HUMAN HEMOGLOBINS and their variants have been the subjects for fruitful clinical and basic research for many years. The knowledge derived from these studies is essential in understanding the relationship between hemoglobin structures and functions. The report on the seminal study of sickle hemoglobin by Pauling et al1 in 1949 provided the first example of molecular disease. With the advances in molecular biology during the past decades, the study of human globin genes and their mutations continues to lead the way in understanding molecular genetics and its clinical relevance.
Presently, there are more than 1,000 known mutations involving the human globin genes. Many of these mutations are associated with clinically significant phenotypes. They include in utero fetal death (homozygous α°-thalassemia), severe anemia requiring life-long transfusions and iron chelation (β-thalassemia major), risk of death during infancy and vaso-occlusive events resulting in multiple organ damage in adults (sickling disorders), hemolytic anemia (unstable hemoglobin variants), cyanosis (methemoglobins), and erythrocytosis (high oxygen affinity hemoglobin variants). New mutations of the globin genes are still being discovered today.
The literature on the human hemoglobin variants and globin gene mutations is vast. Some of this information has been tabulated in monographs2 and in textbooks of hematology.3Updated listings of known variants are published periodically in the journal Hemoglobin. More recently, two comprehensive syllabi were published, one on human hemoglobin variants4 and the other on the thalassemic and related mutations.5
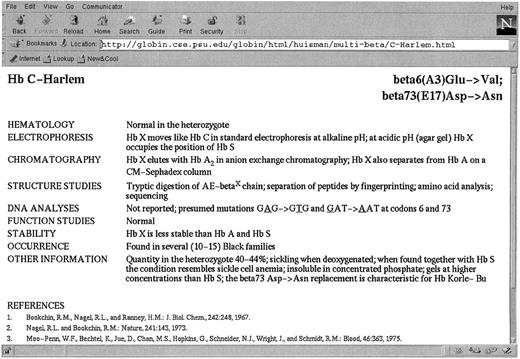
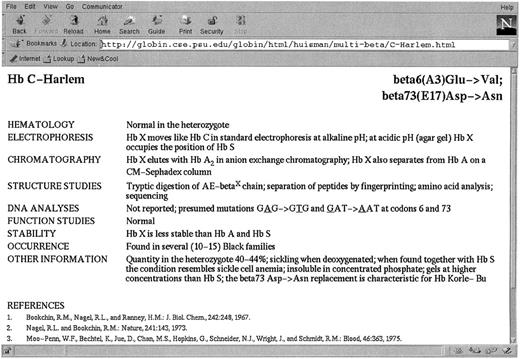
We have recently made available in the World Wide Web the electronic version of the Syllabusof Human Hemoglobin Variants, describing 693 variants. They are arranged according to those resulting from single base changes in the α-, β-, γ-, and δ-globin genes; those with more than one amino acid substitution; those with longer or shorter polypeptide chains; and those producing hybrid globin chains. This Syllabus is accessible at the Globin Gene Server (http://globin.cse.psu.edu). Users can follow the “Browse” link to the Table of Contents, which is hyperlinked to a separate table for each class of globin variants and to the full entry for each variant. Figure 1 shows the entry for Hb C-Harlem, including representative literature references.
The entry for Hb C-Harlem in the on-line version of A Syllabus of Human Hemoglobin Variants. This site is reached via the Globin Gene Server at http://globin.cse.psu.edu.
The entry for Hb C-Harlem in the on-line version of A Syllabus of Human Hemoglobin Variants. This site is reached via the Globin Gene Server at http://globin.cse.psu.edu.
An additional and useful feature of this on-line version of theSyllabus is that it allows simple queries. By using the “Search” link, users can search for matches between a word or phrase they enter and relevant information in all or selected fields of the database. For example, searching for “beta6” will return all hemoglobin entries that mention an alteration at position six of the β-globin chain from all sections of the Syllabus. Some limited flexibility in formulating the query is provided via optional checkboxes, and future developments, such as the use of a table of synonyms, will increase the efficiency of these searches.
This electronic format has advantages that complement the printed material. The World Wide Web provides easy access to users around the world, thereby increasing the utility of the compiled information and facilitating future collaborations. The database can be updated frequently to include information on new discoveries. It also allows queries that can bring out relationships and information that are difficult to find by reading the syllabus with its voluminous data. However, the converse is also true; reading and analysis by humans will bring out relationships that cannot be duplicated by electronic query engines. Thus, the printed book and electronic database provide complementary paths to locate the information needed for either clinical care or research purposes.
Work is in progress to convert the Syllabus on the thalassemic and related mutations, which describes more than 315 mutations, to an electronic form to make it publicly accessible in the near future. Attempts will be made to improve these databases to handle complex queries as required by many biomedical investigators. These globin-gene related databases will be partners to the worldwide alliance6 of “locusspecific databases” administered by the Human Genome Organization.
In the Globin Gene Server, we have applied computational information technology to provide multiple alignments of globin gene DNA sequences from different species and to establish databases on results of laboratory experimentation and manipulation of the globin genes.7 The addition of the human natural mutations and their pathophysiology databases is a logical extension of our efforts to integrate diverse information to better understand the globin gene structure, regulation, expression, and function.
In December 1997, 15 leading clinical investigators from the United States, Canada, France, Hong Kong, and the United Kingdom met in San Diego to discuss the possible establishment of a database recording individuals' globin-genotypes, hematological and clinical findings, pedigrees, and therapeutic responses. If carried out, the depository will be constructed in such a way that individual confidentiality is respected and protected securely. A panel of editors will be assembled to ensure of the accuracy of the entries. This collation of findings from around the globe will be helpful for laboratory diagnosis, genetic counseling, and patient care. It can be useful to plan for treatment strategies, provide insight into the correlation between genotypes and phenotypes in different populations, and help to identify and investigate novel clinical findings of biological importance. When successful, it can also become a useful prototype for the development of databases for other genetic and nonhereditary diseases.
Bioinformatics is becoming fully integrated into basic research in biology and medicine. The development of databases of clinical relevance should make bioinformatics increasingly important as an aid to the practice of medicine.
Supported by Public Health Services Grants No. LM05773, LM05110, and DK27635 and by The Sickle Cell Anemia Foundation (Augusta, GA).
Address reprint requests to Ross Hardison, PhD, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, 206 Althouse Laboratory, University Park, PA 16802; e-mail:rch8@psu.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.