Abstract
We report the fusion of the Huntingtin interactin protein 1(HIP1) gene to the platelet-derived growth factor βreceptor (PDGFβR) gene in a patient with chronic myelomonocytic leukemia (CMML) with a t(5;7)(q33;q11.2) translocation. Southern blot analysis of patient bone marrow cells with a PDGFβR gene probe demonstrated rearrangement of the PDGFβR gene. Anchored polymerase chain reaction using PDGFβRprimers identified a chimeric transcript containing the HIP1gene located at 7q11.2 fused to the PDGFβR gene on 5q33. HIP1 is a 116-kD protein recently cloned by yeast two-hybrid screening for proteins that interact with Huntingtin, the mutated protein in Huntington's disease. The consequence of t(5;7)(q33;q11.2) is an HIP1/PDGFβR fusion gene that encodes amino acids 1 to 950 of HIP1 joined in-frame to the transmembrane and tyrosine kinase domains of the PDGFβR. The reciprocalPDGFβR/HIP1 transcript is not expressed. HIP1/PDGFβR is a 180-kD protein when expressed in the murine hematopoietic cell line, Ba/F3, and is constitutively tyrosine phosphorylated. Furthermore, HIP1/PDGFβR transforms the Ba/F3 cells to interleukin-3–independent growth. These data are consistent with an alternative mechanism for activation of PDGFβR tyrosine kinase activity by fusion with HIP1, leading to transformation of hematopoietic cells, and may implicate Huntingtin or HIP1 in the pathogenesis of hematopoietic malignancies.
MYELODYSPLASTIC syndromes (MDS) and acute leukemias are disorders of hematopoietic progenitor cells characterized by acquired somatic mutations that confer a proliferative advantage. Cloning of chromosomal translocation breakpoints has been a productive strategy for identification of disease genes in MDS. Examples include the AML1-EVI1 fusion associated with t(3;21),1 theMLL-CBP fusion with t(11;16) in therapy-related MDS,2 the TEL-EVI1 fusion in t(3;12),3the NPM-MLF1 fusion associated with t(3;5) in primary MDS,4 and the TEL/JAK25 andTEL/PDGFβR fusions that are associated with t(9;12) and t(5;12) in chronic myelomonocytic leukemia (CMML), respectively.6
Several of the fusion genes associated with hematopoietic disorders involve tyrosine kinases. These include BCR/ABL,7,8TEL/PDGFβR,6TEL/ABL,9TEL/JAK2,5,10 andCEV14/PDGFβR.11 CMML, associated with the TEL/PDGFβR, is a subtype of MDS characterized by dysplastic monocytosis, variable bone marrow fibrosis, and progression to acute leukemia. The clinical phenotype is similar to chronic myelogenous leukemia associated with constitutive activation of the ABL kinase by fusion with BCR. There is also convincing evidence for contribution of tyrosine kinases to pathogenesis of solid tumors. Noteworthy examples include point mutations that constitutively activate the RETtyrosine kinase gene in medullary carcinoma of the thyroid,12 amplification of the HER2/neu receptor tyrosine kinase in breast cancer,13 and the ETV6-NTRK3 gene fusion in congenital fibrosarcoma.14
A subset of patients with CMML have a t(5;12)(q33;p13) that results in fusion of the amino terminal portion of TEL, which contains the pointed (PNT) oligomerization domain, to the transmembrane and tyrosine kinase domains of platelet-derived growth factor β receptor (PDGFβR). The consequence of the fusion is constitutive oligomerization and activation of PDGFβR tyrosine kinase activity leading to transformation of cells.15 PDGFβR kinase activity is required for transformation of Ba/F3 cells as is the PNT domain.15 We report here a novel PDGFβR fusion associated with CMML and t(5;7)(q33;q11.2) involving the Huntingtin Interacting Protein 1 (HIP1).
MATERIALS AND METHODS
DNA isolation and Southern blots.
Leukocytes were isolated by ficoll sedimentation from the peripheral blood and bone marrow of the index patient and normal controls after informed consent had been obtained. DNA was prepared using standard methods.16 After enzymatic digestion with restriction endonucleases and electrophoretic separation of fragments, the genomic DNA was transferred to HYbond N nylon membranes (Amersham, Arlington Heights, IL). The PDGFβR genomic probe was a 1.1-kb HindIII-Xho I fragment prepared from PDGFβR cosmid B.17 Probes were labeled with 32P by random priming, and Southern hybidizations were performed as described.18
Cloning of the t(5;7) breakpoint.
Mononuclear cells were isolated from t(5;7) bone marrow cells by ficoll sedimentation. Anchored polymerase chain reaction (PCR) was performed to clone the human chromosome 7 partner gene according to the method of Frohman19 with minor modifications. In brief, total RNA was prepared with RNA-STAT reagents according to the manufacturer's recommendations (Tel-Test, Inc, Friendswood, TX). RNA (3 μg) was reverse transcribed using avian myeloblastosis virus (AMV) reverse transcriptase and PDGFβR oligonucleotide primer 1873R (5′-CGTAACGTGGCTTCTTCTGC-3′). A poly(A) tail was appended using terminal transferase and dATP at 37°C for 15 minutes. After a single cycle of amplification (94°C for 1 minute, 50°C for 2 minutes, and 72°C for 40 minutes) using primer Qt (5′-TGAGCAGAGTGACTATTACTCGAGCTCAAGCTTTTTTTTTTTT-3′) and internal PDGFβR primer 1848R (5′-AGTCTCGAGCATGATGAGGATGATAAG-3′), 30 cycles of PCR (94°C for 1 minute, 58°C for 2 minutes, and 72°C for 3 minutes) were performed with primers Qo (5′-CCAGTGAGCAGAGTGACG-3′) and 1848R. The PCR products were diluted 20-fold and reamplified with nested primers 1829R (5′-GAGATGATGGTGGAGCACCAC-3′) and Q1 (5′-GAGGACTCGAGCTCAAGC) using the same PCR conditions (30 cycles of 94°C for 1 minute, 58°C for 2 minutes, and 72°C for 3 minutes). Specific bands were not detected by direct visualization after ethidium bromide staining, but were detectable by Southern blot analysis using the 32P–end-labeled PDGFβR 1806R oligo (5′-GGCCAGGATGGCTGAGATCA-3′). The nested PCR product was subsequently diluted 20-fold and reamplified with primer 1806R and Q1 (30 cycles of 94°C for 1 minute, 58°C for 2 minutes, and 72°C for 3 minutes), yielding a specific 500-bp product that was subcloned into pBluescript KS(+) (Stratagene, La Jolla, CA) and sequenced. The DNA sequence was sent via Netscape to the BLAST server at NIH (http://www.ncbi.nlm.nih.gov) to compare to GenBank (blastn).
Library screening.
In light of the high mRNA levels of HIP1 in tumor cell lines, the cDNA sequence isolated by anchored PCR was used to screen a λgt11 SW480 colon cancer cell line cDNA library (Clontech, Palo Alto, CA) to obtain more 5′ sequence. The 500-bp PCR product obtained from cloning of the breakpoint was labeled with32P by random priming and plaque lifts were performed.18 Positive phage clones were subcloned into pBluescript KS(+) and sequenced.
In the longest clone, SW9, there were 2 new potential in frame initiator methionines in the additional 5′ sequence. The first ATG at nucleotides 16-19 has a better Kozak consensus sequence than the second ATG (nucleotides 37-39). The third methionine at position 368-370 (or 248-250 of the previously published sequence20) has the best fit of the 3, because it has a purine at position −3. No upstream stop codons were identified. However, native HIP1 migrates as a protein of 116 kD by Western blot analysis,20 21 which is consistent with a preferred start site at methionine 368-370.
Reconstruction of the fusion cDNA for expression experiments.
The chromosome translocation breakpoint was amplified from patient material using primers HIP1301F (5′-CCTGAAACTGCTAAGAACCA-3′) and PDGFβR 1806R, and the product was digested with Bgl I and Nhe I. TheBgl I-Sac II fragment of thePDGFβR was isolated after Bgl I andSac II digestion of the PDGFβR cDNA and ligated to the Nhe I-Bgl I breakpoint fragment. This ligation reaction was amplified with primers containing the NheI and Sac II sites (5′-AAATTGCTGCTAGCACAGCCCAGCTTG-3′ and 5′-CTGGTCCCGCGGCAGCTCCCACGTGGA-3′ respectively), digested with Sac II, and ligated with the 3′ end ofPDGFβR.22 The reaction mixture was then digested with Nhe I and ligated with the 5′ end ofHIP1 (from SW480 l clone 9) via the unique Nhe I site. The region amplified by PCR was confirmed to be void of PCR generated mutations by sequence analysis.
Stable expression of HIP1/PDGFβR.
The full-length fusion cDNA was subcloned into the pMSCVneo vector (kindly provided by R. Hawley, University of Toronto, Toronto, Ontario, Canada). Bosc cells (the kind gift of W. Pear, University of Pennsylvania, Philadelphia, PA) were transfected via the calcium phosphate technique.18 The 48-hour supernatent (1 mL) was then added to 106 Ba/F3 cells (1 mL) in the presence of polybrene (4 μL) as described previously.23Cells with stable expression were selected in the presence of G418 and interleukin-3 (IL-3) as described.15
RESULTS
Identification of HIP1/PDGFβR in CMML.
The HIP1/PDGFbR fusion was cloned from a single patient with a clinical phenotype of CMML. The patient was a 54-year-old man who presented with fatigue, weight loss, and splenomegaly. Laboratory evaluation showed monocytosis, anemia, and peripheral eosinophilia. Bone marrow biopsy showed a hypercellular marrow with increased myeloid:erythroid ratio, eosinophilia, and dysplastic maturation of monocyte lineage cells. Cytogenetic analysis of the bone marrow cells showed t(5;7)(q33;q11.2) (data not shown).
We hypothesized that the PDGFβR on 5q33 was activated as a consequence of fusion to a novel partner on chromosome 7. This region of chromosome 7 is of particular interest because it is frequently deleted in de novo and therapy-related MDS/AML. Rearrangement of thePDGFβR gene was demonstrated by Southern blot analysis of EcoRI, BamHI, Pst I, EcoRV, and HindIII digests using a 1.1-kbPDGFβR genomic probe localized near theTEL/PDGFβR breakpoint (Fig 1). These data demonstrated that the chromosome 5 breakpoint was at or near the same intron ofPDGFβR as for the t(5;12)(q33;p12) breakpoint.6
Identification and molecular analysis of t(5;7)(q33;q11.2). Southern blot analysis of thePDGFβR gene locus in patient DNA with t(5;7)(q33;q11.2). Genomic DNA of patient t(5;7)-positive cells (lane 1) and control cells (lane 2) was analyzed by Southern blotting with a 1.1-kb HindIII-Xho I PDGFβRprobe.6 Arrows indicate the rearranged bands in theEcoRI, BamHI, Pst I, EcoRV, andHindIII digests.
Identification and molecular analysis of t(5;7)(q33;q11.2). Southern blot analysis of thePDGFβR gene locus in patient DNA with t(5;7)(q33;q11.2). Genomic DNA of patient t(5;7)-positive cells (lane 1) and control cells (lane 2) was analyzed by Southern blotting with a 1.1-kb HindIII-Xho I PDGFβRprobe.6 Arrows indicate the rearranged bands in theEcoRI, BamHI, Pst I, EcoRV, andHindIII digests.
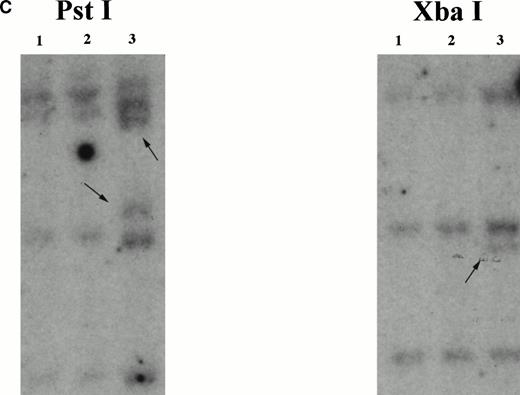
The chromosome 7 fusion partner was identified using anchored PCR withPDGFβR primers to amplify the fusion transcript from the patient's bone marrow cell cDNA (Fig 2A). Analysis of the amplified cDNA clones demonstrated 500 bp of non-PDGFβRsequence encoding an open reading frame fused to the transmembrane and tyrosine kinase encoding regions of thePDGFβR gene (Fig 2B). A database search showed this sequence to be identical to the HIP 1gene20,21 localized by fluorescence in situ hybridization (FISH) to 7q11.2.20 Southern blot analysis with anHIP1 cDNA probe (nucleotides 1141-3041) demonstrated rearranged bands in Pst I and Xba I digests of patient DNA (Fig2C).
Identification of the chromosome 7 fusion partner. (A) Schematic diagram of anchored PCR.6 19 (B) Sequence of theHIP1/PDGFβR breakpoint and schematic of the fusion protein. (C) Southern blot analysis of HIP1 gene locus in control DNA (lanes 1 and 2) and patient DNA (lane 3). Arrows indicate rearranged fragments in the Pst I and Xba I digests.
Identification of the chromosome 7 fusion partner. (A) Schematic diagram of anchored PCR.6 19 (B) Sequence of theHIP1/PDGFβR breakpoint and schematic of the fusion protein. (C) Southern blot analysis of HIP1 gene locus in control DNA (lanes 1 and 2) and patient DNA (lane 3). Arrows indicate rearranged fragments in the Pst I and Xba I digests.
HIP1 is a 116-kD protein that was cloned by yeast two-hybrid screening for proteins that interact with Huntingtin. Huntingtin is the protein mutated in Huntington's disease.24 HIP1 has a leucine zipper motif and homology to talin, a cytoskeletal associated protein, at amino acids 412-433 and 861-900, respectively.20
HIP1 has homology with the SLA2 gene product (Sla2p) fromSaccharomyces cerevisiae,25 an essential cytoskeletal associated protein. The leucine zipper motif and talin homology domain of HIP1 are conserved in Sla2p. HIP1 is also homologous to the Caenorhabditis elegans ZK370.3 gene product that has no known function.26 The degree of homology (40% similarity, 20% identity in both yeast and worm) suggests that HIP1 is the human homologue of these proteins. The highest degree of homology is at the carboxy terminus, where all 3 proteins share homology with talin.27
Tissue expression of HIP1/PDGFβR and HIP1.
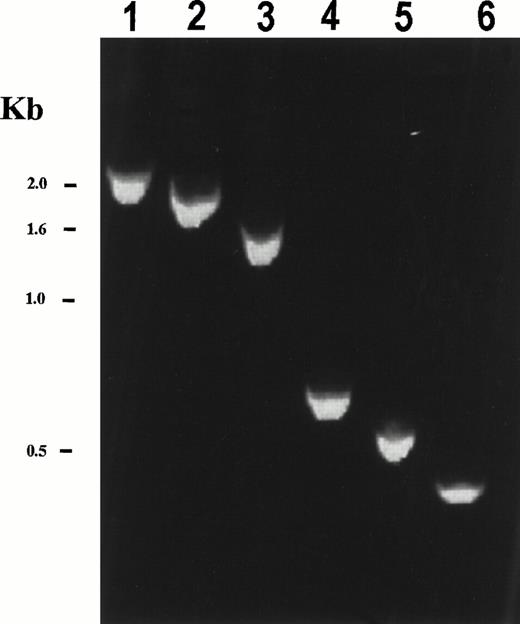
Reverse transcription-PCR (RT-PCR) using PDGFβR 3′ primers and 5′ primers spanning the coding sequence of HIP1 generated the expected size fragments from patient cDNA (Fig 3), but was not detected in mRNA from normal bone marrow. The reciprocal PDGFβR/HIP1fusion could not be detected by RT-PCR analysis (data not shown). Because bone marrow cells from the patient were limited, detection of the fusion transcript required the use of nested PCR primers. In addition, because of this limitation, Northern blot analysis of patient material was not possible. HIP1/PDGFβR protein contained nearly all of the HIP1 coding sequence, including the leucine zipper and talin homology domains, fused in frame to the transmembrane and tyrosine kinase domain of the PDGFβR (Fig 2B). Only 18 C-terminal amino acids of HIP1 were excluded from the fusion protein.
Expression of the chimericHIP1/PDGFβR mRNA in patient bone marrow. RT-PCR analysis of HIP1/PDGFβR was performed using total RNA (2 μg) from t(5;7) patient bone marrow that had been reverse transcribed using the Qt primer. PCR was performed using a HIP301F forward primer and the PDGFβR 1848R primer for 30 cycles (94°C for 1 minute, 60°C for 2 minutes, and 72°C for 3 minutes). Nested PCR was performed on PCR products from the first reaction diluted 20-fold and amplified using the same PCR reaction conditions with HIP1 forward primers HIP721F, HIP1141F, HIP1561F, HIP2372F, HIP2494F, and HIP2613F in lanes 1 through 6, respectively, and the PDGFβR reverse primer 1806R. HIP1 primer numbers correspond to the first nucleotide of a 20-bp primer of HIP1 sequence according to Kalchman et al.20 The expected band sizes are 2,279, 1,859, 1,439, 628, 506, and 387 bp for lanes 1 through 6, respectively. Control experiments with no template or in the absence of reverse transcriptase gave no PCR product (not shown). TheHIP1/PDGFβR fusion was not detected in normal bone marrow and neither was the reciprocalPDGFβR/HIP1 fusion transcript detected in t(5;7) patient bone marrow using a nested PCR reaction with primers PDGFβR1691F, PDGFβR1711F, HIP13071R, and HIP12966R (primer numbers correspond to the first nucleotide of a 20-bp primer of either PDGFβR22 orHIP120 sequence).
Expression of the chimericHIP1/PDGFβR mRNA in patient bone marrow. RT-PCR analysis of HIP1/PDGFβR was performed using total RNA (2 μg) from t(5;7) patient bone marrow that had been reverse transcribed using the Qt primer. PCR was performed using a HIP301F forward primer and the PDGFβR 1848R primer for 30 cycles (94°C for 1 minute, 60°C for 2 minutes, and 72°C for 3 minutes). Nested PCR was performed on PCR products from the first reaction diluted 20-fold and amplified using the same PCR reaction conditions with HIP1 forward primers HIP721F, HIP1141F, HIP1561F, HIP2372F, HIP2494F, and HIP2613F in lanes 1 through 6, respectively, and the PDGFβR reverse primer 1806R. HIP1 primer numbers correspond to the first nucleotide of a 20-bp primer of HIP1 sequence according to Kalchman et al.20 The expected band sizes are 2,279, 1,859, 1,439, 628, 506, and 387 bp for lanes 1 through 6, respectively. Control experiments with no template or in the absence of reverse transcriptase gave no PCR product (not shown). TheHIP1/PDGFβR fusion was not detected in normal bone marrow and neither was the reciprocalPDGFβR/HIP1 fusion transcript detected in t(5;7) patient bone marrow using a nested PCR reaction with primers PDGFβR1691F, PDGFβR1711F, HIP13071R, and HIP12966R (primer numbers correspond to the first nucleotide of a 20-bp primer of either PDGFβR22 orHIP120 sequence).
Northern blot analysis using an HIP1 cDNA probe demonstrated a previously reported 9.4-kb transcript in all tissues tested20 as well as a 2.4-kb transcript present in testis. There are high levels of expression in solid tumor cell lines, including HeLa, SW480, A549, and G861, as well as in testis. Although HIP1 has been implicated in pathogenesis of central nervous system disorders such as Huntington syndrome, there are lower levels of expression in brain and other adult tissues such as bone marrow and peripheral blood (Fig 4 and data not shown).
Northern blot analysis of HIP1 mRNA in various tissues. Blots (Clontech) were probed with with an32P–end-labeled probe from HIP1 nucleotides 2890 to 2930.20 Exposure time was 12 hours. The lower panels are the same blots stripped and reprobed with actin cDNA. (A) Cell lines. RNA sources were HL60, HELA, K562, MOLT4, Raji, SW480, A549, and G361, designated 1 through 8, respectively. (B) Adult tissue RNA sources are spleen, thymus, prostate, testis, ovary, small intestine, colonic mucosa, and peripheral blood, designated 1 through 8, respectively.
Northern blot analysis of HIP1 mRNA in various tissues. Blots (Clontech) were probed with with an32P–end-labeled probe from HIP1 nucleotides 2890 to 2930.20 Exposure time was 12 hours. The lower panels are the same blots stripped and reprobed with actin cDNA. (A) Cell lines. RNA sources were HL60, HELA, K562, MOLT4, Raji, SW480, A549, and G361, designated 1 through 8, respectively. (B) Adult tissue RNA sources are spleen, thymus, prostate, testis, ovary, small intestine, colonic mucosa, and peripheral blood, designated 1 through 8, respectively.
Transformation of Ba/F3 cells by HIP1/PDGFβR.
To characterize the biological properties of the HIP1/PDGFβR fusion, a full-length cDNA encoding HIP1 was obtained by screening a λgt11 SW480 colon cancer cell line cDNA library. The sequence of the longest clone, SW9, from this library is identical to the published sequence, but incorporates and additional 120 nucleotides of 5′ sequence.
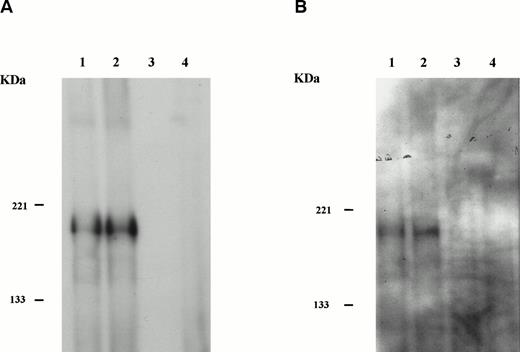
Clone SW9 was then used to reconstruct the full-lengthHIP1/PDGFβR (see the Materials and Methods). Transforming properties of HIP1/PDGFβR were tested by subcloning the cDNA encoding the HIP1/PDGFβRinto the retroviral vector MSCVneo and obtaining stable expression of the fusion protein under control of the LTR in the murine Ba/F3 hematopoietic cell line (Fig 5A). The HIP1/PDGFβR protein was constitutively tyrosine phosphorylated in stably transfected cells (Fig 5B).
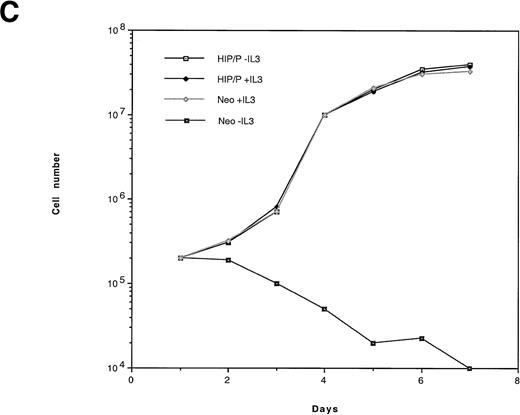
HIP1/PDGFβR transforms Ba/F3 cells to factor independence. Two independent infections of Ba/F3 cells were performed. To assess protein expression and phosphorylation, lysates were immunoprecipitated with anti-PDGFβR antibody (tail; Pharmingen), separated on 8% polyacrylamide gel electrophoresis (PAGE), and blotted onto nitrocellulose. Proteins were detected with anti-PDGFβR peptide antibody directed against the C-terminus (part a) and HRP-conjugated anti-phosphotyrosine 4G10 monoclonal antibody (part b). Lanes 1 and 2 are the HIP1/PDGFβR stable infectants, and lanes 3 and 4 are neomycin-resistant controls. (C) The G418-resistant cells growing in IL-3 were seeded in 96-well trays with 2 × 104 cells per 200 μL per well in RPMI 1640 and 10% fetal calf serum media with or without IL-3. Cells were assessed for number and viability (trypan blue) in triplicate at 24-hour intervals. Each point is the average of the triplicate samples, with standard deviations ranging from 1% to 3% of the number of cells counted each day.
HIP1/PDGFβR transforms Ba/F3 cells to factor independence. Two independent infections of Ba/F3 cells were performed. To assess protein expression and phosphorylation, lysates were immunoprecipitated with anti-PDGFβR antibody (tail; Pharmingen), separated on 8% polyacrylamide gel electrophoresis (PAGE), and blotted onto nitrocellulose. Proteins were detected with anti-PDGFβR peptide antibody directed against the C-terminus (part a) and HRP-conjugated anti-phosphotyrosine 4G10 monoclonal antibody (part b). Lanes 1 and 2 are the HIP1/PDGFβR stable infectants, and lanes 3 and 4 are neomycin-resistant controls. (C) The G418-resistant cells growing in IL-3 were seeded in 96-well trays with 2 × 104 cells per 200 μL per well in RPMI 1640 and 10% fetal calf serum media with or without IL-3. Cells were assessed for number and viability (trypan blue) in triplicate at 24-hour intervals. Each point is the average of the triplicate samples, with standard deviations ranging from 1% to 3% of the number of cells counted each day.
To assay the ability of HIP1/PDGFβR to confer IL-3–independent proliferation, Ba/F3-transfected cells were seeded in 96-well trays at a concentration of 2 × 104 cells/well. Cells infected with insert-free virus failed to proliferate in the absence of IL-3 and died. In contrast, HIP/PDGFβR-expressing cells grew at the same rate in the presence or absence of IL-3 (Fig 5C).
DISCUSSION
Involvement of HIP1 in the pathogenesis of leukemia is a novel finding. As for TEL/PDGFβR, fusion of HIP1 to PDGFβR results in constitutive activation of PDGFβR as assessed by tyrosine autophosphorylation and may be mediated by oligomerization through the HIP1 leucine zipper domain. However, other interactions with this fusion may be relevant to the pathogenesis of leukemia in addition to the protein-protein interation mediated by the leucine zipper. For example, the leucine zipper is not necessary for the interaction of HIP1 with Huntingtin.20 Although the role of HIP1 in Huntingtin's disease is still under investigation, it is conceivable that inhibition of apoptosis mediated by HIP1, as suggested in by Kalchman et al,20 is also relevant in leukemogenesis mediated by the HIP1/PDGFβR fusion. Thus, there may be an additional role for HIP1 in the leukemogenic pathway.
There are several possible consequences of expression of the HIP1/PDGFβR fusion that may be relevant to leukemogenesis. These could include interference with the function of the native HIP1 protein or the Huntingtin protein through HIP1/PDGFβR heterodimerization. In addition, it is possible that, because the HIP1/PDGFβR is an activated tyrosine kinase, heterodimerization with HIP1 or Huntingtin could modify function or protein stability of these associated proteins through tyrosine phosphorylation. Because the physiologic roles of HIP1, Huntingtin, and the HIP1/Huntingtin complex are not well understood, it is difficult to speculate at this time about the relevance of these interactions in the mechanism of transformation. However, it should be possible to directly assay for these interactions and determine whether dysregulation of the apoptotic pathway in hematopoietic cells as a consequence of HIP1/PDGFβR interactions with HIP1 or Huntingtin contributes to leukemogenesis.
Finally, HIP1 localization to 7q11.2 raises the possibility of involvement in 7q− deletions. Loss of chromosome 7 or deletion of the long arm, del(7q), is observed in 10% of MDS or AML de novo and in greater than 50% of therapy-related AML.28 7q− cytogenetics are associated with particularly poor prognosis.28 It has been hypothesized that deleted chromosomal bands contain as yet unidentified myeloid-specific tumor suppressor loci. Most 7q deletions are interstitial and some have been found to be the result of cryptic translocations.29-31 The majority of patients with deletions have proximal breakpoints in bands q11-21 and distal break points in q31-36.29-31 FISH has been used with a panel of YAC clones from 7q to examine patients with deletion breakpoints near 7q22. These data are being used to to narrow down the region of deletion to clone tumor-suppressor genes on 7q involved in myeloid leukemia.29-31 Other data that may be useful in narrowing the critically deleted region is to characterize translocations in this area, such as the t(5;7) described herein, that could result in inactivation or dysregulaton of tumor-suppressor genes. Examples in which translocations that disrupt the function or expression of tumor-supporssor genes include a translocation that disrupts p16.32 In addition, it has been proposed that disruption of PML, a putative growth suppressor, as a consequence of the t(15;17) chromosomal translocation, may contribute to the pathogenesis of acute promyelocytic leukemia.33 The possibility that HIP1 is one of the important genes in this area is currently being investigated.
Analysis of HIP1/PDGFβR should contribute to our understanding of the pathogenesis of CMML and may help to elucidate the function of Huntingtin and HIP1 in normal and neoplastic cells. Furthermore, localization of HIP1 to 7q11.2 warrants an investigation of its relevance in pathogenesis of hematopoietic malignancies with 7q deletions.
ACKNOWLEDGMENT
The authors are grateful to James Griffin, Charis Eng, and Thomas McClean for critical review of this work.
Supported in part by National Institutes of Health Grants No. T32 HL07623 and PO1CA66996-01, the Lawrence Family foundation, the Ligue Nationale Contre le Cancer and the Ligue Nationale Contre le Cancer, Comité de Paris. D.G.G. is a Stephen Birnbaum Scholar of the Leukemia Society of America and an assistant investigator of the Howard Hughes Medical Institute.
Address reprint requests to D. Gary Gilliland, MD, PhD, Division of Hematology/Oncology, Brigham and Women's Hospital, 4 Blackfan Circle, Boston, MA 02115.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.