Abstract
Embryonic stem cells and embryonal carcinoma P19 cells produce erythropoietin (Epo) in an oxygen-independent manner, although lactate dehydrogenase A (LDHA) is hypoxia-inducible. To explore this paradox, we studied the operation of cis-acting sequences from these genes in P19 and Hep3B cells. The Epo gene promoter and 3′ enhancer from P19 cells conveyed hypoxia-inducible responses in Hep3B cells but not in P19 cells. Together with DNA sequencing and the normal transcription start site of P19 Epo gene, this excluded the possibility that the noninducibility of Epo gene in P19 cells was due to mutation in these sequences or unusual initiation of transcription. In contrast, reporter constructs containing LDHA enhancer and promoter were hypoxia inducible in P19 and Hep3B cells, and mutation of a hypoxia- inducible factor 1 (HIF-1) binding site abolished the hypoxic inducibility in both cells, indicating that HIF-1 activation operates normally in P19 cells. Neither forced expression of hepatocyte nuclear factor 4 in P19 cells nor deletion of its binding site from the Epo enhancer was effective in restoring Epo enhancer function. P19 cells may lack an unidentified regulator(s) required for interaction of the Epo enhancer with Epo and LDHA promoters.
MAMMALS RESPOND TO oxygen deficiency in many different ways (reviewed in Bunn and Poyton1). One strategy for survival of the individual cell under hypoxic conditions is the induction of glycolytic enzymes, facilitating ATP production by glycolysis rather than mitochondrial oxidative phosphorylation. Because ATP production is vital for cell survival, such a response is appropriate in all cell types, including undifferentiated cells. In contrast, erythropoietin (Epo), a hypoxia-inducible growth factor, is produced by the specialized cells in liver, kidney, and brain.2-4 Peritubular interstitial fibroblast-like cells have been proposed to produce Epo in kidney,5 whereas in liver the gene is expressed in both hepatocytes and Ito cells.6,7 Hypoxia-stimulated production of Epo in liver and kidney increases formation of red blood cells, leading to better oxygen supply to tissues. Astrocytes are responsible for Epo production in the central nervous system and the hypoxic induction of brain Epo may contribute to protect neurons from ischemia-induced cell death.4 8
Induction of the hypoxia-inducible genes is at least in part due to activation of gene transcription, although prolongation of mRNA half-life may contribute in some cases. Survey of cis-acting DNA sequences required for the hypoxic activation of gene transcription has shown that most of the hypoxia-inducible genes thus far investigated possess hypoxia-response elements (enhancer) to which hypoxia-inducible factor 1 (HIF-1) binds.9-27 HIF-1 is a transcription factor consisting of two basic helix-loop-helix-PAS proteins (HIF-1α and HIF-1β).19-21 Taken together with the wide distribution of HIF-1–like proteins from mammalian to insect cells, it appears that the hypoxia-stimulated transcription of specific genes through HIF-1 activation is a highly conserved and widely operative mechanism responding to cellular oxygen deficiency.13,17 28
The genes encoding glycolytic enzymes, including aldolase A, phosphoglycerate kinase 1, enolase 1, phosphofructokinase L, and lactate dehydrogenase A (LDHA), are hypoxia-inducible.22-25,29 Recent studies ofcis-acting sequences for genes encoding enolase 1 and LDHA indicated that they have multiple sites for HIF-1 binding in the 5′-flanking region and that binding to a single specific site is essential for the hypoxic activation of transcription but not sufficient for the full activation.24,25 The full activation appears to require binding of HIF-1 to multiple sites, if not all.25
Human hepatoma cell lines, Hep3B and HepG2, have been shown to produce Epo in an oxygen-dependent manner and have been widely used for investigation of the mechanism underlying the hypoxic activation of Epo gene.30,31 An important cis-acting sequence required for the hypoxic induction of Epo gene was defined in the 3′-flanking region.9-14,32,33 This enhancer is a 50-bp element consisting of three important segments.12,14,33 The highly conserved sequence near the 5′ end of the enhancer is an HIF-1 binding site.12,15-17 The middle segment containing CA repeats in human gene is not well conserved between human and mouse, but the mutation of this region abolished the inducible behavior of both the human and murine enhancers.12,18 Thus far, the specific protein that binds to this region has not been demonstrated. The third element is the 3′ segment, which is again highly conserved and amplifies the hypoxic signal.12,32,33 Binding of hepatocyte nuclear factor 4 (HNF-4) to this element appears to increase the hypoxic inducibility.33 Whereas the regulatory system for Epo gene expression in liver cells is not fully understood, even less is known of kidney cells and astrocytes.
Mouse embryonic stem (ES) cells express Epo mRNA,34 but oxygen-dependency of its expression is not known. If we assume that some factors required for the hypoxic induction of Epo are expressed only in the cells differentiated for Epo production but not in undifferentiated cells and that the hypoxia-inducible pathway for glycolytic enzymes is complete in most cell types (including undifferentiated cells), we might predict that expression of Epo by ES cells would be oxygen-independent, whereas that of glycolytic enzymes would be hypoxia-inducible. Our finding that multipotential embryonal carcinoma P19 cells produce Epo has made it possible to examine this assumption and the underlying mechanisms. We show here that P19 cells express LDHA in an oxygen-dependent manner, but produce Epo constitutively. We also compare aspects of hypoxic transcriptional activation in P19 cells with those in Hep3B cells (specialized for hypoxia-inducible production of Epo), using reporter constructs that contain promoters and enhancers of the LDHA and Epo genes. In addition, treatment with retinoic acid differentiates P19 cells into astrocytes,35 which is a cell type producing brain Epo in a hypoxia-inducible manner.4 P19 cells thus may provide a useful system for searching for a regulator(s) that is expressed in the differentiated Epo-producing cells and needed for the hypoxic activation of Epo gene transcription.
MATERIALS AND METHODS
Cell culture and Epo production.
Materials for cell culture were obtained from the indicated sources: Dulbecco's modified Eagle medium (DMEM; GIBCO, Grand Island, NY), Minimum Essential Medium α without ribonucleotides and deoxyribonucleotides (αMEM; GIBCO), and fetal calf serum (FCS; Bio Whittaker, Walkersville, MD). P19 cells were maintained in αMEM supplemented with 10% FCS (growth medium for P19 cells) in a gelatin-coated dish. Hep3B cells were maintained in DMEM supplemented with 10% FCS (growth medium for Hep3B cells). The cells were cultured under normoxic conditions (5%CO2, 21% O2, and 74% N2 atmosphere) in a Napco Model 5100 CO2incubator (Wakenyaku Co Ltd, Kyoto, Japan) at 37°C. In the experiments to examine Epo production, P19 cells (5 × 104/well) and Hep3B cells (2 × 105/well) were plated in two 6-well plates (Nunc A/S, Roskilde, Denmark) and cultured for 24 hours in 21% oxygen. Then the medium was replaced with the fresh growth medium. One of the two plates was incubated for a further 24 hours in 21% oxygen, whereas the other was incubated for 24 hours in 2% oxygen. Hypoxia was generated in an oxygen-regulated incubator (MODEL 9200; Wakenyaku Co Ltd, Kyoto, Japan). Epo in the spent medium was measured by enzyme-linked immunoassay (EIA) and its amount was calculated from a standard curve drawn using recombinant Epo as a standard.36 To measure the activity of LDH and the amount of cellular proteins, the cells were washed three times with 10 mmol/L phosphate-buffered saline, pH 7.4 (PBS), and lysed by freeze and thawing. Cell lysates were prepared by centrifugation at 10,000g for 20 minutes. LDH activity in the lysates was measured by using the LDH assay kit following the protocol from the manufacturer (Kyokuto Pharmaceutical Industrial Co Ltd, Tokyo, Japan). Cellular proteins in the lysates were determined with a protein assay kit (Bio-Rad Laboratories, Hercules, CA) using bovine Igs as a standard.
Mouse embryonic stem (ES) cell line, TT2, was maintained on the mitomycin C-treated feeder cells in DMEM containing 10% FCS, 104 U/mL leukemia inhibitory factor, 10−4mol/L β-mercaptoethanol, nonessential amino acid, and sodium pyruvate.37 The feeder cells were prepared from 14-day embryos of YF4 mouse.37 All materials to maintain TT2 cells were purchased from GIBCO. TT2 cells (8 × 105 cells) were seeded on the feeder cells in a 60-mm dish and cultured for 12 hours in 21% oxygen. After being replaced with the fresh medium, the cells were cultured for 24 hours in 2%, 5%, and 21% oxygen. Epo in the spent medium was measured by EIA.
Purification and bioactivity of Epo.
Human recombinant Epo was prepared as described previously.38 P19 cells were cultured in 175-cm2 T-flasks and Epo in the spent medium (2 L) was partially purified with a gel (600 μL) on which Epo-directed monoclonal antibody, R2,36 was fixed. Mouse serum Epo was isolated from phenylhydrazine-treated anemic mice.39 Epo from both sources was stored in PBS containing 0.1% bovine serum albumin. Bioactivity of Epo was assayed using Epo-dependent growth of Ep-FDC-P2 cells measured by the increased absorbance at 600 nm due to 3-(4.5-dimetyl thiazol-2-yl)-2.5-diphenyl tetrazolium bromide (MTT; Sigma, St Louis, MO) reduction.40
Quantification of mRNAs for Epo, LDHA, and β-actin by reverse transcription-polymerase chain reaction (RT-PCR).
Total RNA was prepared from P19 cells cultured for 24 hours under normoxic and hypoxic (5% and 2% oxygen) conditions using a total RNA isolation kit (Promega Corp, Madison, WI). The RT reaction was performed using a random nonamer primer and 1 μg of heat-denatured RNA. PCR primers used in this study were as follows: Epo sense primer (REP62F, 5′-TCCTTGCTACTGATTCCTCTGG-3′) and antisense primer (REP512R, 5′-AAGTATCCGCTGTGAGTGTTCG-3′); LDHA sense primer (LDHA22F, 5′-GTCTCCCTGAAGTCTCTT-3′) and antisense primer (LDHA374R, 5′-ATTCCCCAGAGGGTGTCT-3′); β-actin sense primer (mβ112F, 5′-ATCCTGACCCTGAAGTACCC-3′) and antisense primer (mβ545R, 5′-ATTTCCCTCTCAGCTGTGGT-3′). The primers of Epo, LDHA, and β-actin were based on Nucleotide Sequence Data banks (accession nos. D10763, M17516, and X03765, respectively). The PCR-amplified products derived from Epo competitor DNA, transcribed Epo cDNA, LDHA cDNA, and β-actin cDNA were the DNA fragments of 282, 451, 353, and 434 bp, respectively. We estimated Epo mRNA levels by competitive RT-PCR, as described previously.41 To estimate LDHA mRNA expression, semiquantitative PCR analysis was performed.42 Briefly, 22 to 24 PCR cycles consisting of 1 minute at 94°C for denaturation, 2 minutes at 63°C for annealing, and 3 minutes at 72°C for elongation were performed. The amplified product at each cycle was electrophoresed on a 2.5% agarose gel and stained with ethidium bromide. Quantification of the band intensity was performed on a power Macintosh 7500 computer (Apple Computer Inc, Cupertino, CA) using the public domain NIH image program (written by Wayne Rasband at the US National Institutes of Health [Bethesda, MD] and available from the Internet by anonymous ftp from zippy.nimh.nih.gov). The band intensity was proportional to PCR cycle number. To normalize the efficiency of RT, semiquantitative PCR for β-actin was also performed, using 17 to 19 cycles (1 minute at 94°C, 2 minutes at 64°C, and 3 minutes at 72°C). Because the band intensity of β-actin was proportional to PCR cycle number, the intensity of 17 cycle was used for normalization.
Transient assay of reporter gene expression.
Plasmids were transfected into P19 cells by Trans IT LT1 (PanVera Corp, Madison, WI) and Hep3B cells by LipofectAce (Life Technologies, Inc, MD) following the protocol from the manufacturer. P19 cells (5 × 104/well) and Hep3B cells (2 × 105/well) were seeded in 6-well plate 20 hours before transfection. Cells were cotransfected with 1.5 μg of test plasmid and 0.05 μg of pβactβgal,43 which carried the β-galactosidase gene under the control of the chicken β-actin promoter. For a given test plasmid, two plates were transfected at the same time. The cells were incubated for 6 hours, and the medium was replaced with the fresh growth medium. One of the two plates was incubated for a further 20 hours under normoxic condition, and the other was incubated for 20 hours under hypoxic condition (2% O2, 5% CO2, and balance N2). For transient expression of HNF-4, P19 cells were cotransfected by theTrans IT LT1 reagent with three plasmids, 0.15 μg luciferase (Luc) expression plasmid under control of LDHA promoter or Epo promoter with Epo enhancer, 0.01 μg of pβactβgal for normalization of transfection efficiency, and 1.2 μg pLEn(M4)LPHNF4 or control vector. For the Luc assay, cell extracts were prepared using PicaGene Reporter Lysis Buffer (Toyo Ink MFG CO, LTD, Tokyo, Japan) and assayed for transiently expressed Luc and β-galactosidase. Luc activity was measured using Lumat LB9051 (EG&G Berthold, Bad Widbad, Germany) and Luc Assay Kit (Toyo Ink MFG CO, LTD), and β-galactosidase activity was measured by using chlorophenol red-β-D-galactopyranoside as a substrate.44 Luc activity was divided by β-galactosidase activity for normalization of transfection efficiency. Expression of Luc gene was represented as the ratio of Luc/β-galactosidase.
Promoter and enhancer of Epo gene in P19 cells.
The promoter and enhancer regions of Epo gene were cloned from P19 genomic DNA by PCR. The mouse Epo promoter region was first amplified by PCR in the presence of 1 μg DNA, sense primer (−468F, 5′-AACCCTGACCCTTAGAACAA-3′) and antisense primer (126R, 5′-CCTGGAAGAAAGTGGTCACT-3′) in a total of 20 μL. Each of 35 PCR cycles consisted of 1 minute at 94°C, 2 minutes at 60°C, and 3 minutes at 72°C. The second PCR was performed by adding 1 μL of the first PCR product to reaction mixture containing sense primer (−433F, 5′-GAAGAAAAAGCATCCGAGCC-3′) and antisense primer (53R, 5′-TCGGGAACACAGTGAGACAC-3′) in a total of 100 μL. Each of 35 PCR cycles consisted of 1 minute at 94°C, 2 minutes at 60°C, and 3 minutes at 72°C. The mouse Epo enhancer region was amplified by PCR in the presence of 1 μg DNA, sense primer (3777F, 5′-CTCACCCCATCTGGTCGCAA-3′), and antisense primer (4017R, 5′-GGCTCCTGTTTCCTCAACGG-3′) in a total of 100 μL. The primer number was based on the definition of the transcription start site as +1.45 The amplified products of the mouse Epo promoter and enhancer region were confirmed by direct sequencing.
Determination of the transcription start site of Epo gene.
Total RNA from P19 cells was prepared and the 5′ end of Epo mRNA was determined by Marathon cDNA Amplification Kit (Clontech Laboratories, Inc, Palo Alto, CA).
Preparation of Luc reporter plasmids under control of Epo promoter and enhancer.
An amplified mouse Epo promoter fragment was digested by Acc I and blunted. The resulting 0.2-kb fragment (m0.2k) was subcloned into the HincII site of pUC19 to produce pUCm0.2k. The mouse Epo enhancer (mEn) was blunted and subcloned into the Sma I site of pUC19 to produce pUCmEn. pm0.2kLuc containing m0.2k upstream of Luc gene was prepared by subcloning a BamHI-HindIII fragment of pUCm0.2k into the Bgl II and HindIII sites of basic vector 2, PGV-B2 (Toyo Ink MFG CO, LTD). pm0.2kLucmEn was constructed by insertion of a Bgl II-Sal I fragment from pUCmEn into the BamHI and Sal I sites of pm0.2kLuc. The reporter plasmids containing human Epo promoter and enhancer were described previously.46 A 55 mer (GATCCCCCGGGCTACGTGCTGTCTCACACAGCCTGTCATAAGCTTCGACCTGCA) oligonucleotide and a 47 mer (GGTCGAAGCTTGATGACAGGCTGTGTGAGACAGCACGTAGCCCGGGG) oligonucleotide were synthesized and annealed to prepare the human Epo enhancer fragment that lacks HNF-4 binding site but still retains HIF-1 binding site and CA repeats. The enhancer fragment was subcloned into the BamHI and Sse 8387I sites of p0.2kLucEn to produce p0.2kLucEn(ΔHNF-4).
Preparation of Luc reporter plasmids under control of LDHA promoter preceded with the wild-type or mutant LDHA enhancer.
A Kpn I-BamHI fragment of PGV-B2 was subcloned into theKpn I and BamHI sites of pUC19 to produce pUCLuc. pLEnLPLuc, pLEn(M1)LPLuc, and pLEn(M4)LPLuc were prepared by subcloning a Sac I-Sse8387I fragment of pUCLuc into theSac I and EcoT22I sites of pLDHGH, pLDH(M1)GH, and pLDH(M4)GH,24 respectively. LP, LEn, LEn(M1), and LEn(M4) represent LDHA promoter, LDHA enhancer, LDHA enhancer mutated at the first HIF-1 binding site, and LDHA enhancer mutated at the second binding site, respectively (see also Fig 6).
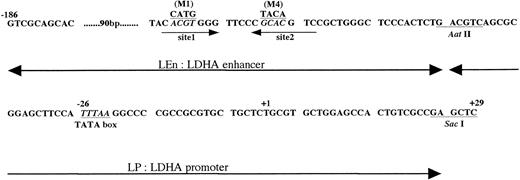
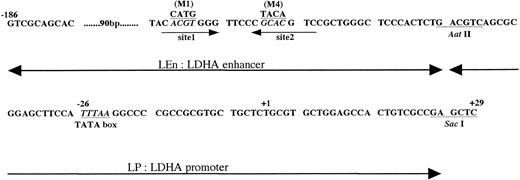
DNA sequence of mouse LDHA enhancer and promoter. +1 is the transcription start site. Sites 1 and 2 are the HIF-1 binding sites.24,25 M1 is the mutant in which ACGT was replaced with CATG, and M4 is the mutant in which GCAC was replaced with TACA.24 In this report, we use the following terms. LEn, LDHA enhancer (−186∼Aat II fragment); LP, LDHA promoter (Aat II∼Sac I fragment); LEnLP, LDHA enhancer plus promoter (−186∼Sac I fragment); LEn(M1 or M4)LP, mutant derivatives of the −186∼Sac I fragment.
DNA sequence of mouse LDHA enhancer and promoter. +1 is the transcription start site. Sites 1 and 2 are the HIF-1 binding sites.24,25 M1 is the mutant in which ACGT was replaced with CATG, and M4 is the mutant in which GCAC was replaced with TACA.24 In this report, we use the following terms. LEn, LDHA enhancer (−186∼Aat II fragment); LP, LDHA promoter (Aat II∼Sac I fragment); LEnLP, LDHA enhancer plus promoter (−186∼Sac I fragment); LEn(M1 or M4)LP, mutant derivatives of the −186∼Sac I fragment.
Preparation of Luc reporter plasmid under control of Epo promoter with LDHA enhancer.
Preparation of Luc reporter plasmid under control of LDHA promoter with Epo enhancer.
An Xba I-Apa I fragment of pEpoP-Luc-E46containing the human Epo enhancer (En) was excised and cloned into theXba I and Apa I sites of pcDNA3 (Invitrogen Co, San Diego, CA) to produce pcEn1. pLEn(M4)LPLucEn containing the mutant LDHA enhancer, LDHA promoter, Luc gene, and human Epo enhancer, in this order, were generated by subcloning anXho I fragment of pcEn1 into the Sal I site of pLEn(M4)LPLuc. An Xba I-EcoRI fragment of pEpoPLE46 containing human Epo enhancer was subcloned into the EcoRI and Xba I sites of pcDNA3 to produce pcEn2. pEnLEn(M4)LPLuc, which contained Epo enhancer, the mutant LDHA enhancer, LDHA promoter, and Luc gene, in this order, was constructed by inserting an Apa I fragment of pcEn2 into the Apa I site of pLEn(M4)LPLuc.
Preparation of HNF-4 expression vector.
An EcoRI fragment of pCMX-HNF433 that contained the full-length HNF-4 coding region was cloned into the EcoRI site of pcDNA3 to produce pCMV-HNF4. pLEn(M4)LPHNF4 was generated by subcloning a Sac I-Not I fragment of pCMV-HNF4 into theSac I and Eco52I sites of pLEn(M4)LPLuc. pLEn(M4)LP that does not contain the HNF-4 gene was prepared by deletion of the Sac I-Eco52I fragment from pLEn(M4)LPLuc and was used as a control vector in the experiments using pLEn(M4)LPHNF4.
RESULTS
Oxygen-independent expression of Epo by ES cells and embryonal carcinoma P19 cells.
Mouse ES cells have been shown to express Epo mRNA,34 but whether they secrete Epo protein into the culture medium is not known. It is also unknown whether, in these cells, the Epo mRNA level is regulated by oxygen. We cultured ES cells in 21%, 5%, and 2% oxygen for 24 hours and assayed Epo in the media by EIA. The amount of Epo protein secreted into the media was similar when cultured in the different oxygen concentrations (data not shown). Immunoreactive Epo was undetectable in the media when the feeder cells were cultured without ES cells, confirming that Epo was produced by ES cells.
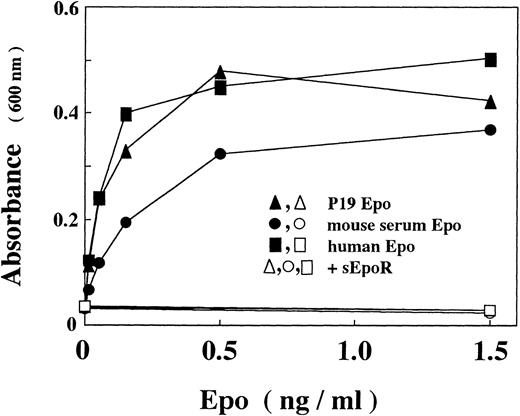
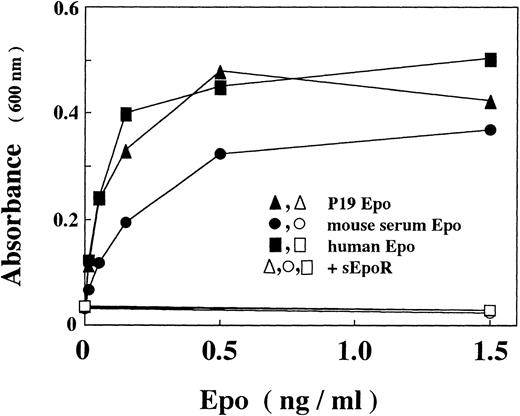
Because the requirement for feeder cells in the maintenance of ES cells complicates assays of gene expression by transient transfection, we examined whether multipotential embyronal cells, that can be cultured without feeder cells, mimic ES cells with respect to the Epo production. We found that P19 cells, which are murine embryonal carcinoma cells with pluripotentiality of differentiation,35 produced Epo. To confirm that the immunoreactive Epo is bioactive, we concentrated the material in the culture media by using a gel on which Epo-directed monoclonal antibody was fixed36 and assayed stimulatory effect of the eluted fraction on the proliferation of an Epo-dependent erythroid precursor cell line. As shown in Fig 1, Epo in the concentrated P19 culture supernatant stimulates proliferation of the cells with an efficiency similar to that of recombinant human Epo or higher than that of Epo isolated from mouse serum. The stimulation was completely abrogated by a soluble form of Epo receptor (sEpoR), an extracellular domain of the receptor capable of binding with Epo.47
In vitro activity of Epo produced by P19 cells. Epo activity was assayed with Epo-dependent proliferation of Ep-FDC-P2 cells measuring the increased absorbance at 600 nm due to reduction of MTT. (▴, ▵) P19 Epo; (•, ○) mouse serum Epo; (▪, □) human recombinant Epo; (▵, ○, □) in the presence of sEpoR capable of binding with Epo. When assayed in the presence of sEpoR, Epo was preincubated with sEpoR (100 μg/mL) for 1 hour. Each point is the mean of duplicate assays.
In vitro activity of Epo produced by P19 cells. Epo activity was assayed with Epo-dependent proliferation of Ep-FDC-P2 cells measuring the increased absorbance at 600 nm due to reduction of MTT. (▴, ▵) P19 Epo; (•, ○) mouse serum Epo; (▪, □) human recombinant Epo; (▵, ○, □) in the presence of sEpoR capable of binding with Epo. When assayed in the presence of sEpoR, Epo was preincubated with sEpoR (100 μg/mL) for 1 hour. Each point is the mean of duplicate assays.
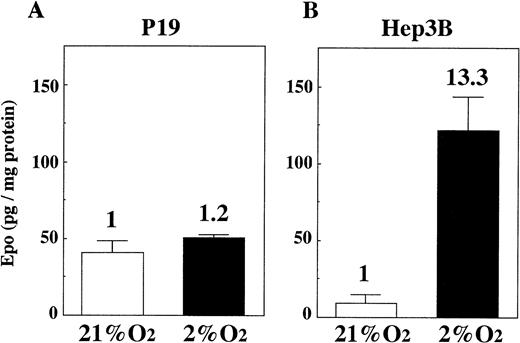
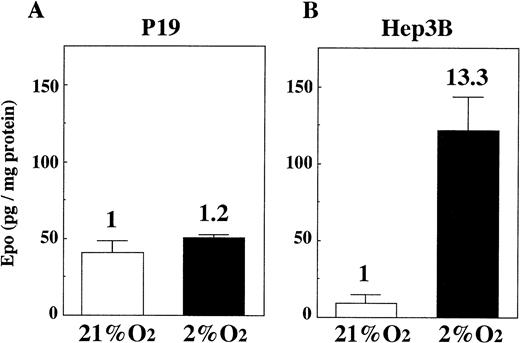
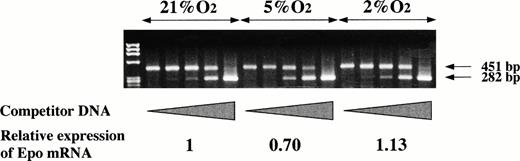
We cultured P19 cells and Hep3B cells in 21% and 2% oxygen for 24 hours and assayed Epo in the culture media. Figure 2 shows that there is only a very small increase of Epo production by P19 cells in 2% oxygen compared with the production in 21% oxygen (Fig 2A), whereas, in keeping with previous results,30 31 the production of Epo by Hep3B cells is greatly increased under the hypoxic condition (Fig 2B). Assay of Epo mRNA with competitive RT-PCR indicated that the mRNA level was almost unchanged when P19 cells were cultured in 21%, 5%, and 2% oxygen (Fig 3).
Oxygen-independent production of Epo by P19 cells. P19 cells and Hep3B cells were cultured in 21% and 2% oxygen for 24 hours. Epo concentration in the spent medium and the total cellular proteins were determined. Epo production is given as Epo (in picograms) per milligram of cellular protein. Inducibility (2% oxygen/21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments.
Oxygen-independent production of Epo by P19 cells. P19 cells and Hep3B cells were cultured in 21% and 2% oxygen for 24 hours. Epo concentration in the spent medium and the total cellular proteins were determined. Epo production is given as Epo (in picograms) per milligram of cellular protein. Inducibility (2% oxygen/21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments.
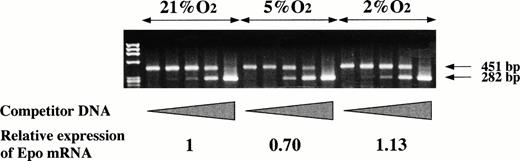
Oxygen-independent expression of Epo mRNA by P19 cells. P19 cells were cultured in 21%, 5%, and 2% oxygen for 24 hours. Total RNA was prepared and competitive RT-PCR was performed to estimate Epo mRNA. Competitor DNA in the PCR reaction mixtures was increased from 0 to 100 fg.41 The amplified products of 451 bp and 282 bp were derived from transcribed Epo cDNA and the competitor DNA, respectively. The band intensity was measured and Epo mRNA was calculated from the equivalence points of the band intensity of the amplified products. The calculation yielded 0.69 ng Epo mRNA/mg total RNA when the cells were cultured in 21% oxygen, 0.48 in 5% oxygen, and 0.78 in 2% oxygen. The content of Epo mRNA relative to that in 21% oxygen is shown.
Oxygen-independent expression of Epo mRNA by P19 cells. P19 cells were cultured in 21%, 5%, and 2% oxygen for 24 hours. Total RNA was prepared and competitive RT-PCR was performed to estimate Epo mRNA. Competitor DNA in the PCR reaction mixtures was increased from 0 to 100 fg.41 The amplified products of 451 bp and 282 bp were derived from transcribed Epo cDNA and the competitor DNA, respectively. The band intensity was measured and Epo mRNA was calculated from the equivalence points of the band intensity of the amplified products. The calculation yielded 0.69 ng Epo mRNA/mg total RNA when the cells were cultured in 21% oxygen, 0.48 in 5% oxygen, and 0.78 in 2% oxygen. The content of Epo mRNA relative to that in 21% oxygen is shown.
The transcription start site for the murine Epo gene in kidney has been shown.48 It was first necessary to know the transcription start site of Epo gene in P19 cells, because unresponsiveness of P19 Epo gene to hypoxia might be due to the use of a cryptic promoter that was unable to interact appropriately with the 3′ enhancer responsible for hypoxic induction. Determination of 5′ end of P19 Epo mRNA by the use of rapid amplification cDNA ends method showed a start site very close to that for kidney Epo gene,48indicating that expression of Epo gene in P19 cells is under control of the promoter functional in the kidney cells.
Transient assay of reporter gene expression to examine if hypoxic-response elements (enhancers) function in P19 cells.
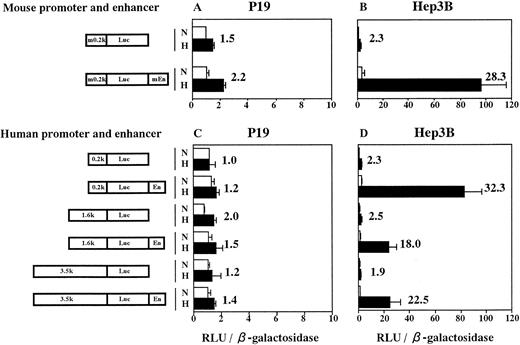
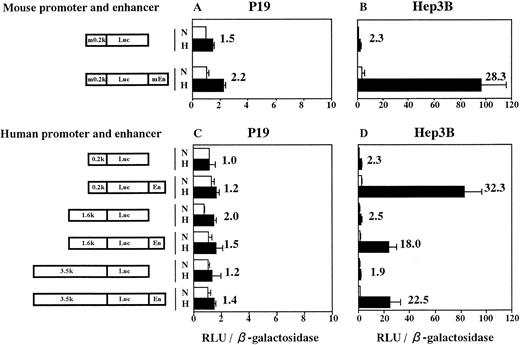
From P19 cell genomic DNA, we cloned both the minimum promoter of mouse Epo gene (283 bp from −230 to +53, numbering nucleotides according to Galson et al45) and the enhancer (213 bp from 3805 to 4017). Sequencing of these fragments showed no mutation. We next constructed plasmids in which Luc reporter gene was under control of the cloned mouse promoter (m0.2k, see Fig 4) with the cloned enhancer (mEn) fused 3′ to Luc gene. Plasmids containing various lengths of 5′-flanking sequence from the human Epo gene fused to Luc gene and human Epo enhancer (En; 116 bp from 3457 to 3572; see Lin et al49), in this order, were also constructed. The plasmids were transfected into P19 cells and Hep3B cells with pβactβgal (to normalize transfection efficiency). Transfected cells were incubated for 6 hours in 21% oxygen and then for 20 hours in either 21% or 2% oxygen. Activities of Luc and β-galactosidase were assayed in cell lysates and the Luc/β-galactosidase ratios were calculated. Results are shown in Fig 4. Transfection of P19 cells with the plasmid containing both mouse Epo promoter and enhancer yielded a 2.2-fold hypoxic induction (Fig 4A). However, the similar (1.5-fold) induction was also seen in the plasmid without the enhancer. The same was true for plasmids constructed with elements from human source (Fig 4C). The extent of Epo 5′-flanking region (0.2 to 3.5 kb) had no influence on the hypoxic inducibility in P19 cells. However, when Hep3B cells were used, large hypoxic induction of Luc expression was observed in the plasmids containing either mouse or human enhancer (Fig 4B and D), which was in good agreement with the results reported previously.9-11 32 Thus, the hypoxia-inducible enhancer of mouse and human Epo genes does not function in P19 cells.
The hypoxia-inducible enhancer of mouse and human Epo gene functions in Hep3B cells but not in P19 cells. Transient expression of reporter constructs in 21% oxygen (N) and in 2% oxygen (H) was assayed. (A and C) P19 cells; (B and D) Hep3B cells. m0.2k, 0.2-kb mouse Epo promoter; mEn, mouse Epo enhancer; En, human Epo enhancer. 0.2k, 1.6k, and 3.5k represent 0.2-kb, 1.6-kb, and 3.5-kb 5′-flanking regions of human Epo gene. Relative light units (RLU; Luc activity) per β-galactosidase activity are shown. Hypoxic inducibility (the ratio of Luc activity in 2% v 21% oxygen) is also indicated. Each value is the mean ± SD of triplicate experiments.
The hypoxia-inducible enhancer of mouse and human Epo gene functions in Hep3B cells but not in P19 cells. Transient expression of reporter constructs in 21% oxygen (N) and in 2% oxygen (H) was assayed. (A and C) P19 cells; (B and D) Hep3B cells. m0.2k, 0.2-kb mouse Epo promoter; mEn, mouse Epo enhancer; En, human Epo enhancer. 0.2k, 1.6k, and 3.5k represent 0.2-kb, 1.6-kb, and 3.5-kb 5′-flanking regions of human Epo gene. Relative light units (RLU; Luc activity) per β-galactosidase activity are shown. Hypoxic inducibility (the ratio of Luc activity in 2% v 21% oxygen) is also indicated. Each value is the mean ± SD of triplicate experiments.
Hypoxic induction of LDHA in P19 and Hep3B cells.
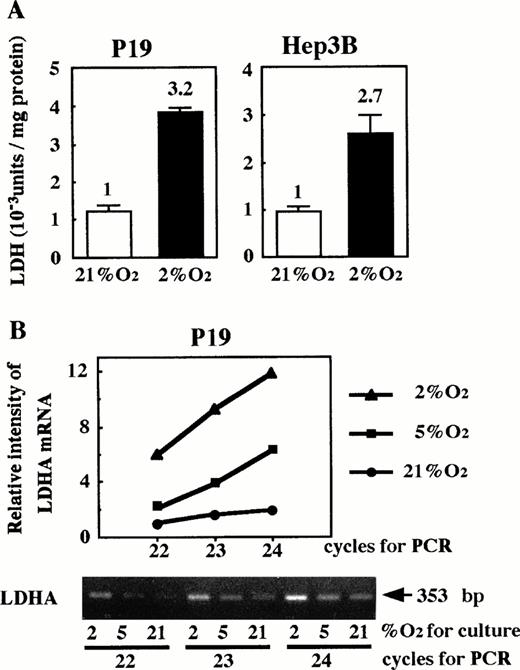
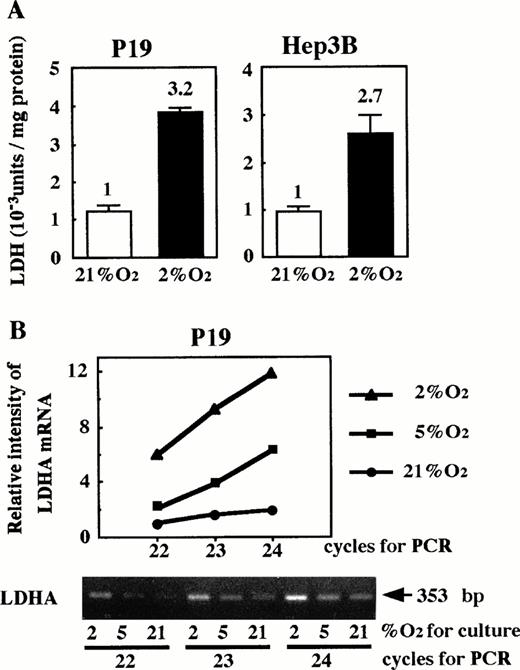
Because HIF-1 is essential for hypoxia-inducible function of the Epo enhancer, we considered the possibility that oxygen-independent production of Epo in P19 cells might be attributable to lack of HIF-1. We examined whether LDHA, a glycolytic enzyme that has been shown to be hypoxia-inducible in a variety of cells,22-25 29 was induced in P19 cells upon hypoxia. P19 and Hep3B cells were cultured in 21% or 2% oxygen, and the intracellular LDH activity was assayed. As shown in Fig 5A, LDH activity in P19 cells increased under the low oxygen in a similar manner to Hep3B cells. LDHA mRNA was assayed by RT-PCR, varying the cycle number for PCR. The amount of product was measured (see the Materials and Methods) and plotted against the cycle number after normalization with the β-actin mRNA-derived product. Figure 5B shows that the amount of LDHA mRNA-derived product is approximately proportional to the cycle number and increases when the cells were cultured under hypoxic conditions. These results indicate that the increase of LDH activity in P19 cells upon hypoxia is due to an elevated LDHA mRNA level.
Hypoxic induction of LDH in P19 cells. (A) LDH activity; (B) LDHA mRNA. In (A), P19 cells and Hep3B cells were cultured in 21% and 2% oxygen for 24 hours and cellular LDH activity was determined. LDH activity (Wro units of the LDH assay kit) per milligram of cellular protein is shown. Hypoxic inducibility (activity ratios in 2% and 21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments. In (B), P19 cells were cultured in 21%, 5%, and 2% oxygen. LDHA mRNA in total RNA was amplified by RT-PCR, varying the PCR cycle number (22, 23, and 24 cycles). LDHA mRNA-derived product of 353 bp is shown. β-Actin mRNA was also amplified to use for normalization of efficiency in RT (see the Materials and Methods). Normalized band intensities relative to that of the product amplified (22 cycle) from LDHA mRNA of the cells cultured in 21% oxygen were plotted against PCR cycle number. (•) 21% oxygen; (▪) 5% oxygen; (▴) 2% oxygen.
Hypoxic induction of LDH in P19 cells. (A) LDH activity; (B) LDHA mRNA. In (A), P19 cells and Hep3B cells were cultured in 21% and 2% oxygen for 24 hours and cellular LDH activity was determined. LDH activity (Wro units of the LDH assay kit) per milligram of cellular protein is shown. Hypoxic inducibility (activity ratios in 2% and 21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments. In (B), P19 cells were cultured in 21%, 5%, and 2% oxygen. LDHA mRNA in total RNA was amplified by RT-PCR, varying the PCR cycle number (22, 23, and 24 cycles). LDHA mRNA-derived product of 353 bp is shown. β-Actin mRNA was also amplified to use for normalization of efficiency in RT (see the Materials and Methods). Normalized band intensities relative to that of the product amplified (22 cycle) from LDHA mRNA of the cells cultured in 21% oxygen were plotted against PCR cycle number. (•) 21% oxygen; (▪) 5% oxygen; (▴) 2% oxygen.
Mutation of HIF-1 binding sites in the LDHA enhancer and hypoxic induction of Luc.
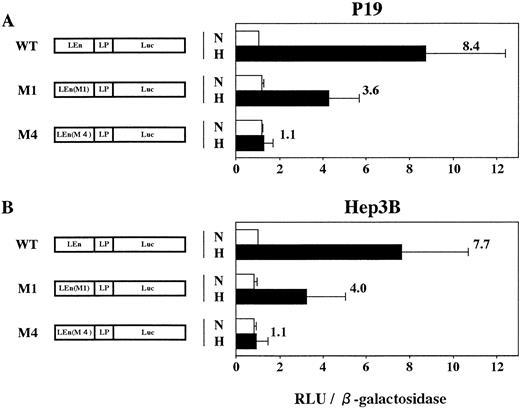
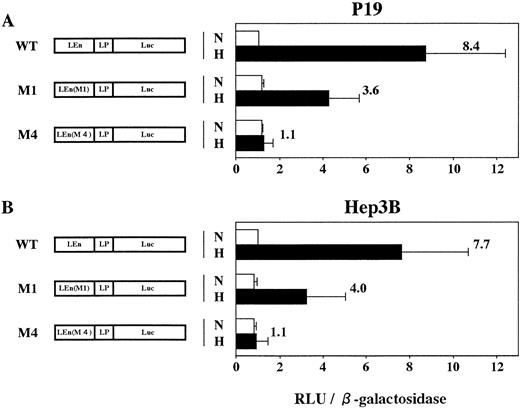
Expression of mouse LDHA is under control of the minimum promoter containing TATA box (−46 to −1 in Fig 6) and the enhancer (−85 to −47).24 The enhancer contains two HIF-1 binding sites (domains defined by site 1 and site 2 in Fig 6). Binding of HIF-1 to site 2 is essential for hypoxic induction but not sufficient. Binding to site 1 is also necessary for the full induction.24,25 To confirm that HIF-1 is present in P19 cells, plasmids in which expression of Luc gene was under control of LDHA promoter and enhancer with or without mutations in HIF-1 binding sites (mutations M1 and M4 in Firth et al24) were constructed and the transient expression of Luc in 21% or 2% oxygen was assayed using P19 and Hep3B cells (Fig 7). Hypoxia induced an 8.4-fold increase of Luc expression in P19 cells (pLEnLPLuc in Fig 7A) when Luc gene was fused to LDHA promoter and wild-type of LDHA enhancer. There was no hypoxic induction when the LDHA enhancer was mutated at HIF-1 binding site 2 [see pLEn(M4)LPLuc]. Mutation of HIF-1 binding site 1 reduced significantly but did not completely abrogate the hypoxic induction [pLEn(M1)LPLuc]. When Hep3B cells were used, similar results were obtained (Fig 7B) that were consistent with the results previously reported using human HeLa cells as host cells and human growth hormone as a reporter.24 P19 cells and Hep3B cells are thus equivalent with respect to the hypoxic induction of LDHA gene through activation of HIF-1, but they were quite different in the hypoxia-inducible response of Epo gene and its cis-acting sequences.
P19 cells possess an intact hypoxia-signaling pathway through HIF-1 activation. Transient expression of reporter constructs in 21% oxygen (N) and in 2% oxygen (H) was assayed. (A) P19 cells; (B) Hep3B cells. WT, wild type; M1, mutant in the HIF-1 binding site 1; M4, mutant in the HIF-1 binding site 2; LEn, LDHA enhancer; LP, LDHA promoter. Relative light units (RLU; Luc activity) per β-galactosidase activity are shown. Hypoxic inducibility (the ratio of Luc activity in 2% oxygen v 21% oxygen) is shown. Each value is the mean ± SD of triplicate experiments.
P19 cells possess an intact hypoxia-signaling pathway through HIF-1 activation. Transient expression of reporter constructs in 21% oxygen (N) and in 2% oxygen (H) was assayed. (A) P19 cells; (B) Hep3B cells. WT, wild type; M1, mutant in the HIF-1 binding site 1; M4, mutant in the HIF-1 binding site 2; LEn, LDHA enhancer; LP, LDHA promoter. Relative light units (RLU; Luc activity) per β-galactosidase activity are shown. Hypoxic inducibility (the ratio of Luc activity in 2% oxygen v 21% oxygen) is shown. Each value is the mean ± SD of triplicate experiments.
Hypoxic interaction between heterologous promoters and enhancers.
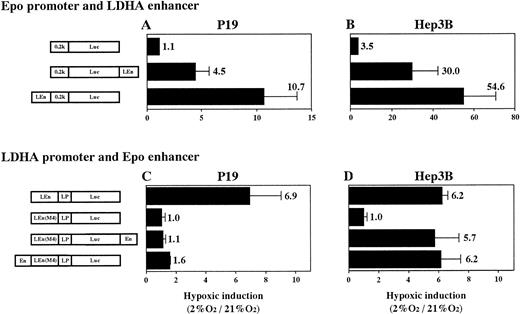
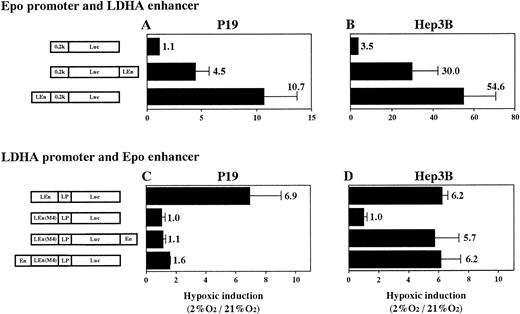
To know the interaction in hypoxia between heterologous promoters and enhancers (Epo promoter v LDHA enhancer and LDHA promoterv Epo enhancer) in P19 and Hep3B cells, we constructed plasmids (shown in Fig 8) and transient expression of Luc in 21% and 2% oxygen was assayed. First, interaction of Epo promoter (0.2k) and LDHA enhancer (LEn) was examined (Fig 8A and B). Epo promoter was activated by LDHA enhancer in the hypoxic condition in both cells, but the hypoxic activation in P19 cells was fivefold to sevenfold lower than in Hep3B cells. Location of the enhancer upstream of the promoter was more efficient for hypoxic induction. Next, interaction of LDHA promoter (LP) and Epo enhancer (En) was examined (Fig 8C and D). For this experiment, the Epo enhancer was inserted into plasmid pLEn(M4)LPLuc (in which the LDHA enhancer was nonfunctional due to mutation at HIF-1 binding site 2), either upstream of the mutant LDHA enhancer or downstream of the Luc gene. In P19 cells, little hypoxia-inducible response was regained when Epo enhancer was inserted (Fig 8C). In contrast, insertion of Epo enhancer resulted in the full recovery of the hypoxia- inducible response in Hep3B cells (Fig 8D). The full retrieval was achieved even when Epo enhancer was cloned 3′ to Luc gene (distant from the promoter).
Hypoxic interaction between heterologous promoters and enhancers. (A and C) P19 cells; (B and D) Hep3B cells. Transient expression of reporter constructs in 21% and 2% oxygen was assayed. Hypoxic inducibility (the ratio of Luc activity in 2% oxygen v21% oxygen) is indicated. pLEn(M4)LPLuc shows little hypoxic inducibility in both P19 and Hep3B cells (see also Fig 7). Each value is the mean ± SD of triplicate experiments.
Hypoxic interaction between heterologous promoters and enhancers. (A and C) P19 cells; (B and D) Hep3B cells. Transient expression of reporter constructs in 21% and 2% oxygen was assayed. Hypoxic inducibility (the ratio of Luc activity in 2% oxygen v21% oxygen) is indicated. pLEn(M4)LPLuc shows little hypoxic inducibility in both P19 and Hep3B cells (see also Fig 7). Each value is the mean ± SD of triplicate experiments.
Cotransfection of HNF-4.
In addition to HIF-1 binding site on Epo enhancer, other two sites play an important role in the hypoxic induction.12,18,32,33HNF-4 expressed in Hep3B cells has been shown to bind to the 3′ portion of the enhancer and expression of HNF-4 in HeLa cells (which do not express HNF-4) elevated the hypoxia-inducible response of a reporter construct containing Epo enhancer.33 Plasmids containing an HNF-4 gene fused to LDHA promoter [LEn(M4)LP] or cytomegalovirus (CMV) promoter were cotransfected into P19 cells to examine whether HNF-4 increases Epo enhancer activity that was otherwise inactive in P19 cells. Transient assay of Luc gene expression in 2% oxygen showed that there was a slight activation by HNF-4 when the ratio of the HNF-4 expression plasmid to the reporter plasmid was increased to 8 (Fig 9). In Fig 9A, interaction with the Epo enhancer in the context of the mutated mouse LDHA promoter-enhancer [pLEn(M4)LPLuc] was examined. The result was very similar when reporter plasmids constructed from human Epo gene elements (0.2k and En) were cotransfected with the HNF-4 expression plasmid (Fig 9B).
Effect of HNF-4 on transient expression of reporter constructs in P19 cells. P19 cells were cotransfected with the reporter plasmids shown in the figure and HNF-4 expression plasmid [pLEn(M4)LPHNF-4] to determine if HNF-4 activates the Epo enhancer in P19 cells. In (A), combination of LDHA promoter (LP) and Epo enhancer (En); in (B), combination of human Epo promoter (0.2k) and En. Transient expression of reporter constructs in 21% and 2% oxygen was assayed. The ratio of HNF-4 expression plasmid versus reporter plasmid is shown. Hypoxic inducibility (the ratio of Luc activity in 2% oxygenv 21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments.
Effect of HNF-4 on transient expression of reporter constructs in P19 cells. P19 cells were cotransfected with the reporter plasmids shown in the figure and HNF-4 expression plasmid [pLEn(M4)LPHNF-4] to determine if HNF-4 activates the Epo enhancer in P19 cells. In (A), combination of LDHA promoter (LP) and Epo enhancer (En); in (B), combination of human Epo promoter (0.2k) and En. Transient expression of reporter constructs in 21% and 2% oxygen was assayed. The ratio of HNF-4 expression plasmid versus reporter plasmid is shown. Hypoxic inducibility (the ratio of Luc activity in 2% oxygenv 21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments.
DISCUSSION
Several cell lines have been reported to produce Epo constitutively (see Goldberg et al30 and the references therein), but the mechanisms underlying the constitutive expression in normoxic cells and the absence of inducibility by hypoxia have not been investigated. We found that multipotential embryonal carcinoma P19 cells produce Epo mRNA and secrete bioactive Epo protein in an oxygen-independent manner. This finding provided an opportunity to compare the operation of Epo gene control sequences in these undifferentiated cells with Hep3B cells, which are specialized for oxygen-dependent Epo production. Sequencing of the Epo promoter and enhancer regions cloned from P19 cells excluded the possibility that the oxygen-independent expression of Epo gene is due to a mutation in these sequences. Determination of mRNA 5′ end also excluded the possibility that transcription of Epo gene in P19 cells might start using an unusual promoter incapable of interacting with the Epo enhancer.
As summarized in Table 1, reporter constructs containing the cloned Epo promoter and enhancer showed very low hypoxia-inducibility in P19 cells, whereas they were highly inducible in Hep3B cells. Similar results obtained using reporter constructs containing varying extents of 5′-flanking sequence from the human Epo gene together with the human Epo 3′ enhancer (Fig 4C) support our conclusion that the cis-acting elements required for the hypoxic induction of Epo gene are structurally intact in P19 cells, but fail to function in a hypoxia-inducible manner. In contrast, the LDHA gene was hypoxia-inducible in P19 cells. Reporter constructs containing the LDHA enhancer were also hypoxia-inducible in P19 cells, as in Hep3B cells. Furthermore, mutational analysis showed the critical importance of an HIF-1 binding site in this response, showing that the cellular components required for oxygen sensing and the subsequent signal transduction pathway leading to HIF-1 are intact in P19 cells. It is one of the possibilities accountable for the noninducibility of the endogenous Epo gene in P19 cells that the 3′ enhancer region is not open for HIF-1 to be accessible. Nevertheless, the fact that the Epo enhancer does not interact with the cognate or LDHA promoter in P19 cells but interacts in Hep3B cells (Table 1) indicates that, in understanding why Epo gene expression is not inducible in P19 cells, further analysis of the Epo enhancer 3′ to the HIF-1 binding site should be informative.
In hepatoma cells, the 3′ region of the Epo enhancer acts to increase the inducible response to hypoxia and enable operation at a distance from a promoter.12,32,33 Analysis of this region has demonstrated that these functions are, at least in part, dependent on binding of HNF-4 to a directly repeated nuclear receptor binding motif in this region. Furthermore, in HeLa cells, heterologous expression of HNF-4 was found to increase the hypoxic inducibility of the cotransfected Epo enhancer.33 This raised the possibility that absent or insufficient function of HNF-4 in P19 cells might account for noninducibility of this sequence. However, forced expression of HNF-4 did not stimulate the function of Epo enhancer in P19 cells (Fig 9), suggesting that this was not the case. Because the COUP family can antagonize HNF-4 by competing the binding site,33 the COUP family may be highly expressed in P19 cells and responsible for the effect. Interestingly, in mouse erythroleukemia cells that are non-Epo producing, expression of a reporter construct containing just the 5′ region of mouse Epo enhancer is hypoxia-inducible, whereas the whole enhancer is inactive, suggesting that these cells contain a suppressive factor interacting with the 3′ region.18 We examined the reporter plasmid consisting of Epo promoter and Epo enhancer in which the HNF-4 binding site was deleted. There was no hypoxic induction of the reporter gene when this plasmid was transfected into P19 cells. In Hep3B cells, the reporter gene was considerably induced in hypoxia, but the inducibility was lower than that of the plasmid containing the whole enhancer (date not shown). These results made it unlikely that the nonfunctional Epo enhancer in P19 cells is caused by the presence of COUP family. Alternatively, factors binding at other sites could be involved. For instance, the sequence just 3′ to the HIF-1 binding site of Epo enhancer is essential for induction by hypoxia, although this sequence is not well conserved.12 18 Hep3B cells may express an unidentified protein bound to this region that is absent in P19 cells.
The LDHA enhancer activated its promoter in hypoxia with a similar efficiency in both P19 and Hep3B cells, whereas the effect of LDHA enhancer on the Epo promoter was less efficient in P19 cells than in Hep3B cells (Table 1). These results suggest that P19 cells may be insufficient in a component that binds to the Epo promoter, causing the full interaction with the enhancer. In this context, it is interesting that the sequence −61 to −45 relative to the transcription start site of the murine Epo gene is required for the 3′ enhancer to be maximally functional and Hep3B nuclear extracts contain proteins that bind to this region.50
P19 cells differentiate into astrocytes upon treatment with retinoic acid.35 In addition to the established physiologic function of Epo in the stimulation of erythropoiesis, we have proposed that Epo functions as a neurotrophic factor in the central nervous system.4,8,41,51-53 In the central nervous system, neurons express Epo receptor51,52,54 and astrocytes produce Epo.4,41,55 Production of Epo in the brain is also hypoxia-inducible.4,55,56 Furthermore, Epo infused into the lateral ventricle by an osmotic minipump protects pyramidal neurons in the hippocampal CA1 region from ischemia-induced death (manuscript submitted), and Epo protects the primary cultured hippocampal and cortical neurons from glutamate toxicity, which is considered to be a major mechanism in ischemic neuron death.8 Conditions that differentiate P19 cells (showing noninducible Epo expression) into astrocytes that have acquired hypoxia-inducible Epo production should therefore be of interest. Thus, P19 cells may be an excellent model for better understanding regulation of Epo gene expression.
Supported by grants-in-aids from the Ministry of Education, Science, Sports and Culture of Japan, from Snow Brand Milk Products Co, Ltd, and from Kato Memorial Bioscience Foundation to S.M.
Address reprint requests to Ryuzo Sasaki, PhD, Division of Applied Life Sciences, Graduate School of Agriculture, Kyoto University, Kyoto 606-01, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 9. Effect of HNF-4 on transient expression of reporter constructs in P19 cells. P19 cells were cotransfected with the reporter plasmids shown in the figure and HNF-4 expression plasmid [pLEn(M4)LPHNF-4] to determine if HNF-4 activates the Epo enhancer in P19 cells. In (A), combination of LDHA promoter (LP) and Epo enhancer (En); in (B), combination of human Epo promoter (0.2k) and En. Transient expression of reporter constructs in 21% and 2% oxygen was assayed. The ratio of HNF-4 expression plasmid versus reporter plasmid is shown. Hypoxic inducibility (the ratio of Luc activity in 2% oxygenv 21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/4/10.1182_blood.v91.4.1185/4/m_blod4040509.jpeg?Expires=1769541387&Signature=L6T3VGsd4PiFAa8Qnq8esNBPC9lNS0cV91eHmv3KP22-W-HNS-3OS61zpNgvOBakXFhFaXXxvZ-dJtpzvb6acCfjS22oBHFC60DYVKC9icM8tZNceRXlftKBNK4wZGQp9-PNTgUhxJsAqWraRPnC7UUJRO6cjKwJVo7Z58MV63lZ8flxCc0qdn3SH6MioAZmcmtmmbY~E1cKtstj1U7qXmzBSMamx78C84lr~Q-4pb8Q8jBcS5kPAR7t8HRAumD07-~CVJTdRe24IeXyZ5Gr~X0PbqvRGmeZuk1nL-C81C9zEbG~PReXKD0Qw13yLudtppTeoVXilczHZ4FrKGdRrg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 9. Effect of HNF-4 on transient expression of reporter constructs in P19 cells. P19 cells were cotransfected with the reporter plasmids shown in the figure and HNF-4 expression plasmid [pLEn(M4)LPHNF-4] to determine if HNF-4 activates the Epo enhancer in P19 cells. In (A), combination of LDHA promoter (LP) and Epo enhancer (En); in (B), combination of human Epo promoter (0.2k) and En. Transient expression of reporter constructs in 21% and 2% oxygen was assayed. The ratio of HNF-4 expression plasmid versus reporter plasmid is shown. Hypoxic inducibility (the ratio of Luc activity in 2% oxygenv 21% oxygen) is indicated. Each value is the mean ± SD of triplicate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/4/10.1182_blood.v91.4.1185/4/m_blod4040509.jpeg?Expires=1769620893&Signature=qbRDIAyq-0TkxgNK0nGRelynIZeuNXcAfdG-fFu84HDTbk9jekJCUAgZBWfSx3Dm~sBtpzJ3zyExugE6rQ5~ynSK5Ld-F-pyIIJ7W8M4voGvReDfqgDa1v2d4CyHKOo8LdSmDRwNEKJ1rElPOPpDNdVESyeBEqTPqdlQLjfIXn43z9dwppCPSQwvspj~AY9msp2pzdvVCZkyaSFFgwmHEsEN6H7ozf3EUudgH~qK1t4wpQHhkiRTWQnJMiGYH5ni206YM0OhAVe69I82XT6B5BRJXfW0GOBanAkVZIHPTB0xgvG66UkEEmcejeD8tZcNCXX23Rpkv5tAbVkFxMvqsw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)