Abstract
Hematopoiesis is tightly controlled by a family of cytokines that signal through a related set of receptors. The pleiotropic and overlapping response of a cell to different cytokines is reflected in the number and complex pattern of activated signal transducers. Of special interest is STAT5, which is stimulated by a large and diverse set of cytokines. In addition to the two highly homologous proteins, STAT5A and STAT5B, encoded by duplicated genes, expression and activation of a dominant-negative, carboxyl-truncated form has also been described in early hematopoietic progenitors. We show here that a protease expressed in early hematopoietic cells cleaves the α forms of STAT5A/5B (p96/p94) to generate carboxyl-truncated β forms (p80/p77). Inhibition studies assigned this protease to the serine class of endopeptidases. Cell fractionation experiments showed that the protease is associated with the nucleus in a constitutively activated form and does not require an activated STAT5 substrate. The ability of a protease to modulate the specificity of an activated transcription factor is unprecedented and underlines the importance of proteases in regulation of cell functions.
CYTOKINES ACT AS extracellular signals in myelopoiesis and lymphopoiesis to activate cell function and to maintain homeostasis in the adult tissue.1,2 These polypeptide ligands bind cell-surface receptors that trigger a cascade of signaling pathways, including the JAK/STAT pathway.3Activation of the receptor-associated Janus protein tyrosine kinases (JAKs) results in the phosphorylation of a unique family of transcription factors termed the signal transducers and activators of transcription (STATs).4,5 Phosphorylation triggers dimerization, transport to the nucleus, and DNA binding. There are seven STATs known (including both proteins encoded from the duplicated STAT5A and STAT5B genes) that can be activated by more than 30 different ligands in various cell systems, resulting in diverse biological effects. It is clear that the specificity of the response is dictated by both direct and indirect interactions with other transcription factors present in the different cell types. In the case of STAT1, STAT3, and STAT5, specificity can also be mediated by short β forms that, unlike the long α forms, can uniquely interact with specific transcription factors or inhibit transcription in a dominant negative fashion.6-10
STAT5 is expressed in a wide range of tissue and is involved in a variety of responses observed in both hematopoietic and nonhematopoietic cells. In addition to its important role in prolactin signaling in mammary glands,11 STAT5 is activated by cytokines that regulate the proliferation and differentiation of myeloid (interleukin-3 [IL-3], IL-5, granulocyte-macrophage colony-stimulating factor [GM-CSF], and thrombopoietin),6,12,13 erythroid (erythropoietin [Epo]),14,15 and lymphoid lineages (IL-2 and IL-15).16,17 Two highly related genes encode the STAT5A and STAT5B proteins, which are greater than 95% identical.6,12,18 These proteins differ at the carboxyl terminus, a region that is highly variable among other STAT proteins and thought to be involved in transcriptional activation.4No evidence has yet been presented that define functional differences between these proteins,12 although differentiation-specific differences in activation patterns have been observed.19 In contrast, the carboxyl-truncated forms, termed STAT5β, have been shown to act in a dominant negative fashion to inhibit transactivation9 and may interact with a unique set of transcription factors, in analogy with STAT3β.7
A number of studies have clearly shown that the truncated β form is the predominant phosphorylated STAT5 form observed after IL-3, GM-CSF, or Epo stimulation in early hematopoietic cells.6,12,14,20Significantly, these results were observed in both mice and humans, and with both primary and established cells. A previous report showed that the β form can be generated from an alternatively spliced message (the last intron remaining unspliced),9 consistent with transcripts detected in rat liver and mammary glands.21,22However, the levels of this alternative message were quite low and inconsistent with the almost exclusive activation of STAT5β reported in early hematopoietic cells.6 This study was initiated to determine if an alternative mechanism is involved in the generation of the truncated STAT5β isoforms. We have identified a nucleus-associated protease present in early hematopoietic cells that cleaves activated STAT5α to generate STAT5β. The importance of this serine endopeptidase in regulating hematopoiesis is discussed.
MATERIALS AND METHODS
Cell culture.
The multipotent FDC-Pmix independent isolates A4 and 15S23,24 and the factor-dependent FDC-P1(M) (clone 4) and FDC-P125 cell lines were used for these studies. The latter two cell lines were obtained from T.M. Dexter (Paterson Laboratories, Manchester, UK) and D. Metcalf (Walter and Eliza Hall Institute, Melbourne, Australia), respectively, and differ in their responsiveness to GM-CSF, their infectivity with ecotropic retroviruses, and cell surface markers (C. Laker and C.S., unpublished results). FDC-Pmix cells were maintained in Iscove's modified Dulbecco's medium (IMDM; GIBCO, Paisley, UK) supplemented with IL-3 and 20% horse serum, whereas FDC-P1 and FDC-P1(M) cells were held in modified Eagle's medium (2×) in 10% fetal calf serum and IL-3. IL-3 was obtained from conditioned medium from cells transfected with a bovine papilloma virus vector carrying the IL-3 gene and was used at concentrations necessary for maximum stimulation.
Preparation of cell extracts.
Cell extracts were prepared from cells under three different conditions: (1) 20 hours (or 12 hours for FDC-Pmix) after removal of IL-3; (2) after stimulation of starved cells (20 or 12 hrs) with IL-3 for 30 minutes; or (3) under normal proliferating conditions in the presence of IL-3 (approximately 24 hours after the last addition of fresh medium and factor). To prepare nuclear and cytosolic extracts, 5 × 107 cells were washed twice with phosphate-buffered saline containing 1 mmol/L orthovanadate and pelleted at 1,000gfor 5 minutes. The cell pellet was resuspended in a hypotonic buffer containing 20 mmol/L HEPES, pH 7.6, 10 mmol/L KCl, 1 mmol/L MgCl2, 0.5 mmol/L dithiothreitol (DTT), 0.1% Triton X-100, 20% glycercol, 1 mmol/L Pefabloc (Boehringer Mannheim, Mannheim, Germany), 5 μg/mL leupeptin (Sigma, Deisenhofen, Germany), 5 μg/mL pepstatinA (Biomol, Hamburg, Germany), 200 KIU/mL aprotinin (ICN, Eschwege, Germany), and 1 mmol/L sodium orthovanadate. Cells were lysed by 20 strokes in a glass Dounce homogenizer. The homogenate was centrifuged at 2,000g for 5 minutes. The supernatant (cytosolic extract) was centrifuged again for 15 minutes at 14,000 rpm to clear lysate and flash frozen in liquid nitrogen. The pelleted nuclei were extracted in a hypertonic nuclear extract buffer (NEB; 20 mmol/L HEPES, pH 7.9, 0.4 mol/L NaCl, 1 mmol/L EDTA, 0.5 mmol/L DTT, 0.1% Triton X-100, 20% glycerol) containing proteinase inhibitors (as listed above) for 15 minutes at 4°C. The extracts were then centrifuged at 14,000 rpm for 5 minutes. Aliquots were frozen in liquid nitrogen and stored at −70°C. Buffer volumes were adjusted so that the protein concentrations of nuclear and cytosolic extracts reflected the cellular ratio. The purity of the cell fractionation was confirmed in Western blot analysis using antisera against laminB (kindly provided by W. Bohn, Heinrich-Pette-Institut, Hamburg, Germany), which is specific for the nucleus and with either anti–c-src (sc-18; Santa Cruz Biotechnology, Santa Cruz, CA) or anti-SHC (#6-203; Upstate Biotechnology Inc [UBI; Lake Placid, NY]). Only low levels of cross-contamination were observed (<1%).
In experiments in which cell extracts from different cells were combined, extracts were incubated on ice for 15 minutes after mixing (1:1 or 1:10) and then frozen in liquid nitrogen. To enhance protease activity, in some cases extracts were incubated for an additional 2 minutes at 37°C before freezing. Freezing was found to destroy proteolytic activity.
The following proteinase inhibitors were screened for inhibition activity: phenylmethylsulfonyl fluoride (PMSF; ≥1 mmol/L; Sigma), Pefabloc (4 mmol/L; Boehringer Mannheim), E64 (10 μg/mL; Boehringer Mannheim); 3,4 Dichloroisocoumarin (DCI; 200 μmol/L; Sigma);N-tosyl-L-phenyalanine chloromethylketone (TPCK; 0.1 mmol/L; Boehringer Mannheim), andNα-p-tosyl-L-lysine chloromethalketone (TLCK; 1 mmol/L; Boehringer Mannheim). In all cases, the given concentration was added to all buffers used during all stages of cell extract isolation, in addition to the above-listed proteinase inhibitors, except Pefabloc. Concentrations chosen were based on the highest recommended dose given by the manufacturer. EDTA (2 mmol/L) was added to one set of buffers.
Electrophoretic mobility shift assays (EMSA).
Oligonucleotides were end-labeled with polynucleotide kinase to a specific activity of 5 × 103 cpm/fmol. The STAT5 binding site of the bovine β-casein promoter was used as a probe (5′-AGATTTCTAGGAATTCAAATC-3′).11 Nuclear extracts (1.2 μg) were incubated at room temperature for 30 minutes in a volume of 20 μL with 16 fmol end-labeled oligonucleotides and 2 μg of poly(dI.dC) in 10 mmol/L HEPES (pH 7.9), 50 mmol/L KCl, 5 mmol/L MgCl2, 1 mmol/L DTT, 1 mmol/L EDTA, and 5% glycerol. Reaction mixtures were then electrophoresed at 10 V/min on a 6% polyacrylamide gel in 0.25× TBE (45 mmol/L Tris-borate, 1 mmol/L EDTA). For supershift assays, antibody (1 μg) was added after 15 minutes and incubated for another 15 minutes at room temperature.
Immunoprecipitations, Western blot analysis, and antibodies.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis as well as immunoprecipitations were performed as described previously.26 Rabbit polyclonal antisera against STAT5b (C-17; Santa Cruz Biotechnology) was used to immunoprecipitate STAT5α. To detect STAT5 in Western blots, antisera (C-17 and N-20) and monoclonal antibody (p89), purchased from Santa Cruz Biotechnolgy and Transduction Laboratories (Lexington, KY), respectively, were used. Antisera against phosphorylated tyrosine residues (4G10) purchased from UBI was used to confirm phosphorylation of STAT5. Filters were developed using the enhanced chemiluminescence (ECL) system according to the manufacturer's protocol (Amersham, Braunschweig, Germany).
Oligonucleotide affinity purification of activated STAT5.
Nuclear extracts (500 μg protein) were incubated with Sepharose beads coupled to multimerized oligonucleotides containing the STAT5 binding sites from the β-casein gene promoter. Binding reactions were performed in the presence of 30 μg poly(dI.dC) and 30 μg poly(dA.dT) for 1.5 hours at 4°C in NEB supplemented with 60 mmol/L NaCl. After two washes with NEB containing 60 mmol/L NaCl and one wash containing 100 mmol/L NaCl, proteins bound to the Sepharose beads were eluted with 400 mmol/L NaCl in NEB. One-third volume 3× SDS loading buffer was added to samples that were subsequently separated by SDS-PAGE (7.5%), blotted onto nitrocellulose membrane, and visualized with antibody.
RESULTS
Truncated STAT5β is the preferential activated isoform observed in multipotent hematopoietic progenitors.
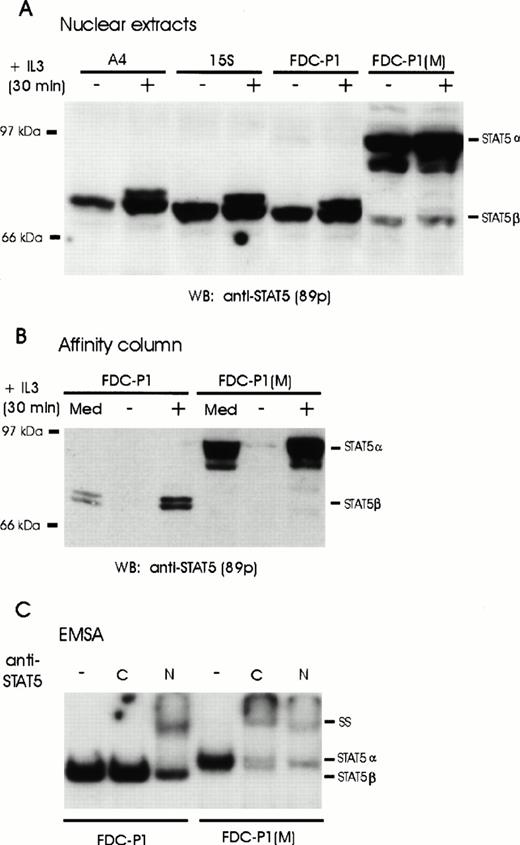
Activated β forms of STAT5 have previously been detected in both primary and established myeloid progenitors and precursors, the expression of which has been correlated with a more immature phenotype. To further investigate this isoform, we screened cells of several IL-3–dependent cell lines for expression of STAT5β. Nuclear extracts were separated by SDS-PAGE electrophoresis and probed with STAT5 antisera after transfer to nylon membranes. Consistent with the hypothesis that the short form is preferentially expressed in early progenitors, only the truncated forms (p80/p77) of STAT5A and STAT5B were detected in the multipotent FDC-Pmix cells (Fig 1A). Similar results were obtained with FDC-P1 cells, which do not express any lineage-specific markers. In contrast, predominantly full-length STAT5α (p96/p94) isoforms were detected in the IL-3–dependent FDC-P1M cells (Fig 1A), which lack markers of early progenitors. No significant difference in the levels of STAT isoforms was observed in the nucleus of IL-3 stimulated as compared with IL-3–deprived cells; however, a slower migrating band could be observed in stimulated cell extracts, indicative of phosphorylation. To confirm that both the activated β forms detected in FDC-P1 and FDC-Pmix cells and the α forms in FDC-P1M cells were able to bind DNA, proteins binding to oligonucleotides containing the STAT5 recognition sequences from the β-casein gene promoter were purified by affinity chromatography (Fig 1B). Consistent with earlier results that phosphorylation is required for DNA binding, STAT5 was only purified in stimulated cell extracts. Significantly, both STAT5α and STAT5β isoforms bound the STAT5 recognition sequence. This was further confirmed by EMSA (Fig 1C). Two distinct complexes were observed in the two types of cells reflecting homodimerization of the short α forms or the β forms. Importantly, the DNA-STAT5 complexes from both cell types could be supershifted with antibodies directed against the amino-terminus of STAT5, but antibody directed against the carboxyl-terminus of STAT5 only interacted with the complex detected in FDC-P1M, in which only α-forms were detected (Fig 1C). This confirms that the short β forms expressed in FDC-P1 and FDC-Pmix cells represent a carboxyl-truncated form of full-length STAT5α. In addition, these results verify that expression of the short β is a hallmark of early hematopoietic progenitors.
Expression of full-length and truncated STAT5 proteins in hematopoietic cell lines. (A) Nuclear extracts were prepared from unstimulated (−) or IL-3–stimulated (+) A4, 15S, FDC-P1 and FDC-P1(M) cells. Full-length (STAT5α) or truncated (STAT5β) STAT5 proteins were detected by Western blot analysis using monoclonal anti-STAT5 antibodies (89p). (B) FDC-P1 and FDC-P1(M) cells were grown in IL-3–containing medium (Med) or starved for 20 hours (−) and stimulated with IL-3 (+) for 30 minutes. Nuclear extracts were prepared and incubated with multimerized β-casein oligonucleotides coupled to Sepharose beads. Bound proteins were eluted and analyzed by Western blotting with anti-STAT5 antibodies (89p). (C) A labeled DNA probe containing the β-casein GAS element was added to nuclear extracts from IL-3–stimulated FDC-P1 and FDC-P1(M) cells and subsequently incubated either without (−) or with antibodies directed against the C-terminus (C) or N-terminus (N) of STAT5. Complexes were analyzed by an EMSA.
Expression of full-length and truncated STAT5 proteins in hematopoietic cell lines. (A) Nuclear extracts were prepared from unstimulated (−) or IL-3–stimulated (+) A4, 15S, FDC-P1 and FDC-P1(M) cells. Full-length (STAT5α) or truncated (STAT5β) STAT5 proteins were detected by Western blot analysis using monoclonal anti-STAT5 antibodies (89p). (B) FDC-P1 and FDC-P1(M) cells were grown in IL-3–containing medium (Med) or starved for 20 hours (−) and stimulated with IL-3 (+) for 30 minutes. Nuclear extracts were prepared and incubated with multimerized β-casein oligonucleotides coupled to Sepharose beads. Bound proteins were eluted and analyzed by Western blotting with anti-STAT5 antibodies (89p). (C) A labeled DNA probe containing the β-casein GAS element was added to nuclear extracts from IL-3–stimulated FDC-P1 and FDC-P1(M) cells and subsequently incubated either without (−) or with antibodies directed against the C-terminus (C) or N-terminus (N) of STAT5. Complexes were analyzed by an EMSA.
Full-length STAT5α is the predominant form in the cytoplasm of early progenitors.
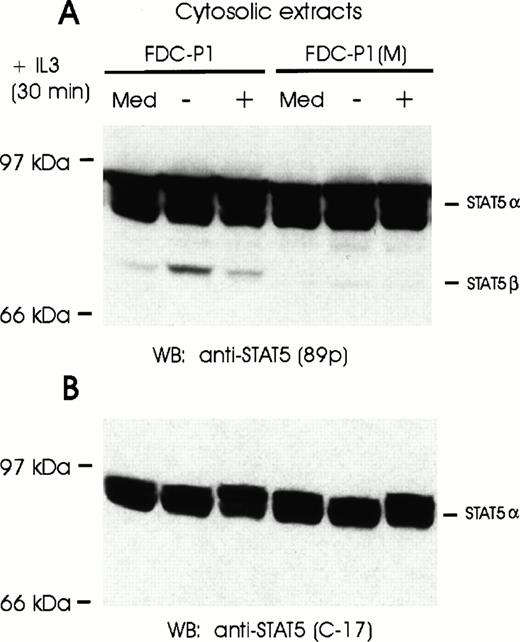
It is conceivable that both α and β forms of STAT5 are present in early progenitors, but only the short form is phosphorylated and transduced to the nucleus. Cytoplasm extracts were thus examined. Despite the exclusive presence of activated STAT5β in the nucleus of IL-3–stimulated FDC-P1, full-length STAT5α was the major form present in the cytoplasm as determined by Western blot analysis (Fig 2). Indeed, only trace amounts of STAT5β were detectable. Significantly, when cytoplasm extracts were analyzed by EMSA, only low levels of both STAT5α and STAT5β were found to bind DNA, the predominant form being the truncated form (data not shown). These results indicate that the majority (if not all) of truncated STAT5β detected in the cytoplasm is phosphorylated and probably enroute to the nucleus. Although it is conceivable that the β form in the FDC-P1 cytoplasm is the preferred substrate for JAK activation, a more likely explanation is that the long-form is altered after phosphorylation or transport to the nucleus.
Full-length STAT5α proteins are expressed in both FDC-P1 and FDC-P1(M) cells. (A) FDC-P1 and FDC-P1(M) cells were grown in IL-3–containing medium (Med) or starved for 20 hours (−) and stimulated with IL-3 (+) for 30 minutes. Cytosolic extracts were analyzed by Western blotting using anti-STAT5 antibodies (89p). (B) The nitrocellulose filter shown in (A) was stripped and reprobed with anti-STAT5 antibodies directed against the C-terminus of STAT5. Locations of full-length (α) and truncated (β) STAT5 proteins are indicated on the right.
Full-length STAT5α proteins are expressed in both FDC-P1 and FDC-P1(M) cells. (A) FDC-P1 and FDC-P1(M) cells were grown in IL-3–containing medium (Med) or starved for 20 hours (−) and stimulated with IL-3 (+) for 30 minutes. Cytosolic extracts were analyzed by Western blotting using anti-STAT5 antibodies (89p). (B) The nitrocellulose filter shown in (A) was stripped and reprobed with anti-STAT5 antibodies directed against the C-terminus of STAT5. Locations of full-length (α) and truncated (β) STAT5 proteins are indicated on the right.
Proteolytic activity in nuclear extracts of FDC-P1 cells cleaves the full-length STAT5α found in FDC-P1(M) nuclear extracts to generate STAT5β.
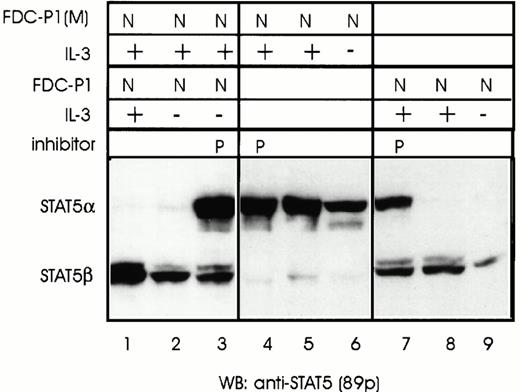
One likely explanation for the presence of different STAT5 isoforms in the nucleus versus cytoplasm is the presence of a protease that either specifically recognizes phosphorylated forms of STAT5 and/or is exclusively associated with the nucleus. Nuclear and cytosolic extracts from FDC-P1 cells were thus incubated with nuclear extracts from FDC-P1(M) that contain full-length phosphorylated STAT5α. In accordance with the hypothesis that a protease associated with the nucleus cleaves STAT5α, Western blot analysis of nuclear extracts of FDC-P1(M) cells incubated with equal amounts of nuclear extracts prepared from FDC-P1 cells showed that all STAT5α from FDC-P1(M) was converted to STAT5β (Fig 3, compare lanes 4 and 5). The levels of conversion of STAT5α to STAT5β were proportional with the amount of nuclear extracts added to the sample (data not shown), consistent with an enzymatic activity. Furthermore, this analysis showed that the shortened form, generated by incubation with nuclear extracts from FDC-P1 cells, was the same length as STAT5β in FDC-P1 cells (Fig 3, compare lanes 2 and 5). In contrast, nuclear extracts incubated with cytosolic extracts of FDC-P1 showed only a slight reduction in levels of STAT5α (Fig 3, compare lanes 4 and 7). To rule out the possibility that the cytoplasm contained a specific protease inhibitor, cytoplasm extracts from both FDC-P1(M) and FDC-P1 cells were added to nuclear extracts of FDC-P1 cells before incubation with FDC-P1(M) nuclear extracts containing the STAT5α template. No inhibition of the protease activity in FDC-P1 nuclear extracts was observed (Fig 3, compare lane 5 with 9 and 10). Thus, the slight levels of protease activity observed with cytosol extracts is most likely due to trace levels of nuclear contamination. In conclusion, a protease activity associated with the nucleus in FDC-Pmix and FDC-P1 cells is able to cleave full-length STAT5α to generate a carboxyl-truncated STAT5β.
Cleavage of full-length STAT5α in FDC-P1(M) cells by a nuclear-associated protease from FDC-P1 cells. Unstimulated (−) or IL-3–stimulated (+) FDC-P1 and FDC-P1(M) cells were lysed and nuclear (N) and cytoplasmic (C) extracts were prepared. Extracts from FDC-P1 and FDC-P1(M) cells were analyzed either separately (lanes 1 through 4) or mixed and incubated for 15 minutes on ice (lanes 5 through 10). STAT5 proteins were detected by Western blotting using anti-STAT5 antibodies (89p). The trace levels of STAT5α observed in lane 10 are due to the addition of high levels of STAT5α in FDC-P1(M) cytosolic extracts (see Fig 2). This could be eliminated by raising the incubation temperature for 2 minutes at 37°C (data not shown).
Cleavage of full-length STAT5α in FDC-P1(M) cells by a nuclear-associated protease from FDC-P1 cells. Unstimulated (−) or IL-3–stimulated (+) FDC-P1 and FDC-P1(M) cells were lysed and nuclear (N) and cytoplasmic (C) extracts were prepared. Extracts from FDC-P1 and FDC-P1(M) cells were analyzed either separately (lanes 1 through 4) or mixed and incubated for 15 minutes on ice (lanes 5 through 10). STAT5 proteins were detected by Western blotting using anti-STAT5 antibodies (89p). The trace levels of STAT5α observed in lane 10 are due to the addition of high levels of STAT5α in FDC-P1(M) cytosolic extracts (see Fig 2). This could be eliminated by raising the incubation temperature for 2 minutes at 37°C (data not shown).
Protease activity is independent of IL-3 stimulation and does not require an activated substrate.
The predominant localization of the protease in the nuclear extract could imply that, in analogy with STAT family members, IL-3 stimulation leads to its activation and translocation to the nucleus. We thus determined if FDC-P1 nuclear extracts prepared from starved cells also contained proteolytic activity. Significantly, nuclear extracts from either stimulated or IL-3–deprived cells converted STAT5α isolated from FDC-P1(M) cells to STAT5β (Fig 3, compare lanes 5 and 6). Thus, the protease is in an active form in the nucleus of both stimulated and unstimulated cells.
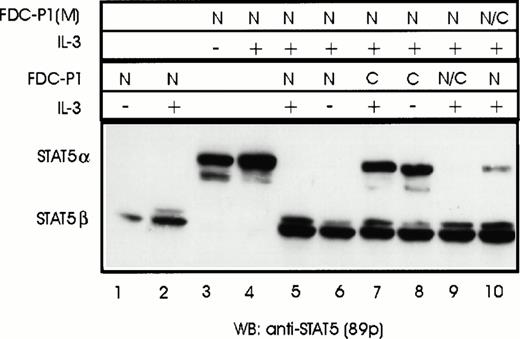
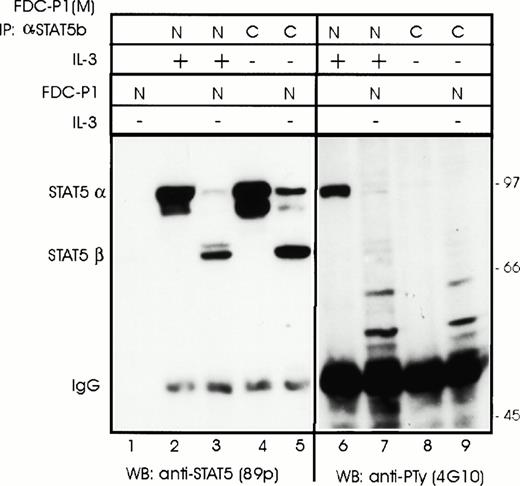
We next asked the question if nuclear transport of STAT5α was the only prerequisite for cleavage (cellular colocalization) or if activation by IL-3 (eg, phosphorylation or dimerization) was also required. STAT5α was isolated by immunoprecipitation from either the nuclei of stimulated FDC-P1(M) cells (active form) or from the cytoplasm of starved FDC-P1(M) cells (inactive form) and incubated with nuclear extracts of FDC-P1 cells. Western blot analysis after SDS-PAGE showed that both forms could be used as a substrate by the protease in FDC-P1 nuclear extracts (Fig 4, lanes 1 through 5). Under the conditions used, not all of the inactive form was cleaved. This may indicate a preference by the protease for the active form or be due to the higher levels of STAT5α in the cytosolic extracts (compare lanes 2 and 4 in Fig 4). To confirm the active state of the nuclear STAT but not the cytosolic STAT, PTy antisera was used to detect phosphorylated tyrosine residues. As expected, STAT5α isolated from the nucleus but not cytosol was recognized by the antisera (Fig 4, compare lanes 6 and 8). These results confirm that colocalization of the protease with the STAT5 substrate is sufficient for cleavage; modification of STAT5 by IL-3 stimulation is not required.
Both activated and nonactivated forms of STAT5α are cleaved by the protease in FDC-P1 nuclear extracts. The different forms of STAT5α were isolated from either stimulated FDC-P1(M) nuclear extracts (N) or unstimulated cytosol extracts (C) by immunoprecipitation using antisera that recognizes the carboxyl terminus of STAT5b (C-17). The Sepharose pellet was dissolved in nuclear buffer and aliquots were either mixed with 1/10 volume of nuclear extract from unstimulated FDC-P1 cells or nuclear extract buffer. Extracts were incubated on ice for 15 minutes and then transferred to 37°C for 2 minutes before freezing. STAT5 proteins were detected by Western blotting using anti-STAT5 antibodies (89p) or anti–P-Ty (4G10).
Both activated and nonactivated forms of STAT5α are cleaved by the protease in FDC-P1 nuclear extracts. The different forms of STAT5α were isolated from either stimulated FDC-P1(M) nuclear extracts (N) or unstimulated cytosol extracts (C) by immunoprecipitation using antisera that recognizes the carboxyl terminus of STAT5b (C-17). The Sepharose pellet was dissolved in nuclear buffer and aliquots were either mixed with 1/10 volume of nuclear extract from unstimulated FDC-P1 cells or nuclear extract buffer. Extracts were incubated on ice for 15 minutes and then transferred to 37°C for 2 minutes before freezing. STAT5 proteins were detected by Western blotting using anti-STAT5 antibodies (89p) or anti–P-Ty (4G10).
The FDC-P1 protease is an endopeptidase and its activity is inhibited by a serine protease inhibitor.
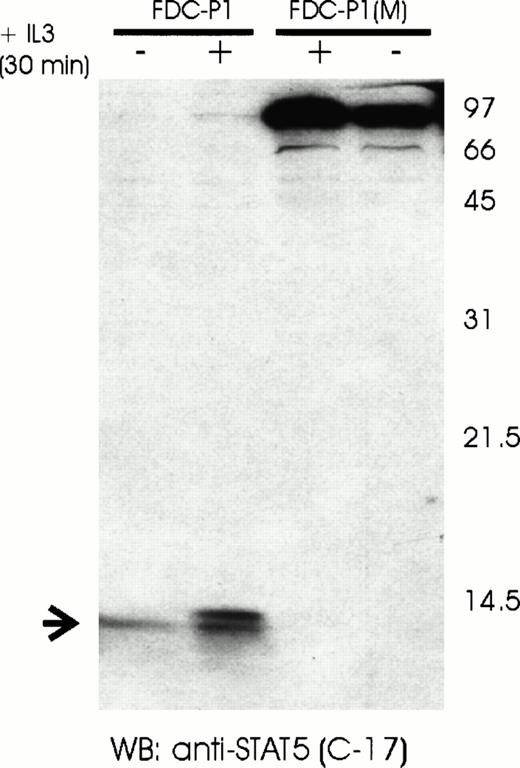
Proteolytic activity can be due to either exopeptidases or endopeptidases. The latter can further be classified according to essential catalytic residues at their active sites, as well as their dependency on cofactors, such as Ca+2 or Zn+2(reviewed in Bond and Butler27). Cleavage by an endopeptidase should lead to the generation of two or more peptides, depending on the number of cleavage sites. To determine if the observed proteolytic activity was due to an endonuclease or exonuclease, we separated FDC-P1 nuclear extracts by SDS-PAGE under conditions that would allow the detection of lower molecular weight peptides. Consistent with internal cleavage, a band of approximately 14 kD was observed in nuclear extracts of FDC-P1 cells, but not FDC-P1(M) cells when probed with antisera recognizing an epitope near the carboxyl terminus (Fig 5). In contrast, probing the same extract with antisera recognizing the area around the SH2 domain detected only the STAT5β bands (77/80 kD; data not shown). In conclusion, the nucleus-associated protease found in early hematopoietic cells cleaves a specific internal peptide bond.
Detection of a proteolytic C-terminal STAT5 fragment in FDC-P1 cells. Nuclear extracts of unstimulated (−) or IL-3–stimulated (+) FDC-P1 and FDC-P1(M) cells were analyzed by Western blotting using a C-terminal anti-STAT5 antibody (C-17). The full-length STAT5α protein is indicated on the right. The proteolytic C-terminal fragment of STAT5 is indicated on the left.
Detection of a proteolytic C-terminal STAT5 fragment in FDC-P1 cells. Nuclear extracts of unstimulated (−) or IL-3–stimulated (+) FDC-P1 and FDC-P1(M) cells were analyzed by Western blotting using a C-terminal anti-STAT5 antibody (C-17). The full-length STAT5α protein is indicated on the right. The proteolytic C-terminal fragment of STAT5 is indicated on the left.
We thus sought to further classify the protease detectable in FDC-P1 and FDC-Pmix nuclear extracts. Although a battery of protease inhibitors were routinely added to all buffers during the preparation and analysis of cell extracts (see the Materials and Methods for details), none of these effectively inhibited the observed proteolytic activity. We thus added one of several other protease inhibitors to our basic buffer systems, and in one case EDTA, and analyzed the FDC-P1 nuclear extracts either alone or in mix experiments as described above. Taken together, this series of experiments can be summarized as follows. Neither the site-directed inhibitor of active-site cysteine residues (E64), the specific inhibitor of aspartic proteinases (pepstatin), nor the addition of EDTA altered the protease activity of FDC-P1 cells, indicating that the protease is, most likely, not a member of either the cysteine, aspartic, or metallo-proteinases. In support of its classification as a serine protease, the protease activity could be inhibited with saturating (>1 mmol/L) concentrations of PMSF, which is a relatively specific and broad inhibitor of serine proteases (Fig 6, compare lanes 2 and 3). Interestingly, other proteases that inhibit serine proteases, including DCI, TPCK, TLCK, leupeptin, and Pefabloc, did not inhibit the protease, although the highest recommended concentrations were used. However, this is perhaps not unexpected, because these compounds inhibit distinct classes of serine proteases.
The protease activity in FDC-P1 nuclear extracts can be inhibited by PMSF. Both unstimulated (−) or IL-3–stimulated (+) FDC-P1 and FDC-P1(M) cells were lysed in the presence or absence of the protease inhibitor PMSF (P) and nuclear (N) extracts were prepared. Extracts from FDC-P1 and FDC-P1(M) cells were analyzed either separately (lanes 9 through 14) or mixed and incubated for 15 minutes on ice (lanes 1 through 8). STAT5 proteins were detected by Western blotting using anti-STAT5 antibodies (89p).
The protease activity in FDC-P1 nuclear extracts can be inhibited by PMSF. Both unstimulated (−) or IL-3–stimulated (+) FDC-P1 and FDC-P1(M) cells were lysed in the presence or absence of the protease inhibitor PMSF (P) and nuclear (N) extracts were prepared. Extracts from FDC-P1 and FDC-P1(M) cells were analyzed either separately (lanes 9 through 14) or mixed and incubated for 15 minutes on ice (lanes 1 through 8). STAT5 proteins were detected by Western blotting using anti-STAT5 antibodies (89p).
The presence of protease activity in the cell extracts raised the question if cleavage occurred in vitro, ie, during the preparation of the extracts, but not in vivo. To address this, nuclear extracts of FDC-P1 cells were analyzed that had been exposed to saturating concentrations of PMSF at all stages of extraction. In contrast to nuclear extracts prepared with 0.2 mmol/L PMSF or 4 mmol/L Pefabloc, in which only STAT5β was detectable, approximately equal levels of STAT5α and STAT5β were detectable in Western blot analysis (Fig 6, compare lanes 7 and 8). Although these results clearly show that in vitro cleavage does occur, a significant portion of STAT5β could not be inhibited by PMSF. This is in contrast to our results obtained in mixing experiments in which incubation with PMSF entirely inhibited STAT5α cleavage (Fig 6, compare lanes 2 and 3, and see figure legend), confirming that the concentration of PMSF used is sufficient for complete inhibition. In conclusion, these results show that STAT5α present in the nucleus of FDC-P1 cells is converted to STAT5β in vivo.
DISCUSSION
The cell-specific response to a particular cytokine stimulation reflects various parameters of the cell, largely, but not exclusively, determined by an endogenous program of differentiation.28One of the important consequences of cytokine-receptor stimulation is the transcriptional activation of previously quiescent genes. The identification of the STAT family of transcription factors, which are directly activated by cytokine stimulation, has provided an important tool to identify mechanisms by which differential gene expression is achieved through signaling of a common receptor. Various mechanisms have been shown, including the differential activation of STAT family members due to specificity determined by receptor subunits (reviewed in Darnell29), quantitative differences in STAT transcriptional activation modulated by the duration or strength of receptor signaling or degree of serine phosphorylation,30and functional interactions with other transcription factors.31 32 Modification of the STAT proteins themselves, due either to transcriptional or posttranscriptional mechanisms, is clearly another mechanism by which the target gene specificity can be altered.
The work presented here has functionally identified a protease that modifies the action of STAT5 in early hematopoietic cells by cleavage of the carboxyl terminus. We propose the name MSA (Modulator of Stat Activity) for this protease. During the course of this work, this protease activity was also reported by Azam et al.33 Our findings extend their work by determining the localization of the protease, defining its substrate, and determining the role of cytokine stimulation for its activity. Although a previous report has proposed that truncated STAT5 isoforms are generated at the level of transcript splicing,9 we were unable to detect the proposed alternative transcripts in the cells we examined (J.M. and C.S., unpublished results). However, we cannot exclude that this mechanism is used to generate STAT5β in other cell types.
MSA is found in nuclear extracts and most probably has a serine catalytic domain, thus belonging to one of the larger family of proteases. Although a large number of protease inhibitors were screened, we were able to only identify one, PMSF, that inhibited its activity. Although serine proteases are generally found in secretory or specialized granules, MSA is unique in that it is associated with the nucleus. To date, only a limited number of proteases have been localized to the nucleus.34-36 Molecular cloning of the gene encoding this protease may thus lead to the identification of a unique family of proteases that regulate the activity of transcription factors.
MSA is in an activated state in both IL-3 stimulated and nonstimulated cells; thus, cytokine receptor stimulation is not necessary for its activation. However, stimulation involving one of the JAK family members is necessary to activate STAT5 and induce its translocation into the nucleus, where it is cleaved by MSA. Interestingly, neither phosphorylation nor dimerization is necesary for STAT to be used as a substrate of MSA. JAK stimulation is thus only necessary for the translocation of STAT5α to the nucleus and not for inducing conformational changes required for MSA recognition.
Earlier reports have shown that early hematopoietic progenitors preferentially express STAT5β.6,20 This was clearly shown in human primary monocyte cultures in which differentiation induction leads to a switch from the coexpression of both STAT5α and STAT5β to the exclusive expression of the full-length form. The work presented here extends this observation. Of particular interest is the high expression levels of STAT5β in the two independent isolates of the multipotent FDC-Pmix cells. These cells are able to differentiate into both erythroid and myeloid lineages (G, M, Eo, and Meg) and exhibit an open-chromatin structure for lymphoid-specific genes24,28,37 and thus clearly represent an early hematopoietic progenitor or stem cell. The high expression levels of MSA in these cells may reflect a mechanism by which the cell is able to translate cytokine stimulation into a mitogenic signal coupled with self-renewal versus differentiation. A role of proteases and protease inhibitors in early hematopoietic differentiation has been suggested by other studies in which the downregulation of genes encoding granzyme B and serpin2A during differentiation of FDC-Pmix cells was reported.38,39 Interestingly, upregulation of MSA has occurred in 10 of 15 factor-independent mutants of FDC-P1(M) cells (J.M., W.O., and C.S., unpublished results). The ability of MSA to modulate the activity of a pivotal transcription factor in cytokine stimulation suggests a possible mechanism by which proteases can regulate differentiation and self-renewal. Although at this point purely speculative, it could be envisaged that truncated STAT5β in synergy with other transcription factors upregulates genes important in the mitogenic response, whereas STAT5α is more effective in the regulation of genes involved in differentiation. This would explain the conflicting reports of STATs role in these two important functions of cytokine stimulation.40-42
STAT5β could differentially regulate gene expression by exerting a dominant negative effect on the transcription of a subset of target genes9,30 or by interacting with a unique subset of transcription factors.7,31 Previous studies have shown that carboxyl-truncated STAT5 proteins are unable to transactivateCis and Osm,9,41 two target genes shown to be upregulated by STAT5 in early hematopoiesis.43,44 Azam et al33 have also verified that cells expressing exclusively STAT5β showed a delayed and reduced activation of these two genes. Importantly, IL-3 stimulation of FDC-P1(M) cells (and consequent activation of STAT5α) induces the Osm expression to levels more than fivefold higher than in stimulated FDC-P1 cells, in which activated STAT5β is predominantly present (J.M. and C.S., unpublished results). Taken together, these results are consistent with the idea that MSA is able to modulate STAT activity. However, we were unable to detect significant difference between levels of Cis transcripts between the two cells types after IL-3 stimulation. This discrepancy most likely reflects interactions with different transcription factors that may also be activated by IL-3 stimulation or that are able to interact with both STAT5α and STAT5β. The identification of other target genes of STAT5 is necessary to clarify the role of STAT5β in early hematopoietic cells.
Proteases have long been known to play important roles in regulating cell proliferation, differentiation, and apoptosis. There are several examples in which proteases play an important role in the regulation of transcription factors, either by directing degradation (reviewed in Ciechanover45) or by activating latent transcription factors in the cytoplasm.46-48 We have described here a novel protease that modulates the activity of a pivotal transcription factor of cytokine regulation. Although we have shown that activated STAT5α is a substrate of MSA, we cannot rule out that other transcription factors in the nucleus are also regulated by this protease. Purification and molecular cloning of this protease should provide valuable insight into its regulation and specificity.
ACKNOWLEDGMENT
The authors are indebted to Bernd Groner and Fabrice Gouilleux for introducing us to the world of STATs. We also thank Drs B. Groner, J. Heukeshoven, and J. Ihle for stimulating discussions.
This work is a part of the doctoral thesis of J.M. at the Faculty of Biology, University of Hamburg and was supported by grants from the Thyssen Foundation and the Deutsche Forschungsgemeinschaft (SFB545). The Heinrich-Pette-Institut is financially supported by Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
Address reprint requests to Carol Stocking, PhD, Heinrich-Pette-Institute, Martinistr. 52, D-20251 Hamburg, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.