Abstract
Complete remission is achieved in a high proportion of patients with acute promyelocytic leukemia (APL) after all-trans retinoic acid (RA) treatment, but most patients relapse and develop RA-resistant APL. We have previously reported that both RA-resistant HL-60 (HL-60R) and APL cells express P-glycoprotein and MDR1 transcripts; and these cells differentiate to mature granulocytes after culture with RA and P-glycoprotein antagonist. Ribozymes have been shown to be able to intercept a target RNA by catalytic activity. To address the role of MDR1 in overcoming RA-resistance in APL cells, we investigated the biologic effects of ribozymes against the MDR1 transcript in HL-60R cells. These ribozymes efficiently cleaved MDR1 mRNA at a specific site in vitro. The 196 MDR1 ribozyme was cloned into an expression vector, and stably transfected (HL-60R/196Rz) cells were obtained. Expression of MDR1 transcripts was decreased in HL-60R/196Rz cells compared with parental HL-60R and empty vector-transfected (HL-60R/neo) cells. Interestingly, RA inhibited cellular proliferation and induced differentiation of HL-60R/196Rz cells in a dose-dependent manner, suggesting reversal of drug resistance in HL-60R cells by the MDR1 ribozyme. These data are direct evidence that P-glycoprotein/MDR1 is responsible in part for acquired resistance to RA in myeloid leukemic cells. The MDR1 ribozyme may be a useful tool for investigating the biology of retinoid resistance and may have therapeutic potential for patients with RA-resistant APL.
RETINOIC ACID (RA) has a critical role in proliferation and differentiation of a wide variety of cell types, especially hematopoietic cells.1,2 All-trans RA and its stereoisomer, 9-cis RA, induce differentiation and inhibit proliferation of HL-60 cells and fresh leukemic cells from patients with acute promyelocytic leukemia (APL) in vitro.3-5 Recent clinical studies have shown that a high proportion of patients with APL achieve complete remission after treatment with all-transRA.6-9 However, most patients who receive continuous treatment with RA relapse and develop RA-resistant disease.10 Although the mechanisms for development of RA-resistance in APL are unclear, several hypotheses are possible. We have previously reported that RA-resistant HL-60 (HL-60R) and RA-resistant APL cells express P-glycoprotein and MDR1 transcripts unlike wild-type HL-60 and fresh APL cells.11 RA-resistant cells differentiate to mature granulocytes after culture with all-trans RA combined with verapamil, a P-glycoprotein antagonist, but not with either agent alone.11 Resistance to multiple chemotherapeutic agents is related to the expression of P-glycoprotein, a transmembrane drug efflux pump that is encoded by the MDR1 gene.12 13 Therefore, we speculated that P-glycoprotein may be important in the development of RA-resistance in myeloid leukemic cells.
Studies have shown that ribozymes are able to target RNA by catalytic activity in a specific manner.14-16 The hammerhead ribozyme can be engineered to cleave any triplet of NUX (N = any nucleotide, X = A, C, or U), by changing the flanking sequence.16 We have designed two hammerhead ribozymes targeted against the MDR1 transcript, 179 MDR1 ribozyme and 196 MDR1 ribozyme, which cause recovery of drug sensitivity in multidrug-resistant tumor cells.17 18Ribozyme-mediated inactivation of MDR1 mRNA may overcome RA-resistance in myeloid leukemic cells. Therefore, we investigated the biologic effects of ribozymes against MDR1 transcript in RA-resistant HL-60 cells.
MATERIALS AND METHODS
Cells and chemicals.
Wild-type and RA-resistant HL-60 cells (HL-60R: a generous gift from Dr R.E. Gallagher, Montefiore Medical Center, Bronx, NY), were maintained in RPMI 1640 medium (GIBCO-BRL, Gaithersburg, MD) with 10% fetal bovine serum (Cytosystems, New South Wales, Australia), 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified atomosphere with 5% CO2. All-trans RA and 9-cis RA were purchased from Sigma Chemical Co (St Louis, MO) and Wako Pure Chemical Industries Ltd (Tokyo, Japan), respectively. Both were dissolved in 100% ethanol to a stock concentration of 1 mmol/L, stored at −20°C, and protected from light. All-trans RA was added to the maintenance culture medium of HL-60R cells.19The morphology of differentiated-cells was evaluated from cytospin slide preparations stained with Giemsa.
Synthesis of MDR1 ribozymes.
Production of the 179 and 196 MDR1 ribozymes and its RNA substrate are described elsewhere.17 18 Briefly, the 196 MDR1 ribozyme (Fig 1) was produced from two synthetic oligonucleotides, 5′-TCT TTC AGT TTC GTC CTC ACG GAC TCA TCA GAA TGG CAA CCC CTA TAG TGA GTC GTA TTA CAT G-3′, and 5′-CAT GTA ATA CGA CTC ACT ATA GGG-3′. The oligonucleotides were mixed and heated at 80°C for 2 minutes and slowly cooled to room temperature to form a hemiduplex. They were incubated at 37°C for 4 hours with 5 U/μL T7 RNA polymerase (New England Biolabs, Beverly, MA), 1.2 U/μL RNase inhibitor (Promega, Madison, WI), and 2 mmol/L each adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triphosphate (UTP). After incubation with 0.1 U/μL RQ1 RNase-free DNase (Promega) for 15 minutes, ribozyme was extracted, and the pellet was dissolved with diethyl pyrocarbonate (DEPC)-treated water.
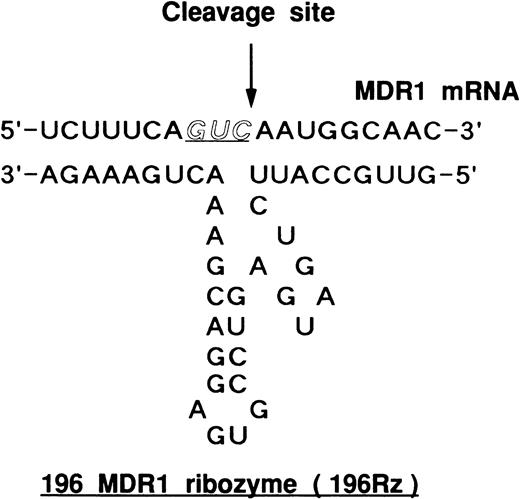
Hammerhead ribozyme targeted against MDR1 RNA. 196 MDR1 ribozyme (196Rz) cleaved MDR1 transcript at codon 196, as indicated by the arrow. Codon 196 is one of the substrate cleavage sites.
Hammerhead ribozyme targeted against MDR1 RNA. 196 MDR1 ribozyme (196Rz) cleaved MDR1 transcript at codon 196, as indicated by the arrow. Codon 196 is one of the substrate cleavage sites.
Ribonuclease protection assay.
To address the cleavage activity of MDR1 ribozymes, MDR1 mRNA was analyzed by ribonuclease protection assay RPAII (Ambion, Austin, TX) using total cellular RNA extracted from MOLT-3/trimetrexate (TMQ) 800 cells.20 After cDNA was synthesized from this RNA with reverse transcription (RT), exons 6 (5′-TTC ATG CTA TAA TGC GAC AGG AGA TA-3′) through 8 (5′-TTC TTT ATC AGT AAA TGA AGA TAG TA-3′) of the MDR1 gene was amplified by polymerase chain reaction (PCR). The [α-32P]CTP-labeled MDR1 RNA probe used in this assay was synthesized from the opposite orientation of the PCR product of MDR1 in pT7Blue T-vector by T3 RNA polymerase.
196 MDR1 ribozyme transfection in HL-60R cells.
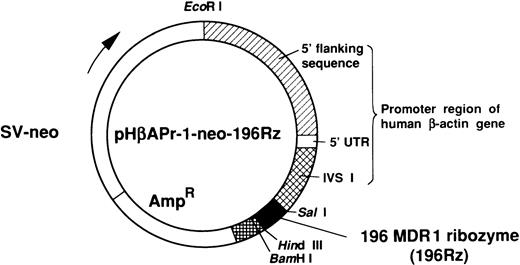
The cDNA of ribozyme with Sal I and HindIII restriction sites on both ends was cloned into the expression vector pHβAPr-1-neo (kindly provided by Dr L. Kedes, University of Southern California, Los Angeles, CA) (Fig 2).21 HL-60R cells were transfected with plasmid containing 196 MDR1 ribozyme or an empty vector by electroporation using an Electroporator II (Invitrogen, San Diego, CA) at 350 V, 960 μF. A pulse was delivered to a 0.5-mL suspension containing 5 × 106 cells in RPMI 1640 without fetal bovine serum and 20 μg of plasmid DNA. After an additional 10 minutes incubation at room temperature, cells were diluted with prewarmed RPMI 1640 with 10% fetal bovine serum. Three days after transfection, cells were selected by culture with G418 (1 mg/mL; GIBCO-BRL). Selective medium was replaced every 5 days, and stably transfected polyclonal cell populations were isolated after 4 weeks of selection with G418.
Reverse transcriptase (RT)-PCR assay for MDR1 transcript.
Expression of the MDR1 gene was investigated by the RT-PCR method. The RT reaction was performed using 1 μg of total cellular RNA, 100 pmol of random hexamer (Boehringer-Mannheim, Indianapolis, IN), 10 U RNase inhibitor (Promega), 200 U moloney murine leukemia virus-RT (GIBCO-BRL), and deoxynucleotides (final concentration, 0.5 mmol/L each; Pharmacia, Tokyo, Japan). After cDNA synthesis, 30 cycles of PCR were sequentially performed using MDR1 and β-actin as follows: for MDR1, sense, 5′-ATG TTG AGC CGG GCA GTG TGC-3′ and antisense, 5′-CTG AAG AGC TGT CTG GGC TGT-3′,12and for β-actin, sense, 5′-ATG GAT GAT GAT ATC GCC GCG-3′ and antisense, 5′-AAA GAA CAC GGC TAA GTG TGC-3′.22 PCR was performed using denaturing steps at 95°C for 1 minute, annealing steps at 55°C for 1 minute, and extending steps at 72°C for 2 minutes. At the end of 30 cycles, further extension for 7 minutes at 72°C was included. All reactions were performed in a total volume of 50 μL containing 1 μL of cDNA (of 10 μL total RT reaction), 1 mmol/L of each primer, 200 mmol/L of each deoxynucleotide, 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 2.5 mmol/L MgCl2, and 0.5 U Taq DNA polymerase (Perkin Elmer-Cetus, Norwalk, CT). PCR products were electrophoresed on a 2% NuSieve/1% Seakem agarose (FMC, Rockland, MI) gel and visualized by staining with ethidium bromide. The specificity of PCR amplification was also examined by Southern blotting of the amplified cDNA with hybridization to nonradioactive probes (ECL 3′-oligolabeling and detection system; Amersham Japan, Tokyo, Japan).
Assays for cellular proliferation.
Cellular proliferation was measured by cell viability and a nonradioactive cell proliferation assay system (MTT assay) according to the manufacturer's specifications (Boehringer-Mannheim).
Flow cytometric analysis.
For analysis of cellular differentiation, expression of cell surface antigens was measured by direct immunofluorescence staining technique. Cells were incubated for 30 minutes with human AB serum (Sigma) to block Fc receptors and then were stained with phycoerythrin (PE)-conjugated mouse antihuman CD11b antibodies (Becton-Dickinson, Mountain View, CA). For P-glycoprotein detection, cells were stained with monoclonal antibody MRK16 (Kyowa Medex Co Ltd, Tokyo, Japan), followed by staining with goat antimouse antibody conjugated to fluorescein isothiocyanate (FITC). Control studies were performed with nonbinding control mouse IgG isotype antibody (Becton-Dickinson). Cells were analyzed by a Cytoron Absolute Flow Cytometer (Ortho Diagnostic Systems, Raritan, NJ).
RESULTS
Cleavage of MDR1 mRNA by ribozymes.
We have designed two hammerhead ribozymes to cleave the GUC sequence at codons 179 and 196 of MDR1 mRNA.17,18 We already reported that the ribozymes efficiently cleaved target RNA substrates in a cell-free system.18 Furthermore, we analyzed the cleavage activity of ribozymes in mRNA extracted from MOLT-3/TMQ 800 cells using ribonuclease protection assay (Fig 3). The 196 MDR1 ribozyme cleaved MDR1 mRNA into two fragments of 124 bases of 5′ fragment (5′F) and 133 bases of 3′ fragment (3′F) by using 32P-labeled MDR1 RNA probe (Fig 3). The 196 MDR1 ribozyme was more active than the 179 MDR1 ribozyme (Fig3); thus, we used the 196 MDR1 ribozyme in this study.
Cleavage reaction of MDR1 mRNA with ribozymes. Ribonuclease protection assay was performed to study for the cleavage of MDR1 mRNA by the ribozyme expressed in MOLT-3/TMQ 800 cells. Lane 1, probe only; lane 2, probe hybridized with yeast tRNA, digested with ribonucleases; lane 3, protected fragment of MDR1 mRNA; lane 4, 1 μg of mRNA extracted from MOLT-3/TMQ 800 cells plus 179 MDR1 ribozyme; lane 5, 1 μg of mRNA extracted from MOLT-3/TMQ 800 cells plus 196 MDR1 ribozyme.
Cleavage reaction of MDR1 mRNA with ribozymes. Ribonuclease protection assay was performed to study for the cleavage of MDR1 mRNA by the ribozyme expressed in MOLT-3/TMQ 800 cells. Lane 1, probe only; lane 2, probe hybridized with yeast tRNA, digested with ribonucleases; lane 3, protected fragment of MDR1 mRNA; lane 4, 1 μg of mRNA extracted from MOLT-3/TMQ 800 cells plus 179 MDR1 ribozyme; lane 5, 1 μg of mRNA extracted from MOLT-3/TMQ 800 cells plus 196 MDR1 ribozyme.
Expression of P-glycoprotein and MDR1 mRNA in ribozyme-transfected HL-60R cells.
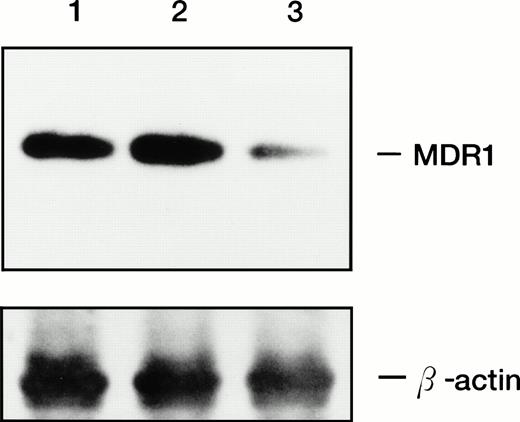
The 196 MDR1 ribozyme was cloned into the mammalian expression vector (pHβAPr-1-neo) (Fig 2) and transfected into HL-60R cells by electroporation. After G418 selection, stably transfected polyclonal cells were obtained (HL-60R/neo and HL-60R/196Rz cells). Morphologic changes did not occur in HL-60R/neo and HL-60R/196Rz cells and a similar phenotype was observed in all transfected cells; and these cells were resistant to 1 mg/mL G418, and their growth rate was similar to parent HL-60R cells (data not shown). Expression of P-glycoprotein was low in parental HL-60R, HL-60R/neo and HL-60R/196Rz cells (data not shown). Therefore, to test the ability of the 196 MDR1 ribozyme to cleave MDR1 mRNA in HL-60R/196Rz cells, we investigated the expression of MDR1 transcripts in both parental and ribozyme-transfected HL-60R cells by the RT-PCR technique. The expression of MDR1 transcripts decreased in HL-60R/196Rz cells compared with parental HL-60R and HL-60R/neo cells, and this was confirmed by subsequent Southern blotting of the RT-PCR products using a probe specific for the MDR1 gene (Fig 4). β-actin was amplified simultaneously and showed comparable amounts of RNA in the cells. The disabled ribozyme is considered to be an appropriate control to determine the possible effects of functional ribozymes. However, we have already shown that a disabled 196 MDR1 ribozyme was neither capable of specific cleavage in vitro nor of decreasing the MDR1 transcripts in MOLT-3/TMQ 800 cells.18 These data therefore suggest that the 196 MDR1 ribozyme specifically inhibits the expression of the MDR1 transcripts in HL-60R cells.
Expression of MDR1 transcript in parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells by RT-PCR analysis and subsequent Southern blotting. 196 MDR1 ribozyme was cloned into the expression vector, pHβAPr-1-neo and transfected into HL-60R cells by electroporation (HL-60R/196Rz). Control clone was derived by transfection of empty pHβAPr-1-neo alone (HL-60R/neo). A total of 1 μg of total RNA was reverse transcribed to cDNA and amplified by PCR with primers specific for MDR1 and β-actin as an internal control; the amplification products were electrophoresed, transferred, and hybridized with probes specific for MDR1 (upper bands) and β-actin (lower bands) genes. Lane 1, HL-60R cells; lane 2, HL-60R/neo; lane 3, HL-60R/196Rz cells.
Expression of MDR1 transcript in parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells by RT-PCR analysis and subsequent Southern blotting. 196 MDR1 ribozyme was cloned into the expression vector, pHβAPr-1-neo and transfected into HL-60R cells by electroporation (HL-60R/196Rz). Control clone was derived by transfection of empty pHβAPr-1-neo alone (HL-60R/neo). A total of 1 μg of total RNA was reverse transcribed to cDNA and amplified by PCR with primers specific for MDR1 and β-actin as an internal control; the amplification products were electrophoresed, transferred, and hybridized with probes specific for MDR1 (upper bands) and β-actin (lower bands) genes. Lane 1, HL-60R cells; lane 2, HL-60R/neo; lane 3, HL-60R/196Rz cells.
Effects of ribozyme on cellular proliferation of HL-60R cells.
Cell growth was assessed by absorbance in the MTT cellular proliferation assay system. We have already reported that neither all-trans RA nor its stereoisomer, 9-cis RA, inhibited proliferation or induced differentiation of HL-60R cells into either mature granulocytes or monocytes.11 23 Parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells were cultured for 4 days in the presence of various concentrations of retinoids (10-10 to 10-6 mol/L) (Fig 5). All-trans RA did not affect the number of viable cells or the absorbance of MTT by parental HL-60R and HL-60R/neo cells (Fig 5 and data not shown). However, all-trans RA and 9-cis RA inhibited cellular proliferation in HL-60R/196Rz cells in a dose-dependent manner (10-10 to 10-6 mol/L), suggesting reversal of drug resistance in HL-60R cells by 196 MDR1 ribozyme. 9-cis RA showed slightly more inhibition of the growth of HL-60R/196Rz cells.
Effects of retinoids on proliferation of parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells. Cells (2 × 103) were incubated in 96-well plates with various concentrations (10−10 to 10−6 mol/L) of all-transRA or 9-cis RA for 4 days, and MTT incorporation was then measured. The absorbance at 570 nm (optical density [OD] 570) was recorded using an enzyme-linked immunosorbent assay plate reader. Results are expressed as percent of control absorbance of three experiments, and standard deviation (SD) was within 10% of the mean.
Effects of retinoids on proliferation of parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells. Cells (2 × 103) were incubated in 96-well plates with various concentrations (10−10 to 10−6 mol/L) of all-transRA or 9-cis RA for 4 days, and MTT incorporation was then measured. The absorbance at 570 nm (optical density [OD] 570) was recorded using an enzyme-linked immunosorbent assay plate reader. Results are expressed as percent of control absorbance of three experiments, and standard deviation (SD) was within 10% of the mean.
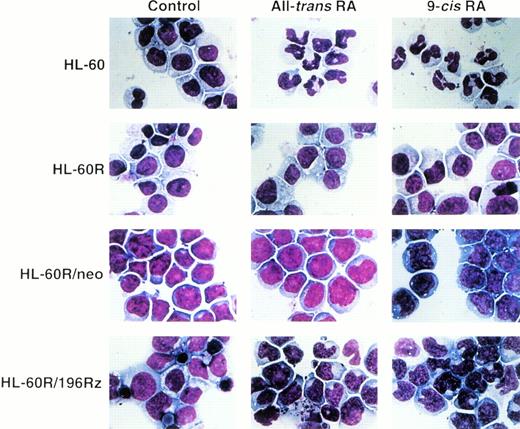
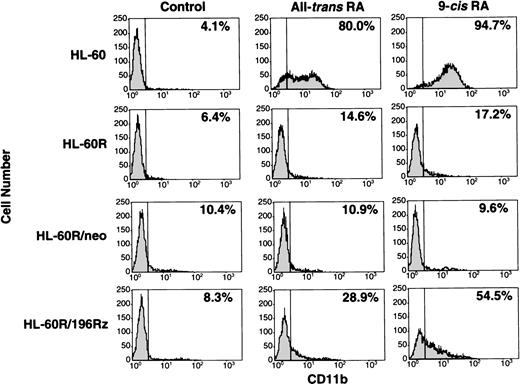
Effects of ribozyme on differentiation of HL-60R cells.
Induction of differentiation of parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells into mature granulocytes by retinoids was assessed by morphology (Fig 6) and expression of CD11b antigen (Fig 7). Morphologically, exposure of wild-type HL-60 cells to either 10-7 mol/L all-trans RA or 9-cis RA for 4 days resulted in differentiation towards mature granulocytes (Fig 6) and increased expression of CD11b (Fig 7). However, retinoid treatment did not change the morphology or expression of CD11b antigen in parental HL-60R and HL-60R/neo cells (Figs 6 and 7). In contrast, expression of CD11b antigen in HL-60R/196Rz cells was increased by 10-7 mol/L all-trans RA or 9-cis RA (Fig 7). Incubation with either retinoid resulted in differentiation of HL-60R/196Rz cells as evidenced by nuclear maturation and lobulation (Fig 6), suggesting that retinoids could partially differentiate HL-60R/196Rz cells, but not parental HL-60R and HL-60R/neo cells, toward granulocytes. 9-cis RA was slightly more potent than all-trans RA in inducing differentiation of HL-60R/196Rz cells.
Morphologic changes induced in wild-type HL-60, parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells by either all-transRA or 9-cis RA. Cells were treated with 10−7mol/L of either retinoid for 4 days, and cytospin slides were then prepared and stained with Giemsa. Original magnification × 1,000.
Morphologic changes induced in wild-type HL-60, parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells by either all-transRA or 9-cis RA. Cells were treated with 10−7mol/L of either retinoid for 4 days, and cytospin slides were then prepared and stained with Giemsa. Original magnification × 1,000.
Expression of CD11b antigen in wild-type HL-60, parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells by fluorescence-activated cell sorting (FACS) analysis. Cells were cultured with 10-7 mol/L of either all-trans RA or 9-cisRA for 4 days. Cells were incubated for 30 minutes with human AB serum to block Fc receptors, and then stained with PE-conjugated mouse antihuman CD11b antibody. Control studies were performed with control mouse IgG isotype antibody.
Expression of CD11b antigen in wild-type HL-60, parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells by fluorescence-activated cell sorting (FACS) analysis. Cells were cultured with 10-7 mol/L of either all-trans RA or 9-cisRA for 4 days. Cells were incubated for 30 minutes with human AB serum to block Fc receptors, and then stained with PE-conjugated mouse antihuman CD11b antibody. Control studies were performed with control mouse IgG isotype antibody.
DISCUSSION
All-trans RA is now widely used in the treatment of patients with APL as a differentiation-inducing therapy.6-9 Although a high proportion of patients with APL achieve complete remission with RA, a minority of patients are primarily resistant to retinoids, and relapsed patients frequently develop RA-resistant disease.10 Relapse and acquired resistance to treatment with all-trans RA have been hypothesized to result from alteration of drug metabolism and increased expression of proteins related to retinoid metabolism.24,25 HL-60 and APL cells have different genetic changes. HL-60 cells do not have an APL-specific 15;17 translocation resulting in a PML/RARα chimeric gene, however, both cells can be induced to differentiate into mature granulocytes by all-trans RA. Therefore, HL-60 cells can provide a model to help study the mechanisms of induction of differentiation in response to retinoids. We have previously reported that RA-resistant HL-60 (HL-60R) and APL cells, but not RA-sensitive cells, express MDR1 gene transcripts and P-glycoprotein.11 Moreover, the combination of retinoids and a P-glycoprotein antagonist, verapamil, but neither of them alone, induced morphologic and functional differentiation of RA-resistant leukemic cells.11 P-glycoprotein is an energy-dependent drug efflux pump that decreases intracellular accumulation of various lipophilic compounds12; we speculated that retinoid, a fat soluble vitamin, may be a substrate for P-glycoprotein. Recent studies have shown significantly lower P-glycoprotein expression in APL than in other types of AML.26 27 Therefore, a metabolic pathway for retinoids may exist that depends on P-glycoprotein expression; P-glycoprotein induction may be responsible in part for retinoid resistance in myeloid leukemic cells.
Ribozymes inhibit the expression of a variety of genes and may be a novel therapeutic approach in infectious disease and cancer.28,29 Ribozymes appear to act in an antisense fashion and as catalytic enzymes that cleave their substrate.15,16 One molecule of ribozyme can cleave many molecules of target RNA, then dissociate from the products, bind to a new target molecule, and inactivate it. Thus, ribozymes are thought to be more potent than antisense oligonucleotides at suppressing the expression of target genes.30 Several studies have used ribozymes to specifically target the bcr-abl, PML/RARα, andAML/MTG8 fusion genes in leukemic cells.31-35 These studies have shown that ribozymes efficiently cleaved target RNA.36
Because the MDR1 product seems to be responsible in part for retinoid resistance in RA-resistant myeloid leukemic cells,11 we designed hammerhead ribozymes that can cleave the GUC sequence in MDR1 mRNA. In our study, the 196 MDR1 ribozyme produced more efficient cleavage than the 179 MDR1 ribozyme, thus, we used the 196 MDR1 ribozyme in this study. We showed that ribozyme against MDR1 could cleave the substrate RNA extracted from MOLT-3/TMQ 800 cells using ribonuclease protection assay . More importantly, after transfection into RA-resistant HL-60 cells, the ribozyme inhibited expression of MDR1 transcripts, partially inhibited proliferation, and induced differentiation of these cells. The data described in our study suggests that ribozyme-mediated inactivation of the MDR1 gene will partially overcome the block of leukemic cell differentiation in HL-60R cells and provide direct evidence that the MDR1 gene is responsible for acquired resistance to retinoids in myeloid leukemic cells.
The partial response to retinoids after inhibiting MDR1 transcription indicates a complex mechanism of RA-resistance in these cells. In addition, pharmacologic levels of retinoids were required to overcome RA-resistance even when MDR1 gene was targeted by the ribozyme, suggesting the other defect existed in HL-60R cells. Several mechanisms have been proposed to explain resistance to retinoids in leukemic cells,25 including molecular alterations of the RA receptor (RAR) gene,37,38 and pharmacologic alterations in the metabolism of retinoids.24,25 Based on these studies, several strategies may overcome RA resistance in APL patients. To date, however, most approaches have not yet been successful in overcoming retinoid resistance in vivo.25 Therefore, further studies are needed to clarify the detailed molecular mechanisms of retinoid-resistance in APL; the combination of retinoids and 196 MDR1 ribozyme is one possible strategy to cure patients who fail the initial differentiation-inducing therapy with all-trans RA.
In summary, retinoid resistance is a serious and important clinical problem of differentiation-inducing therapy for the patients with APL. However, there is redundancy and complexity of the biologic mechanisms of RA-resistance in vivo. Our data presented in this study have direct evidence that the MDR1 gene is responsible in development of RA-resistance in myeloid leukemic cells. In addition, inhibition of MDR1 transcripts by ribozymes restored the sensitivity to RA. Taken together, we conclude that this technology may be useful as a new therapy in the treatment of patients with RA-resistant APL and other cancers.
ACKNOWLEDGMENT
We thank Dr T. Ohnuma, Mount Sinai School of Medicine, New York, NY for providing MOLT-3/TMQ 800 cells.
Supported by grants from the Ministry of Education, Science, and Culture in Japan, the National Grant-in-Aid for the Establishment of High-Tech Research Center in a Private University, and the Keio University Special Grants.
Address reprint requests to Masahiro Kizaki, MD, Division of Hematology, Keio University School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


![Fig. 5. Effects of retinoids on proliferation of parental HL-60R, HL-60R/neo, and HL-60R/196Rz cells. Cells (2 × 103) were incubated in 96-well plates with various concentrations (10−10 to 10−6 mol/L) of all-transRA or 9-cis RA for 4 days, and MTT incorporation was then measured. The absorbance at 570 nm (optical density [OD] 570) was recorded using an enzyme-linked immunosorbent assay plate reader. Results are expressed as percent of control absorbance of three experiments, and standard deviation (SD) was within 10% of the mean.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/7/10.1182_blood.v91.7.2452/3/m_blod4073605.jpeg?Expires=1769290967&Signature=MXk7w2g4K9jMbygPmvosclyJHzoQynzSSt2XT-6xNsDVyi-vZFbwY2S79j8-UgcOnMrLN-SPVBBpmtnDXOTPcg6l57ZVe52Hii1gQEdwepLrzaDFYTLt1Rxfe3pCcKgTW~FT4kQsdM~8uc9JJBRrTzhBiOcw9DNXGt70wklXZSDvS-C0bg0spZhipusRjMQfZ1xiCkSZQ6VILCku1PwNmKqB1l3yydiDKPAREaJnkrw0W3YPvO9wrYcyA2YSihm005m54S1Mum-CM04kvnX-IIgBPDJ5R~heNvTtX~geZbrt78TqTb1SWQNyGQXOj1BLn34ZrCUpt85FK~vJNZOK6w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)