Abstract
Signals from transforming growth factor-β (TGF-β), a bifunctional regulator of the proliferation of hematopoietic progenitor cells, have been recently shown to be transduced by five novel human genes related to a Drosophila gene termed MAD (mothers against the decapentaplegic gene). We showed by reverse transcriptase polymerase chain reaction that the RNA from one homologue gene, Smad5, was present in the immortalized myeloid leukemia cell lines, KG1 and HL60, in bone marrow mononuclear and polymorphonuclear cells, as well as in purified CD34+ bone marrow cells. Therefore, we studied the role of this gene in the regulation of human hematopoiesis by TGF-β. TGF-β1 and TGF-β2 significantly inhibited myeloid, erythroid, megakaryocyte, and multilineage colony formation as assayed in semisolid culture systems. The levels of Smad5 mRNA in CD34+ cells were decreased by antisense but not sense oligonucleotides to Smad5. Preincubation of CD34+ marrow cells with two sense oligonucleotides to Smad5 did not reverse the inhibitory effects of TGF-β on hematopoietic colony formation. However, preincubation with two antisense oligonucleotides to Smad5 reversed the inhibitory effects of TGF-β. These data show that the Smad5 gene is involved in the signaling pathway by which TGF-β inhibits primitive human hematopoietic progenitor cell proliferation and that Smad5 antisense oligonucleotides can interrupt this signal.
THE HUMAN HEMATOPOIETIC system is regulated by the interaction between the hematopoietic microenvironment, a complex network composed of stem cells, progenitor cells, stromal cells, endogenous growth factors, extracellular matrix proteins, and various exogenous cytokines.1-5 Several cytokines, which exhibit both in vitro and in vivo effects on hematopoietic cells, have been shown to be involved in the regulation of hematopoiesis.6-25 A number of cytokines are known to promote hematopoietic cell proliferation, whereas other factors have been shown to actually inhibit this biological process.6-25
Transforming growth factor-β (TGF-β) has been shown to exhibit unique biological properties because it acts as a bifunctional regulator of in vitro hematopoietic progenitor cell proliferation. Several groups have shown that TGF-β is a potent inhibitor of myeloid (CFU-GM), erythroid (BFU-E), megakaryocytic (CFU-MK), and multilineage (CFU-MIX) progenitor cells.8,26-33 By contrast, Ottmann and Pelus32 and Jacobsen et al33 have reported that TGF-β can also significantly enhance the growth of human CFU-GM stimulated by granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-3 (IL-3). Recently five novel human genes related to a Drosophila gene termed MAD (mothers against the decapentaplegic gene) have been identified and shown to transduce intracellular signals elicited by the interaction of TGF-β family members with their respective receptors.34-37 One of these MAD-homologues recently designated as Smad5,34 also known as JV5-1 or MAD5, has been shown to be located on human chromosome 5q31, which has been proposed to be the site of a tumor suppressor gene in human myeloid leukemia.38
The above studies showing an involvement of the Smad5 gene with the signaling pathway regulating the multipotential effects of TGF-β encouraged our laboratory to study this gene to determine if it indeed plays a significant role in mediating TGF-β signaling in primitive human hematopoietic cells. Antisense oligonucleotides were designed to a coding region unique to the Smad5 gene and subsequently used to study the intracellular pathway by which TGF-β exerts its inhibitory affect on human hematopoietic cells in vitro. Our studies indicate that the inhibitory effect of TGF-β on human hematopoietic progenitor cells is clearly mediated through the Smad5 gene.
MATERIALS AND METHODS
Bone marrow (BM) aspirate specimens were obtained under local anesthesia from healthy, hematologically normal volunteer donors. Informed consent was obtained from donors according to guidelines established by the Institutional Review Board of the University of Illinois at Chicago.
Cell separation.
BM aspirates were diluted 1:1 with Iscove's modified Dulbecco's medium (IMDM) containing 20 U/mL of preservative-free sodium heparin (GIBCO-BRL, Grand Island, NY) and layered over an equal volume of Ficoll-Paque (specific gravity 1.077 g/mL; Pharmacia Biotech, Piscataway, NJ).2,6-10,17 Density centrifugation was performed at 500g for 25 minutes at 4°C. The interface low density BM (LDBM) mononuclear cells were collected and washed with IMDM containing no defined growth factors.2,6-10 17
Positive selection of CD34+ BM cells.
Washed LDBM cells were pre-enriched for a CD34+ cell population using the Miltenyi Magnetic Cell Sorting System (Miltenyi Biotech, Auburn, CA). Briefly, the CD34+ subpopulation of LDBM cells were indirectly magnetically labeled using an hapten-conjugated primary monoclonal antibody (QBEND/10) and an antihapten antibody coupled to colloidal super-paramagnetic MACS Microbeads (Miltenyi Biotech). The magnetically labeled cells are then enriched on positive selection columns. The average purity of CD34+ BM cells was 85% to 90% (data not shown).
Isolation and detection of Smad5 RNA.
Total RNA was isolated from cells using the guanidine-acid phenol method (TRIZOL; Life Technologies, Gaithersburg, MD).39 After isopropanol precipitation, RNA was resuspended in water and quantified by ultraviolet absorbance. For isolation of RNA from CD34+ cells, 20 μg glycogen was added to the initial homogenate as a carrier and coprecipitant.39
To detect Smad5 mRNA, RNA isolated both from cell lines and normal bone marrow-derived cells were used in a coupled reverse transcription polymerase chain reaction (RT-PCR) assay (Titan RT-PCR system, Boehringer Mannheim, Indianapolis, IN) using those oligonucleotides designed to span the entire coding region of Smad5 (f-TGAGTTACAGGAAGGTCTCCGA, r-TCCAAATTCTTCTCAGGAATAAGACC).34Approximately 200 ng total RNA was added to a master mix containing 1X Titan RT-PCR buffer, 1.5 mmol/L MgCl2, 0.2 mmol/L dNTPs, 5 mmol/L DTT, 10 U Rnasin, AMV reverse transcriptase and Expand PCR enzyme mix, and 0.3 μmol/L of each primer. The samples were incubated as follows: 50°C for 30 minutes, then cycled 34 times at 94°C for 30 seconds, 57°C for 30 seconds, and 68°C for 1 minute. Samples were then incubated at 68°C for 7 minutes and loaded onto a 1.5% agarose gel for analysis by EtBr staining.
The presence of Smad5 mRNA in CD34+ BM cells after treatment with oligonucleotides was assayed by RT-PCR as follows: Briefly a master mix containing all components except the primers of the coupled RT-PCR system was assembled as above.38 The master mix was divided, and oligonucleotides for amplification were added to 0.3 μmol/L of each aliquot and divided into individual assay tubes. This primer-master mix was then aliquoted into individual assay tubes. Total RNA from approximately 10,000 cells for experimental samples was added to each tube containing the RT-PCR mix. Using similar numbers of CD34+ BM cells (10,000), control experiments determined that at this RNA concentration and cycle number, samples were still within the exponential phase of amplification and, therefore, should be representative of the starting amount of mRNA.38 Samples were amplified and analyzed as above. Amplification primer sequences were as follows: Smad5, f-GAAGCTTGCTGGTAATCTTAAGAATTTTC, r-GCTTGTATCCATAGGCTGGGAA; Smad2, f-CGAAATGCCACGGTAGAAAT, r-CGGCTTCAAAACCCTGATTA; Smad4, f-GTGAAGATCAGGCCACCT, r-TGTCTGAGCATTGTGCATAG; TGF-βR1, f-CTATATCTGCCACAACCGCACTGTC, r-CGCCACTTTCCTCTCCAAACTTCTC; TGF-βR2, f-CTGCAAGATACATGGCTCCA, r-CTCGATCTCTCAACACGTTGTC; and transferrin receptor (TFR), f-AGCATTTGCAACCTTTT, r-CTCAGAGCGTCGGGATATC (as a nonspecific control).
Sense and antisense oligonucleotide synthesis.
Sense and antisense phosphothioate oligonucleotides overlapping the translational initiation codon of the Smad5 gene were synthesized (Research Genetics Inc, Huntsville, AL). Two sense oligonucleotides, S-1 (5′-GATTTGTGTCAAATGACG-3′) and S-2 (5′-TGTCAAATGACGTCAATG-3′) and two antisense oligonucleotides, AS-1 (5′-CGTCATTTGACACAAATC-3′) and AS-2 (5′-CATTGACGTCATTTGACA-3′) were used in the progenitor cell assays. All oligonucleotides were resuspended in sterile distilled H2O to a stock concentration of 1,750 μg/mL and diluted in the culture assay to yield a final concentration of 70 μg/mL. Distilled H2O was used as a negative control in all assays performed.
Progenitor cell assays.
The effects of oligonucleotides to the MAD-homologue, Smad5, on the ability of TGF-β1 and TGF-β2 to inhibit the proliferation of multiple classes of hematopoietic progenitor cells (CFU-GM, BFU-E, CFU-MIX, and CFU-MK) cloned from BM CD34+ cells were studied. Oligonucleotides were either added directly to cultures containing CD34+ BM cells plus selected cytokines with or without the addition of TGF-β or CD34+ cells were preincubated with oligonucleotides according to the method of Methia et al.40 After pretreatment with oligonucleotides, the cells were washed with IMDM and cultured in both a standard methylcellulose assay6,10 and a serum-depleted fibrin-clot culture system.2,7-9,17,23 24 CFU-GM, BFU-E, and CFU-MIX–derived colony formation in the methylcellulose culture system was stimulated by the addition of erythropoietin (EPO), IL-3, GM-CSF, and stem cell factor (SCF; kindly provided by Amgen Inc, Thousand Oaks, CA) at plateau level concentrations (100 ng/mL), previously shown to be capable of stimulating optimal numbers of colonies in vitro. The proliferation of CFU-MK–derived colonies in the fibrin-clot system was stimulated by the addition of IL-3 alone.
Immunofluorescent identification of human MK colonies.
Statistical analysis.
Results are expressed as mean ± standard error of the mean (SEM) obtained from multiple experiments. Statistical significance was determined using the Student's t-test.
RESULTS
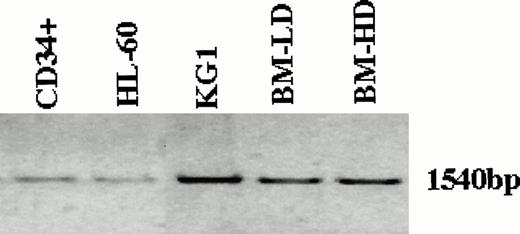
Because the expression of Smad5 RNA in human hematopoietic tissues was essential for its possible role in the signaling pathway by which TGF-β affects hematopoietic cells, we first attempted to detect Smad5 RNA in various cell populations. The entire coding region of Smad5 was amplified by PCR from RNA isolated from two immortalized human myeloid leukemia cell lines, KG1 and HL-60 as well as from human low and high density BM and CD34+ BM cells. The amplification product was subsequently analyzed on an agarose gel. As can be clearly seen in Fig 1, in all cases, a single band representing the full length coding region was found in all cell populations. Smad5 mRNA levels in antisense oligonucleotide-treated CD34+ cells were analyzed to study the effects of the oligonucleotides on Smad5. Antisense oligonucleotides were carefully designed to correspond to sequences unique to the Smad5 coding region, and had no homology with sequences of other Smad family members. Figure 2 shows a significant reduction in Smad5 mRNA levels in those CD34+ cells treated for 22 hours with antisense oligonucleotides. Although samples treated with sense oligonucleotides or mock-treated controls contained similar amounts of Smad5 message, samples treated with antisense to Smad5 contained barely detectable levels of Smad5 mRNA (Fig 2). Transferrin receptor mRNA, used as a nonspecific control, was unaffected in all samples. To evaluate the effect of Smad5 antisense oligonucleotides on other members of the TGF-β signaling pathway we also analyzed the mRNA levels of two Smad family members known to transmit the TGF-β signal, Smad2 and Smad4 (also known as DPC4), and the two TGF-β receptor subunits, TGF-βR1 and TGF-βR2. No change was found in the levels of any of these mRNA species.
Detection of Smad5 mRNA in the immortalized myeloid leukemia cell lines KG1 and HL-60, as well as low density mononuclear, high density polymorphonuclear and CD34+ selected cells isolated from normal human BM. The entire coding region of Smad5 was amplified by PCR from RNA isolated from the various sources and analyzed on an agarose gel. In all cell populations analyzed, a single band representing the full length coding region was found.
Detection of Smad5 mRNA in the immortalized myeloid leukemia cell lines KG1 and HL-60, as well as low density mononuclear, high density polymorphonuclear and CD34+ selected cells isolated from normal human BM. The entire coding region of Smad5 was amplified by PCR from RNA isolated from the various sources and analyzed on an agarose gel. In all cell populations analyzed, a single band representing the full length coding region was found.
Analysis of Smad5 mRNA levels in antisense-treated CD34+ cells. RNA from approximately 10,000 CD34+ BM cells preincubated with antisense oligonucleotides to the Smad5 gene were analyzed by RT-PCR for the presence of Smad5, Smad2, Smad4, TGF-β receptor subunit 1 (TGF-βR1), TGF-β receptor subunit 2 (TGF-βR2) and TFR (as a nonspecific control) mRNA. Amplifications were done under identical conditions and RNA concentrations and represent time points within the exponential phase of the assay. Samples treated with antisense oligonucleotides to Smad5 (AS1 and AS2) contain barely detectable or undetectable levels of Smad5 mRNA, whereas samples treated with sense oligonucleotides (S1 and S2) or mock-treated samples (control) contain similar amounts of Smad5 message. The levels of all other mRNA species were unaffected for all treatments.
Analysis of Smad5 mRNA levels in antisense-treated CD34+ cells. RNA from approximately 10,000 CD34+ BM cells preincubated with antisense oligonucleotides to the Smad5 gene were analyzed by RT-PCR for the presence of Smad5, Smad2, Smad4, TGF-β receptor subunit 1 (TGF-βR1), TGF-β receptor subunit 2 (TGF-βR2) and TFR (as a nonspecific control) mRNA. Amplifications were done under identical conditions and RNA concentrations and represent time points within the exponential phase of the assay. Samples treated with antisense oligonucleotides to Smad5 (AS1 and AS2) contain barely detectable or undetectable levels of Smad5 mRNA, whereas samples treated with sense oligonucleotides (S1 and S2) or mock-treated samples (control) contain similar amounts of Smad5 message. The levels of all other mRNA species were unaffected for all treatments.
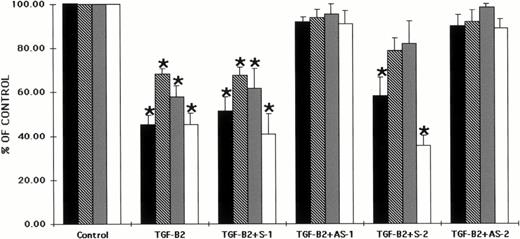
Several isoforms of TGF-β have been recently identified. Therefore, we studied the effects of antisense oligonucleotides to the Smad5 coding sequences on the ability of two forms of TGF-β, TGF-β1, and TGF-β2 to inhibit hematopoietic progenitor cell proliferation. CD34+ BM cells were preincubated with oligonucleotides to the Smad5 gene using previously reported methods.40Preincubation of CD34+ BM cells with each of the oligonucleotides had no significant effect on colony formation when compared with control values (data not shown). The cells were then plated in both methylcellulose and fibrin-clot assays in the presence of TGF-β2. Figure 3 shows that TGF-β2 had a significant inhibitory effect on human hematopoietic colony formation in vitro as has been reported previously.8 26-32When TGF-β2 was added to either methylcellulose or fibrin-clot assays at 5 ng/mL, CFU-GM, BFU-E, CFU-MIX, and CFU-MK–derived colony formation was significantly reduced (P < .5; Fig 3). Preincubation of CD34+ BM cells with sense oligonucleotides to the Smad5 gene maintained the inhibition, whereas preincubation with antisense oligonucleotides to Smad5 reversed the incubation by TGF-β2 back to control values (Fig 3). The direct addition of either sense or antisense oligonucleotides to cultures containing TGF-β2 produced results similar to those studies in which CD34+ cells were preincubated with the oligonucleotides (data not shown).
Preincubation of CD34+ cells with antisense oligonucleotides to the Smad gene reverses the inhibitory effects of TGF-β2 on human hematopoietic colony formation. CD34+BM cells were preincubated with oligonucleotides to the Smad5 gene and plated at a concentration of 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β2 was added at 5 ng/mL. The results are expressed as the mean ± SEM of data taken from three separate experiments performed in duplicate. *P ≤ .5 when compared with control values. Control CFU-GM = 45.0 ± 3.4 colonies, control BFU-E = 39.8 ± 4.8 colonies, control CFU-MIX = 11.2 ± 1.5 colonies, and control CFU-MK = 27.0 ± 5.2 colonies. (▪) CFU-GM; (□) CFU-E; (▩) CFU-MIX; (□) CFU-MK.
Preincubation of CD34+ cells with antisense oligonucleotides to the Smad gene reverses the inhibitory effects of TGF-β2 on human hematopoietic colony formation. CD34+BM cells were preincubated with oligonucleotides to the Smad5 gene and plated at a concentration of 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β2 was added at 5 ng/mL. The results are expressed as the mean ± SEM of data taken from three separate experiments performed in duplicate. *P ≤ .5 when compared with control values. Control CFU-GM = 45.0 ± 3.4 colonies, control BFU-E = 39.8 ± 4.8 colonies, control CFU-MIX = 11.2 ± 1.5 colonies, and control CFU-MK = 27.0 ± 5.2 colonies. (▪) CFU-GM; (□) CFU-E; (▩) CFU-MIX; (□) CFU-MK.
The effects of oligonucleotides to the Smad5 gene on the ability of TGF-β1 to inhibit hematopoietic progenitor cell proliferation is shown in Fig 4. TGF-β1 also significantly inhibited (P < .5) human CFU-GM, BFU-E, CFU-MIX, and CFU-MK–derived colony formation in vitro (Fig 4). As was seen in Fig 3with TGF-β2, the inhibitory effect of TGF-β1 was maintained by preincubation with sense oligonucleotides and reversed when CD34+ BM cells were preincubated with antisense oligonucleotides to Smad5 (Fig 4). Although all oligonucleotides used to preincubate CD34+ cells were at a concentration of 70 μg/mL, preincubation of CD34+ cells with an increased concentration of oligonucleotides (105 μg/mL) failed to further reverse the inhibitory effect of either TGF-β1 or TGF-β2 (data not shown). Again, the direct addition of oligonucleotides to cultures containing TGF-β1 gave similar results (data not shown).
Preincubation of CD34+ cells with antisense oligonucleotides to the Smad5 gene reverses the inhibitory effects of TGF-β1 on human hematopoietic colony formation. CD34+BM cells were preincubated with oligonucleotides to the Smad5 gene and plated at a concentration of 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β1 was added at 2 ng/mL. The results are expressed as the mean ± SEM of data taken from three separate experiments performed in duplicate. * P ≤ .5 when compared with control values. Control CFU-GM = 63.6 ± 8.0 colonies, control BFU-E = 61.5 ± 8.4 colonies, control CFU-MIX = 21.0 ± 3.6 colonies, and control CFU-MK = 26.0 ± 5.0 colonies. (▪) CFU-GM; (□) CFU-E; (▩) CFU-MIX; (□) CFU-MK.
Preincubation of CD34+ cells with antisense oligonucleotides to the Smad5 gene reverses the inhibitory effects of TGF-β1 on human hematopoietic colony formation. CD34+BM cells were preincubated with oligonucleotides to the Smad5 gene and plated at a concentration of 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β1 was added at 2 ng/mL. The results are expressed as the mean ± SEM of data taken from three separate experiments performed in duplicate. * P ≤ .5 when compared with control values. Control CFU-GM = 63.6 ± 8.0 colonies, control BFU-E = 61.5 ± 8.4 colonies, control CFU-MIX = 21.0 ± 3.6 colonies, and control CFU-MK = 26.0 ± 5.0 colonies. (▪) CFU-GM; (□) CFU-E; (▩) CFU-MIX; (□) CFU-MK.
The reversal of the inhibitory effect of TGF-β1 was inversely proportional to the concentration of TGF-β1, as shown in Fig 5. Although TGF-β1 at 5 ng/mL exhibited a greater degree of inhibition than TGF-β1 at 2 ng/mL, the antisense oligonucleotides to the Smad5 gene were better able to reverse the inhibitory effect of TGF-β1 at the lower concentration of 2 ng/mL (Fig 5). The inhibitory effect of TGF-β1 on in vitro human hematopoietic colony formation was also significantly greater than the inhibitory effect of TGF-β2, as shown in Fig 6.
The reversal of the inhibitory effects of TGF-β1 on hematopoietic colony formation by antisense oligonucleotides to the Smad5 gene is inversely related to the concentration of TGF-β1. CD34+ BM cells were preincubated with oligonucleotides to the Smad5 gene and plated either at 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β1 was added either at 2 ng/mL or at 5 ng/mL. The results are expressed as the mean ± SEM of total colony numbers. The data was taken from three separate studies performed in duplicate. *P ≤ .5 when compared with control values. (▪) TGF-β1 at 2 ng/mL; (□) TGF-β1 at 5 ng/mL.
The reversal of the inhibitory effects of TGF-β1 on hematopoietic colony formation by antisense oligonucleotides to the Smad5 gene is inversely related to the concentration of TGF-β1. CD34+ BM cells were preincubated with oligonucleotides to the Smad5 gene and plated either at 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β1 was added either at 2 ng/mL or at 5 ng/mL. The results are expressed as the mean ± SEM of total colony numbers. The data was taken from three separate studies performed in duplicate. *P ≤ .5 when compared with control values. (▪) TGF-β1 at 2 ng/mL; (□) TGF-β1 at 5 ng/mL.
The inhibitory effect of TGF-β1 on hematopoietic colony formation is more profound than that exhibited by TGF-β2. CD34+ BM cells were preincubated with oligonucleotides to the Smad5 gene and plated either at 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β1 and TGF-β2 were both added at 5 ng/mL. The results are expressed as the mean ± SEM of total colony numbers. The data was taken from three separate studies performed in duplicate. *P ≤ .5 when compared with control values.
The inhibitory effect of TGF-β1 on hematopoietic colony formation is more profound than that exhibited by TGF-β2. CD34+ BM cells were preincubated with oligonucleotides to the Smad5 gene and plated either at 2 × 103/mL in the presence of EPO (5 U/mL), IL-3 (100 ng/mL), GM-CSF (100 ng/mL), and SCF (100 ng/mL) in a methylcellulose culture assay or at 2 × 104/mL in the presence of IL-3 (100 ng/mL) in a fibrin-clot assay system. TGF-β1 and TGF-β2 were both added at 5 ng/mL. The results are expressed as the mean ± SEM of total colony numbers. The data was taken from three separate studies performed in duplicate. *P ≤ .5 when compared with control values.
DISCUSSION
TGF-β is a secretory polypeptide growth factor that is expressed and released by several different cell types while being stored in human blood platelets.27,28,42,43 TGF-β was originally purified from human platelets, human placenta, and bovine kidney27and identified as a disulfide-linked dimer of two usually identical chains of 112 amino acids.27,28,42 However, heterodimers of TGF-β isotypes have also been identified.42 TGF-β is recognized as a multifunctional regulator of hematopoietic cellular activity because of its ability to either stimulate (cells of mesenchymal origin) or inhibit (cells of epithelial or neuroectodermal origin) cell proliferation and growth.27,28,42 Although specific receptors for TGF-β are present on almost all mammalian cells, its effects on hematopoiesis varies depending on the cell type, growth conditions, the state of cell differentiation, and the presence of other growth factors.27,28 42
TGF-β1, the originally described isoform of TGF-β, is but one member of a superfamily of regulatory proteins, including the activins, inhibins, bone morphogenetic proteins, and a number of more closely related proteins designated TGF-β2, TGF-β3, TGF-β4, and TGF-β5.43 Although most studies analyzing the effects of TGF-β on hematopoiesis used TGF-β1, additional isoforms including TGF-β2 and TGF-β3 have also been reported to exhibit biological activity equivalent to that of TGF-β1.44,45 However, both Jennings et al46 and Ohta et al47 have reported that TGF-β1 exhibits a higher potency of biological activity in vitro than TGF-β2. Therefore, we used both TGF-β1 and TGF-β2 in the studies described here in this report.
The exact mechanism by which TGF-β inhibits human hematopoiesis is not presently known. The retinoblastoma gene (RB1), a nuclear phosphoprotein, exhibits growth-suppression activity similar to that of TGF-β.48 Using both mink lung epithelial cells and human lung adenocarcinoma cells, several groups have linked the growth inhibition by TGF-β to its ability to keep RB1 in its unphosphorylated, growth-suppressive state.48,49 They show that RB1 can regulate TGF-β gene expression and suggest that both TGF-β and RB1 function together in a common growth inhibitory pathway.48 49
The signaling pathway that regulates the multipotential effects of TGF-β is only partially understood. Although the cellular effects of TGF-β are known to be mediated by transmembrane receptors with cytoplasmic serine-threonine kinase activity, most other components of the TGF-β signaling pathway, including the targets of receptor kinase activity and components of the signal transduction pathway, remain unknown.36,50 However, recently several groups have linked the transduction of signals from TGF-β family members to five novel human genes.34-37 These genes are related to a Drosophila gene called MAD and apparently function downstream of the TGF-β receptors.34 Two MAD-homologues, Smad4 (DPC4)51and Smad2 (JV18-1),34 have recently been shown to be present on chromosome 18q21.1 as candidate tumor suppressor genes whose inactivation may lead to pancreatic, colon, and other human cancers. Smad5 is another MAD-homologue gene bound on human chromosome 5q31,38 the site of a putative tumor suppressor gene in acute myeloid leukemia. Therefore, we studied the involvement of this gene in the signaling pathway by which TGF-β is able to exert an inhibitory effect on human hematopoiesis in vitro. Smad5 RNA was shown by reverse transcriptase PCR to be present in two immortalized human myeloid leukemia cell lines, KG1 and HL60, in human BM mononuclear and polymorphonuclear cells, as well as in human BM cells positively selected for the CD34 antigen. CD34+ cells, which are enriched for human hematopoietic progenitor cells, were incubated with sense and antisense oligonucleotides designed to a unique Smad5 region encompassing the translational start site. The level of Smad5 mRNA was significantly decreased by antisense oligonucleotides to Smad5, whereas sense oligonucleotides or control treated samples contained similar amounts of mRNA. These antisense oligonucleotides to Smad5 had no effect on the mRNA levels of Smad2, Smad4, TGF-βR1, or TGF-βR2 suggesting that the other components of the TGF-β signaling pathway are intact. The effects of these oligonucleotides on the ability of both TGF-β1 and TGF-β2 to inhibit human hematopoietic colony formation was studied in vitro. As was previously reported,8,26-32 TGF-β1 and TGF-β2 both significantly inhibited CFU-GM, BFU-E, CFU-MK, and CFU-MIX–derived colony formation. TGF-β1 exhibited a greater degree of inhibition than TGF-β2, as also reported by different laboratories.46 47 Preincubation of human CD34+ adult bone marrow (ABM) cells with two sense oligonucleotides to the Smad5 gene maintained the inhibitory effect of TGF-β1 and TGF-β2. However, preincubation of ABM CD34+ cells with antisense oligonucleotides to Smad5 reversed the inhibitory effects of TGF-β1 and TGF-β2, allowing colony numbers to return to control values. Although TGF-β1 exerted a higher degree of inhibition than TGF-β2, the reversal of TGF-β1 inhibition by antisense oligonucleotides was significantly less. In addition, the reversal of the inhibitory effect of TGF-β1 by the antisense oligonucleotides was inversely proportional to the concentration of TGF-β1 used in culture. The incomplete reversal at high TGF-β concentrations by antisense oligonucleotides may be caused by residual Smad5 activity or to the presence of alternative signaling pathways.
Overall, these studies suggest that the Smad5 gene plays a critical role in the signaling pathway controlling the inhibitory effect of TGF-β on human hematopoietic progenitor cells. It remains possible that disruption of the function of Smad5 or of other members of this signaling pathway could lead to unrestrained hematopoietic cell proliferation and, thus, provide one of the important steps toward human leukemogenesis. This hypothesis is currently being explored in our laboratory.
Erythrophagocytosis. Kupffer cell containing phagolysosomes is shown on a stained Epon section (A, original magnification ×10,500). Erythrophagocytosis is confirmed by an ultrathin frozen section which is labeled by a polyclonal rabbit antibody against human hemoglobin and immunogold particles showing that the electron dense bodies represent phagocytosed erythrocytes (B, original magnification ×41,000). Unstained Epon section reveals ferritin particles in the cytoplasm, a sidersome, and a lysosome of an adjacent hepatocyte which indicates iron overload (C, original mangification, ×58,000). H, hepatocyte; N, nucleus; G, Golgi complex; PE, phagocytosed erythrocyte; C, cytoplasm; G, glycogen particles; S, siderosome; Lys, lysosome. (Courtesy of K.-P. Zimmer, MD, Universitätskinderklinik, AlbertSchweitzer-Str. 33, D-48149 Münster, Germany.)
Erythrophagocytosis. Kupffer cell containing phagolysosomes is shown on a stained Epon section (A, original magnification ×10,500). Erythrophagocytosis is confirmed by an ultrathin frozen section which is labeled by a polyclonal rabbit antibody against human hemoglobin and immunogold particles showing that the electron dense bodies represent phagocytosed erythrocytes (B, original magnification ×41,000). Unstained Epon section reveals ferritin particles in the cytoplasm, a sidersome, and a lysosome of an adjacent hepatocyte which indicates iron overload (C, original mangification, ×58,000). H, hepatocyte; N, nucleus; G, Golgi complex; PE, phagocytosed erythrocyte; C, cytoplasm; G, glycogen particles; S, siderosome; Lys, lysosome. (Courtesy of K.-P. Zimmer, MD, Universitätskinderklinik, AlbertSchweitzer-Str. 33, D-48149 Münster, Germany.)
Supported in part by Grant No. PO1HL 53762-03 from the National Institutes of Health and by a generous gift from the W. M. Keck Foundation.
Address reprints requests to Ronald Hoffman, MD, University of Illinois at Chicago, MBRB 3150, M/C 734, 900 S Ashland Ave, Chicago, IL 60607.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.