytogenetic rearrangements. These abnormalities are associated with either leukemias of similar phenotype to the t(8; 16) AML, such as the translocations t(8; 22)(p11; q13) and t(8; 19)(p11; q13),5,6 or with an unusual myeloproliferative syndrome associated with the translocations t(8; 13)(p11; q12), t(8; 9)(p11; q32) or t(6; 8)(q27; p11), collectively known as the 8p11 myeloproliferative disorder.7 8 The prevalence of these other translocations is unknown, but they are almost certainly rarer than the t(8; 16)(p11; p13).
Recently, the t(8; 16)(p11; p13) was cloned and shown to fuse the CBP gene at 16p13 to a novel gene, termed MOZ, at 8p11.9 CBP codes for a multidomain transcriptional coactivator,10 and both genes are implicated in histone acetylation, suggesting a novel mode of leukemogenesis by MOZ-CBP.9,11,12 It has been postulated that MOZ may be involved in other 8p11 translocations associated with leukemic syndromes; CBP, presumably, can be replaced by analogous genes at other chromosomal locations.9 The molecular pathogenesis of the 8p11 myeloproliferative disorder is entirely unknown, apart from the preliminary mapping of the t(8; 13)(p11; q12) chromosome 13 breakpoint to a single YAC clone.13
In this study, we have used a combination of fluorescence in situ hybridization (FISH), Southern blotting, and reverse transcriptase-polymerase chain reaction (RT-PCR) to refine the location of 8p11 breakpoints and evaluate the role of MOZ in three cases of t(8; 13)(p11; q12) and in a case of AML-M5 with an inv(8)(p11q13).
MATERIALS AND METHODS
Patients
8p11 myeloproliferative disorder. Three patients with a t(8; 13)(p11; q12) were studied. The clinical phenotype in each case was consistent with the 8p11 myeloproliferative disorder.7 Patients UPN 01 and UPN 02 have been described previously.14 15 Patient UPN 03 was a 48-year old female who presented with a brief history of a mass in the left groin, lethargy, and left upper quadrant pain. On examination she had a 10 cm splenomegaly and prominently enlarged inguinal lymph nodes. The blood count was as follows: hemoglobin (Hb), 15.1 g/dL; white blood cell (WBC), 142.2 × 109/L with marked eosinophilia (17%) and no blasts; platelets, 81 × 109/L. The bone marrow aspirate was extremely hypercellular and eosinophils/eosinophilic precursors were prominent. Cytogenetic studies showed a 47,XX, +21,t(8; 13)(p11; q12) in all bone marrow-derived metaphases. Six months later the disease progressed to acute myeloid leukemia.
AML-M5 with inv(8)(p11q13). A 15-year old girl (UPN 04) was diagnosed with AML-M5. A bone marrow aspirate showed a hypercellular marrow, with 98% blasts and prominent erythrophagocytosis. Cytogenetic analysis showed an inv(8)(p11q13) in 70% of bone marrow metaphases.
FISH. CEPH YAC clones 829D12, 782H11, 847B12, and 953H12 were obtained from the MRC HGMP Resource Centre (Hinxton, UK). The relative positions of these 8p11 clones have been described elsewhere16,17 and are shown in Fig 1. Yeast containing individual YACs were grown in AHC medium for 24 to 72 hours and DNA extracted using a glass-bead method.13. Biotin was incorporated into total yeast DNA using the BioNick Labelling System (Gibco BRL, Paisley, UK). After G50 column purification, 400 ng of labeled DNA was precipitated with 10 μg of human Cot-1 DNA (Gibco BRL), 5 μg of sonicated salmon sperm DNA, and dissolved in 11 μL of hybridization buffer (50% formamide, 2× SSC, 10% dextran sulphate, pH 7.0). In some preparations, whole chromosome painting probes (Scotlab, Coatbridge, UK) were also included. The slides were washed in 2× SSC, dehydrated, and denatured in 70% formamide/2× SSC, at 70°C, for 3 minutes. The probe was then denatured and allowed to reanneal for 15 minutes, after which, it was applied to the surface of the slides. All further hybridization steps and immunologic detection were performed as described.18 Slides were examined using an Olympus Vanox microscope equipped with a fluorescence unit, a CCD camera and image analysis software (SmartCapture, Vysis, UK).
Schematic map of the chromosome 8p11 region. The YACs used in the study are shown. The YACs sizes are drawn to scale although the distances between the genes and markers mapping to 8p11 are arbitrary. Arrows indicate approximate positions of the breakpoints in the t(8; 13) and inv(8)(p11q13) cases.
Schematic map of the chromosome 8p11 region. The YACs used in the study are shown. The YACs sizes are drawn to scale although the distances between the genes and markers mapping to 8p11 are arbitrary. Arrows indicate approximate positions of the breakpoints in the t(8; 13) and inv(8)(p11q13) cases.
Southern blot. Southern blot analysis was performed as described.19 Two probes were used: MOZ 0.9, was a 910-bp MOZ fragment amplified by PCR from normal peripheral leukocyte blood cDNA (nucleotides 4640 to 5550 of the MOZ cDNA sequence, GenBank accession number U47742) and cloned into the pCR2.1 plasmid vector (Invitrogen, Leek, Holland). 5′ CBP (nucleotides 55 to 1280 of the CBP cDNA sequence; GenBank accession number U47741) was constructed in a similar way. Both probes span the t(8; 16) breakpoints as described.9
PCR analysis. Both nested and single step RT-PCR assays were used to search for MOZ-CBP and/or reciprocal CBP-MOZ fusion mRNA in the case with inv(8)(p11q13). RNA was extracted and cDNA synthesized as previously described.20 cDNA quality was confirmed by amplification of the normal BCR gene.20 To amplify any MOZ-CBP transcripts, the first round PCR primers were MOZ-6: 5′-CAGAaTTcCACCCAGGCTGA-3′ and CBP-1: 5′-TGCTCTGTTTGCTGGCTAAC-3′. A total of 1 μL was removed from the first step as template for the second round of amplification using internal primers. Second round primer sequences were MOZ-4: 5′-CCAGAACAAGGATCCCTGT-3′ and CBP-2: 5′-AGCTTGACTAAAGGGCTGTC-3′. To amplify CBP-MOZ, the first round primer sequences were CBP-4: 5′-TGggaTCCCGAGCAGGTGAA-3′ and MOZ-7: 5′-GGAattCTCTCCACCACGCA-3′ (MOZ). Internal primer sequences were CBP-5: 5′-TACCCTACTCCAGCCATGCA-3′ and MOZ-5: 5′-TGGaTcCTCATAGTTTTCAG-3′. Small case letters in the primer sequences denote base substitutions introduced to create restriction enzyme recognition sites. As a positive control for MOZ-CBP, approximately 1 pg of a 764-bp RT-PCR product encompassing the fusion from a patient with a t(8; 16) (kindly provided by Dr J. Borrow, Massachusetts Institute of Technology, Cambridge, MA) was diluted into 1 μg normal peripheral blood leukocyte cDNA. No positive control was available for CBP-MOZ fusion transcripts, however normal MOZ or normal CBP products were amplifiable by single step PCR from normal peripheral blood leukocyte cDNA using both MOZ and both CBP sense primers in combination with both antisense primers from the same gene.
Single step PCRs were used to determine the presence or absence of the 5′ and 3′ ends of the MOZ gene in YACs 829D12 and 782H11. The 5′ primer sequences were MOZ-8: 5′-GGACTTAGGCCAGAAAACTC-3′ and MOZ-9: 5′-TATCCCTTATCCTGATGCTG-3′ (nucleotides 259 to 362; GenBank accession number U47742). The 3′ primer sequences were MOZ-3′-F: 5′-TTGTTGGTGGATGACTGTAC -3′ and MOZ-3′R: 5′-CTCGTTGTGTACTTGAACGC-3′ (nucleotides 7471 to 7786). Control for YAC DNA integrity was performed by PCR amplification of the URA3 gene in the YAC vector using primers URA+: 5′-CATTGGATGTTCGTACCACCAAG-3′ and URA-: 5′-GAGCCCTTGCATGACAATTCTG-3′. To determine if YACs 829D12 and 782H11 overlapped, DNA was amplified with primers to 782H11R, derived from the centromeric end of 782H11.17 All PCRs were run in 25 μL reactions with 0.5 μmol/L of each primer, 1.5 mmol/L MgCl2 , 100 μmol/L of each 2′-deoxynucleoside 5′-triphosphates (dNTP), and 0.625 U Taq polymerase. Conditions were 95°C, 1 minute; 60°C, 50 seconds; 72°C, 2 minutes for 30 cycles.
RESULTS
FISH analysis of 8p11 breakpoints. YAC 782H11 was tested on normal metaphases and produced a single hybridization signal on both chromosomes 8 and no other chromosomes (data not shown). On metaphases from UPN 03, YAC 782H11 mapped proximal to the breakpoint, with hybridization signals on the normal chromosome 8 and der(8) (Fig 2A). In contrast, YACs 847B12 and 953H12 both displayed hybridization signals on the normal chromosome 8 and der(13). As above, hybridization to normal metaphases resulted in a signal on 8p11 only. These data placed the t(8; 13) breakpoint between YACs 847B12 and 782H11 (Fig 1). Conversely, on metaphases from UPN 04, YAC 782H11 hybridized to the normal chromosome 8 and distal to the p-arm breakpoint on the inv(8). This indicated that the 8p11 breakpoints in these two disorders are separated by at least the size of YAC 782H11 (1,050 kb). Further FISH analysis of this case showed that YAC 829D12 was split in UPN 04, generating three hybridization signals, one on the normal chromosome 8 and two signals on the inv(8) (Fig 2B).
FISH analysis. (A) UPN 03 hybridized with YAC 782H11. Large arrow indicates the der(8) and the small arrow indicates the normal chromosome 8. (B) UPN 04 hybridized with YAC 829D12 and a chromosome 8 painting probe. Large arrow shows the normal chromosome 8; the small arrows show the split signal on the inv(8).
FISH analysis. (A) UPN 03 hybridized with YAC 782H11. Large arrow indicates the der(8) and the small arrow indicates the normal chromosome 8. (B) UPN 04 hybridized with YAC 829D12 and a chromosome 8 painting probe. Large arrow shows the normal chromosome 8; the small arrows show the split signal on the inv(8).
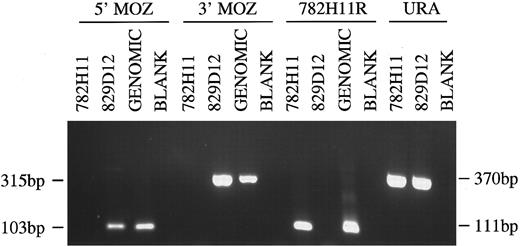
PCR analysis of YACs 782H11, 829D12 and human genomic DNA. Primer combinations used were to the 5′ and 3′ end of the MOZ gene, the centromeric end of YAC 782H11 and the URA gene. Sizes of PCR products are indicated.
PCR analysis of YACs 782H11, 829D12 and human genomic DNA. Primer combinations used were to the 5′ and 3′ end of the MOZ gene, the centromeric end of YAC 782H11 and the URA gene. Sizes of PCR products are indicated.
Mapping of MOZ gene to YAC clones. Southern blot analysis with the MOZ 0.9 probe, derived from the 3′ end of the MOZ coding sequence, showed hybridization to YAC 829D12, but not YAC 782H11 (not shown). Using PCR to amplify both the 5′ and 3′ ends of the MOZ gene, we observed positive signals of the expected sizes for 829D12, but not for 782H11. Amplification of the centromeric end of 782H11 (782H11R) showed that the two YACs did not overlap. Amplifiable DNA for each clone was confirmed by amplification of a fragment of the URA3 gene from the YAC vector (Fig 3).
Rearrangement of MOZ in the inv(8)(p11q13) case. Southern blot analysis using the MOZ 0.9 cDNA probe resulted in novel bands for UPN 04 after digestion with Bgl II and Hind III (Fig 4). Conversely, only the germline configuration was detected in the three t(8; 13) patients studied, after digestion of their DNA with five different restriction endonucleases (data not shown and Fig 4). To exclude the presence of a rare polymorphism, Bgl II digested DNA from 20 patients with chronic myeloid leukemia (CML) was probed with MOZ 0.9. In each case, only a single 7-kb band was observed (data not shown).
Southern blot of DNA from a control (CML patient without an 8p11 abnormality), UPN 03 and UPN 04 probed with MOZ 0.9. Restriction endonucleases used are indicated. Germline and rearranged bands are indicated by size and an arrow, respectively.
Southern blot of DNA from a control (CML patient without an 8p11 abnormality), UPN 03 and UPN 04 probed with MOZ 0.9. Restriction endonucleases used are indicated. Germline and rearranged bands are indicated by size and an arrow, respectively.
No evidence for CBP involvement. Southern blot analysis using the 5′ CBP probe did not show any abnormally migrating bands in UPN 04 in comparison with patients with CML (not shown). In addition, single step and nested RT-PCR assays failed to amplify either the MOZ-CBP or CBP-MOZ fusions from this patient (Fig 5).
PCR analysis of normal peripheral blood leukocyte (PBL) cDNA, cDNA from patient UPN 04 [inv(8)] and a MOZ-CBP positive control [t(8; 16)]. Primer combinations used were: MOZ-CBP, MOZ-6 + CBP-1; CBP-MOZ, CBP-4 + MOZ-7; MOZ, MOZ-6 + MOZ-7; CBP, CBP-4 + CBP-1. Sizes of PCR products are indicated.
PCR analysis of normal peripheral blood leukocyte (PBL) cDNA, cDNA from patient UPN 04 [inv(8)] and a MOZ-CBP positive control [t(8; 16)]. Primer combinations used were: MOZ-CBP, MOZ-6 + CBP-1; CBP-MOZ, CBP-4 + MOZ-7; MOZ, MOZ-6 + MOZ-7; CBP, CBP-4 + CBP-1. Sizes of PCR products are indicated.
DISCUSSION
Chromosome band 8p11 is one of the regions of the human genome in which there is a cluster of malignancy-associated breakpoints.21 In this report, we have attempted to clarify the molecular background of two distinct leukemia syndromes with 8p11 abnormalities.2,3 7
The translocation (8; 16)(p11; p13) in AML M4/M5 results in a rearrangement of the MOZ gene at 8p11 and the creation of a MOZ-CBP fusion transcript.9 Using FISH and Southern blotting, we have shown involvement of the MOZ gene in a case of AML-M5 with an inv(8)(p11q13) and a clinical phenotype apparently identical to that described in patients with a t(8; 16)(p11; p13). This is the first case without a t(8; 16) for which rearrangement of the MOZ gene has been found. The MOZ cDNA probe we used spans the t(8; 16) breakpoints described previously,9 suggesting that the position of inv(8) and t(8; 16) breakpoints are similar. We have not found MOZ sequences (either the 5′ or the 3′ ends) in YAC 782H11. Interestingly, this YAC, which is telomeric to the breakpoint in UPN 04 (Fig 1 and data not shown), was split in two cases with a t(8; 22)(p11; q13), considered to be another variant of the t(8; 16).22 Although we cannot exclude the possibility of a deletion in the YAC 782H11 clone we used, these data suggest that the t(8; 22) breakpoint may be distinct from the t(8; 16) and the inv(8).
In leukemia, a number of translocation-associated genes have been isolated that are capable of becoming fused to several different partner genes. Notable examples include the MLL gene at 11q2323,24 and the TEL gene at 12p1325-28; variant fusions have also been described that involve other genes such as E2A, RARα, CAN, and ABL.29 Typically, these various fusions are visible cytogenetically as distinct translocations. Occasionally, however, cytogenetic findings can be misleading, for example variant or complex translocations that obscure the presence of BCR-ABL in CML, or PML-RARα in acute promyelocytic leukemia.30 31 We sought, therefore, to establish whether the inv(8)(p11q13) obscured a typical t(8; 16)(p11; p13) and MOZ-CBP fusion, or whether it potentially indicated a novel fusion between MOZ and an uncharacterized gene at 8q13. Using Southern blotting and RT-PCR, we found no evidence for a rearrangement of the CBP gene, or the presence of MOZ-CBP or CBP-MOZ fusion transcripts. Taken together, the cytogenetic and molecular findings argue against the presence of a cryptic t(8; 16) in this case; definitive proof of existence of a new fusion gene in the inv(8), however, awaits the molecular cloning of the putative MOZ inversion partner at 8q13.
CBP is a multidomain transcriptional coactivator and histone acetyltransferase that shares structural and functional homology with the p300 protein.12 Interestingly, the gene encoding p300 maps to chromosome band 22q13,32 cytogenetically the same position as the breakpoint in the t(8; 22)(p11; q13).22 It is possible that CBP and p300 are part of a larger family of genes that become fused to MOZ in phenotypically similar cases of AML with abnormalities of 8p11.3 At present, no genes with such functional characteristics, or any other obvious candidate genes, have been mapped to chromosome band 8q13.
The myeloproliferative disorder with t(8; 13)(p11; q12) is part of a group of leukemic syndromes associated with 8p11 translocations. They are characterized by an initial myeloproliferative phase with marked eosinophilia, lymphadenopathy, and a high incidence of T- or B-cell non-Hodgkin's lymphoma with progression to acute myeloid leukemia.7,8 While the phenotype of the 8p11 myeloproliferative disorder and AML with a t(8; 16) differ considerably, this does not necessarily mean that the underlying molecular pathogenesis is different. For example, TEL, when fused to different translocation partners and also BCR-ABL, are associated with several distinct leukemia phenotypes.25-28 33
FISH analysis of one case with t(8; 13) showed that the 8p11 breakpoint was flanked by YACs 782H11 and 847B12, a region of approximately 4 cM.17 This indicates that the t(8; 13) breakpoint, at least in this case, is at least 1 Mb telomeric to the MOZ breakpoint found in UPN 04. While the genomic structure of MOZ is unknown, it is very unlikely that it spans such a large region. Furthermore, we did not find MOZ coding sequences within YAC 782H11, and Southern blot analysis of all three t(8; 13) cases showed no rearrangement of the MOZ gene.
In summary, we have made a preliminary characterization of the molecular basis of two distinct leukemia syndromes involving chromosome band 8p11. We have shown that the MOZ gene is not involved in the t(8; 13) myeloproliferative disorder and have narrowed down the region within which the breakpoint lies. We have shown that MOZ is disrupted in a new variant of the t(8; 16), an inv(8)(p11q13) and that this abnormality does not result in a cryptic MOZ-CBP fusion.
NOTE ADDED IN PROOF
After submission of this manuscript, we have studied two additional cases of 8p11 myeloproliferative disorder with t(8;13)(p11;q12). Using FISH, we have found that YAC 782H11 maps centromeric to the breakpoint at 8p11 in both cases. These data reinforce the notion that the chromosome 8 breakpoint in this leukemia syndrome consistently maps to an area at least 1 Mb distal to the MOZ gene, and support the idea that MOZ is not involved in its pathogenesis.
ACKNOWLEDGMENT
We would like to thank the Medical Research Council Human Genome Mapping Project (MRC HGMP) Resource Centre (Hinxton, UK) for providing the CEPH megaYAC clones used in this study, Dr J. Borrow (Massachusetts Institute of Technology, Cambridge, MA) for providing the MOZ-CBP RT-PCR product, Dr G. Jackson and N. Bown (Newcastle, UK) for providing patient samples.
Supported by CNDq (Conselho Nacional de Desenvolvimento Cientifico e Tecnologico) Grant No. 200995/94-4, Brazil (R.C.T.A.), the Leukaemia Research Fund (UK), the Dr Mildred Scheel Stiftung (Germany), and the Lady Tata Memorial Fund (UK).
Address reprint requests to Nicholas C.P. Cross, PhD, LRF Centre for Adult Leukaemia, Royal Postgraduate Medical School, Hammersmith Hospital, Du Cane Rd, London W12 0NN, U.K.



![Fig. 5. PCR analysis of normal peripheral blood leukocyte (PBL) cDNA, cDNA from patient UPN 04 [inv(8)] and a MOZ-CBP positive control [t(8; 16)]. Primer combinations used were: MOZ-CBP, MOZ-6 + CBP-1; CBP-MOZ, CBP-4 + MOZ-7; MOZ, MOZ-6 + MOZ-7; CBP, CBP-4 + CBP-1. Sizes of PCR products are indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/8/10.1182_blood.v90.8.3130/4/m_bl_0012f5.jpeg?Expires=1769331966&Signature=KB01-XdWhE6njDfjhpZaTMBBHykBFcZTrCLKVeXqBP421hzYIaufeh-bt4CU1OdZU-4ZxVjwBPBNPJnt2sJLci5zGPpMf43zcZ12AdQc6von-QsNsFOrmUOW2iX5G6hjBy5usrHOUOcEZ2Uy3txzaLV-s4xH-CBmiG7N6rUxU9sZrwXsDJagL5QyUMaRdze~iIHlEr~~XzbsJXj~mhSGD1tNDrdxSwVPjv98tGc0mJ8gDlLqKzwRZntIiAvzWAxre1IJtUJ6Jo5-vZhvhONdXBOQtIZOo6CkEt64IU4KE05UKiiWEu8-gKJbZ87WSHWwEQTrD7ppSCu~h3npTaa9zA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
