To explore the biological and clinical implications of the structure/function relationships in factor XIII, mutations in two patients with type II deficiency were identified and characterized in a mammalian expression system. Nucleotide sequence analysis of the A subunit gene showed that case no. 1 had a deletion of 4 bp (AATT) in exon XI and that, in case no. 2, Gly562 (GGG) had been replaced by Arg(AGG). The deletion in case no. 1 leads to a premature termination at codon 464. Restriction digestion of amplified DNAs confirmed that both cases were homozygous for their respective mutations. Reverse transcription-polymerase chain reaction analysis demonstrated that the level of mRNA was greatly reduced in case no. 1, whereas the level of mutant mRNA expressed in case no. 2 was normal. Molecular modeling calculated that Arg562 changed the conformation of the A subunit, suggesting misfolding and/or destabilization of the molecule. To determine how these mutations impaired synthesis of the A subunit, recombinant A subunits bearing the mutations were expressed in mammalian cells. Pulse-chase experiments showed that the mutants were synthesized normally but disappeared rapidly, whereas the wild-type remained. These results indicate that both mutant proteins with an altered conformation become prone to rapid degradation, resulting in factor XIII deficiency in these patients.
COAGULATION FACTOR XIII is a plasma transglutaminase consisting of two catalytic A and two noncatalytic B subunits.1-3 A combination of cDNA cloning and amino acid sequence analysis established the primary structures of both subunits.4,5 The A subunit contains an active site Cys314, and the B subunit is composed of 10 tandem repeats termed Sushi domains.6 We previously characterized the genes for both the A and B subunits of human factor XIII.7,8 The gene for the A subunit spans more than 160 kb and consists of 15 exons interrupted by 14 introns. The gene for the B subunit is approximately 28 kb in length and is composed of 12 exons interrupted by 11 introns. Determination of the genomic organization and sequence for both subunits has made it possible to characterize factor XIII deficiency at the DNA level.9
Factor XIII deficiency has been classified into two categories10: type I deficiency, characterized by the lack of both the A and B subunits; and type II deficiency, characterized by the lack of the A subunit alone. To clarify the genetic bases of these diseases, two cases of type I deficiency were examined at the DNA level,11 and a patient with complete B subunit deficiency was also studied.12 The insertion of a triplet created a stop codon in the gene for the B subunit in the former cases, and a single base deletion caused the splicing defect in its mRNA in the latter case. An additional substitution of T for G in the latter case replaced Cys430 with Phe in the seventh Sushi domain and resulted in impaired intracellular transport of the B subunit.13Accordingly, it was shown that type I deficiency can be caused by defects in the gene for the B subunit.14
Mutations in the gene for the A subunit have also been identified in patients with type II factor XIII deficiency (ie, the A subunit deficiency).9,15-26 These mutations are highly heterogeneous and include a variety of nonsense mutations, small deletions, and insertions with or without out-of-frame shift/premature termination, splicing abnormalities, etc. Various missense mutations in the A subunit will also result in type II deficiency. The mechanisms of defective A subunit biosynthesis caused by the missense mutations have not been clarified, mainly because in vitro expression systems were unavailable using mammalian cells. Recently, we succeeded in synthesizing both the A and B subunits in baby hamster kidney (BHK) cells and an abnormal B subunit of type I deficiency in COS cells.13 27 However, no abnormal A subunit of type II deficiency has been characterized in a mammalian expression system to date.
It is important to closely examine amino acid substitutions and deletion of peptide regions to understand the structure/function relationships of the factor XIII molecule as well as its clinical implications in factor XIII deficiency. In this study, we identified a 4-bp deletion and a Gly562-Arg mutation in the gene for the A subunit of factor XIII in two patients (cases no. 1 and 2) with A subunit deficiency28 and predicted altered structures of the mutant A subunits by molecular modeling. In addition, we present for the first time the successful expression of novel mutant A subunits in BHK cells and provide evidence indicating that the mutants are degraded rapidly in the synthesizing cells.
MATERIALS AND METHODS
Venous blood was drawn from normal individuals and from two patients with A subunit deficiency28 and their family members after informed consent had been obtained. Both cases had essentially no enzymatic activity for factor XIII and the A subunit antigen (<5% of normal by Laurell's method). The A subunit was also absent in the patients' platelets.11 Genomic DNA and RNA samples were prepared from peripheral blood cells by standard techniques, as described previously.11 12
Southern blotting.
Ten micrograms of genomic DNA was digested with 20 U of EcoRI restriction enzyme at 37°C for 2 hours and applied to a 0.4% Agarose gel. The DNA fragments were transferred to a nylon membrane. The membrane was hybridized overnight at 65°C with a32P-labeled cDNA encoding the A subunit5 and exposed to Kodak XAR-5 film (Eastman Kodak, Rochester, NY) at −80°C.
Polymerase chain reaction (PCR).
One microgram of genomic DNA was amplified using 5.0 U of Thermus aquaticus DNA polymerase (Promega, Madison, WI) in a 100 μL reaction mixture. A total of 17 pairs of gene-specific primers were designed from the nucleotide sequence of the normal A subunit gene7 (Fig 1, top). After 30 to 35 cycles of amplification, 9 μL of each reaction mixture was applied to a 1.5% Agarose gel.
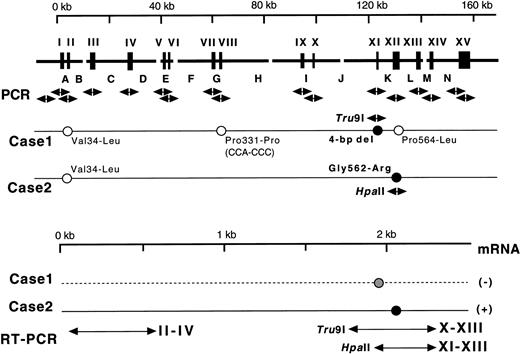
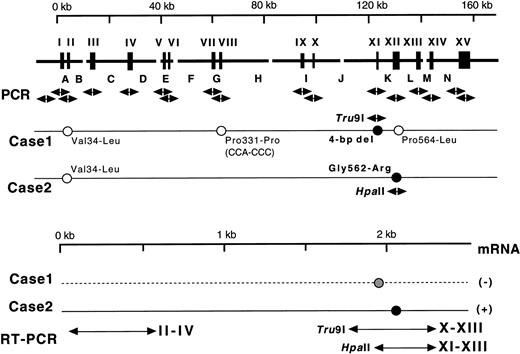
(Top) Gene for the A subunit and mutations identified. Exons are indicated by wide vertical bars and Roman numerals, and introns are indicated by capital letters. Each exon and its boundaries and the 5′-flanking regions were amplified one by one using 17 pairs of primers (arrows under exons). Solid and open circles with sequences represent homozygous causative mutations and changes known as common polymorphisms, respectively. Normal sequences are followed by those corresponding to substitutions found in the probands' DNAs. For analysis of the two mutations identified, two amplified fragments were digested with appropriate endonucleases as indicated by their names and arrows. (Bottom) mRNAs for the A subunit in the probands. Three regions of exons II-IV, X-XIII, and XI-XIII were amplified by RT-PCR. A solid line represents the expressed mRNA, whereas a broken line indicates a possible mRNA that was not detected.
(Top) Gene for the A subunit and mutations identified. Exons are indicated by wide vertical bars and Roman numerals, and introns are indicated by capital letters. Each exon and its boundaries and the 5′-flanking regions were amplified one by one using 17 pairs of primers (arrows under exons). Solid and open circles with sequences represent homozygous causative mutations and changes known as common polymorphisms, respectively. Normal sequences are followed by those corresponding to substitutions found in the probands' DNAs. For analysis of the two mutations identified, two amplified fragments were digested with appropriate endonucleases as indicated by their names and arrows. (Bottom) mRNAs for the A subunit in the probands. Three regions of exons II-IV, X-XIII, and XI-XIII were amplified by RT-PCR. A solid line represents the expressed mRNA, whereas a broken line indicates a possible mRNA that was not detected.
Nucleotide sequence analysis.
Amplified DNA samples were digested with restriction enzymes to generate proper ends for ligation into sequencing vectors. The DNA sequence of an insert was obtained using the dideoxynucleotide method29 with ABI sequence analyzer 373A (Perkin-Elmer, Norwalk, CT). To minimize the possibility of obtaining the DNA sequences with misincorporation by the DNA polymerase, 10 or more samples of each amplified region of the A subunit were examined.
Detection of mutations by restriction digestion.
For restriction enzyme analysis to detect the presence of the 4-bp deletion in exon XI and the replacement of G by A in exon XII (Fig 1, top), 15 μL of the amplified DNA samples was incubated withTru9I and Hpa II endonucleases (Boehringer Mannheim GmbH, Mannheim, Germany), respectively. After digestion for 2 hours, half of each sample was applied to a 2% Agarose gel.
Reverse transcription-PCR (RT-PCR) assay of mRNA for the A subunit.
RT of the total RNA (1.0 μg) was performed using an oligo dT (dT18) primer and Superscript II RNase H−(GIBCO-BRL, Grand Island, NY), as described previously.11 One fiftieth of the synthesized first-strand cDNA was used for PCR in a reaction mixture of 50 μL by employing two pairs of primers separately; for exons II-IV of the A subunit (Fig 1, bottom), 5′-ATGTCAGAAACTTCCAGGACC-3′ (sense) and 5′-CGTGTCTGTTTCTGGGTTTCG-3′ (antisense); for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control, 5′-CATCACCATCTTCCAGGAGC-3′ (sense) and 5′-TAAGCAGTTGGTGGTGCAGG-3′ (antisense). After 25 cycles, the reaction mixtures were electrophoresed in a 2% Agarose gel. The gel was subjected to quantitation of the amplified products by Densitograph 3.01 Imaging Analyzer (Atto Inc, Tokyo, Japan). The fluorescence intensity of the PCR product for the A subunit was normalized to that for GAPDH.
One fiftieth of the synthesized first-strand cDNA was also used for PCR in a reaction mixture of 50 μL to amplify two regions (Fig 1, bottom) by employing two pairs of primers: for exons X-XIII, 5′-CATCAAGCACGGCCATGTCTGC-3′ (sense) and 5′-CTTGATGATGATCTCAGG-3′ (antisense); for exons XI-XIII, 5′-TCCGGAACAACAGCCACAACCG-3′ (sense) and 5′-CTTGATGATGATCTCAGG-3′ (antisense). To detect extremely small amounts of cDNAs, prolonged amplification was performed using 35 to 40 cycles. Amplified fragments were subjected to digestion by eitherTru9I or Hpa II to detect expressed mutations.
Molecular modeling.
To predict the tertiary structure of the mutant molecule, molecular modeling was performed by using the coordinates of the A subunit defined by X-ray crystallography.30 To discuss the structural change caused by the Gly562-Arg mutation, we used a partial molecule from residues 190 to 628 consisting of domains II (residues 190-319), III (320-513), and IV (514-628) according to the domain structure defined by Kurochkin et al.31 Hydrogen atoms, except for those belonging to hydrocarbons in the native XIIIA (190-628), were energetically optimized, as were the mutated Arg562 residue and hydrogens in the mutant XIIIA (190-628). The initial conformation of the Arg residue was determined by using the chimera system for homologous protein modeling32 and was inserted into the model for the mutant. Energy optimization was performed using a program called APRICOT33 with a united atom force field of AMBER.34 All the coordinates, except the mutated Arg562 residue and hydrogen atoms that bound to nitrogens and oxygens, were fixed (constrained) to those experimentally determined by Yee et al30 (identifier 1ggt in the Protein Data Bank). Therefore, the omission of the C-terminal domain (domain V) does not affect the calculated results for the wild-type and the Gly562-Arg mutant. In addition, domain V exists a distance from domains II, III, and IV where the mutation is located.
Construction of expression vectors.
A Pst I cDNA fragment of 2.3 kb coding for the A subunit of human factor XIII5 was subcloned into pBluescriptII SK(+) (Stratagene, La Jolla, CA) for mutagenesis. In vitro mutagenesis was performed by recombinant PCR technique35using oligonucleotide primers: for 4-bp deletion, 5′-AGTTTCTTAATGTCACGAGCGTTCACCTGTTCAA-3′ (sense), from the middle 5′-GCCACCCACATTGGGAAATTGTGACCAAACAAATTG-3′ (sense), and 5′-CTTGATGATGATCTCAGG-3′ (antisense); for Gly562-Arg mutation, 5′-(CATCAAGCACGGCCATGTCTGC)-3′ (sense), from the middle 5′-GCCTTCAGGACCCTGGTGTAGAAGGTG-3′ (antisense) and 5′-CACCTTCTACACCAGGGTCCTGAAGGC-3′ (sense), and 5′-GGAAACAGCTATGACCATG-3′ (antisense). The mutations in the cDNAs were confirmed by dideoxy sequencing. The cDNAs for the wild-type and the mutants were released by HindIII/XbaI from the pBluescriptII vector and inserted into expression vector pcDNA3 (Invitrogen, San Diego, CA).
Cell culture and transfection.
BHK cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics (penicillin, streptomycin, and neomycin, GIBCO BRL). Approximately 3 × 106 BHK cells were transfected with 10 μg of an expression plasmid by the calcium phosphate method. Two days after transfection, cells were grown in DMEM containing 0.5 mg/mL G418 (GIBCO-BRL) as a selective agent. All cell cultures were maintained in an incubator humidified with 5% CO2 at 37°C.
Pulse-chase experiments.
For metabolic radiolabeling, 2 × 106 BHK cells in 6-well (37-mm) plates were incubated in Met-free DMEM for 1 hour at 37°C. The cells were then labeled with 50 μCi/well of [35S]Met in Met-free DMEM with 10% dialyzed FBS for 30 minutes (pulse) and further incubated in DMEM supplemented with unlabeled Met (chase) and 10% FBS. The chase was terminated at various time intervals; culture media and cell lysates were harvested and stored at −80°C.
Immunoprecipitation of the A subunit was performed by incubating the supernatant of the cell lysates and culture medium with the anti-A subunit antiserum, as described previously.13 Bound antigen was applied to electrophoresis on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel. The gel was dried and the radioactive band for the A subunit was quantified using a FLA-2000 Fluoroimage Analyzer (Fuji Photo Film, Tokyo, Japan).
RESULTS
Genomic Southern blotting analysis and in vitro amplification of the A subunit gene.
To test for the presence of large deletion(s), insertion(s), and recombination(s), genomic Southern blotting analysis was performed first. EcoRI-digested genomic DNAs of two Italian cases and normal individuals showed an identical pattern (data not shown). To search for possible smaller mutation(s) in and around the exons by PCR, 17 pairs of amplification primers were used, and these produced 17 different DNA fragments, respectively (data not shown). The sizes of these fragments were the same as predicted from the normal genomic sequence.7 These results confirmed that there was no gross change(s) in the probands' genes.
Identification of the 4-bp deletion and Gly562-Arg mutation.
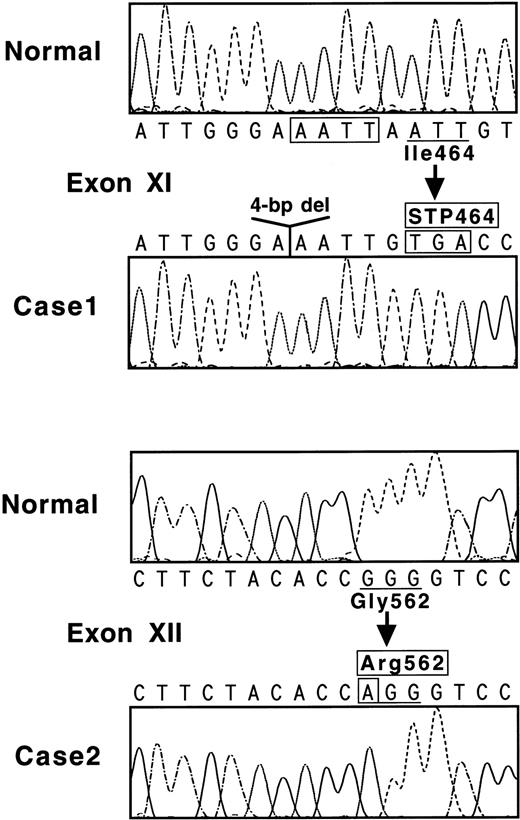
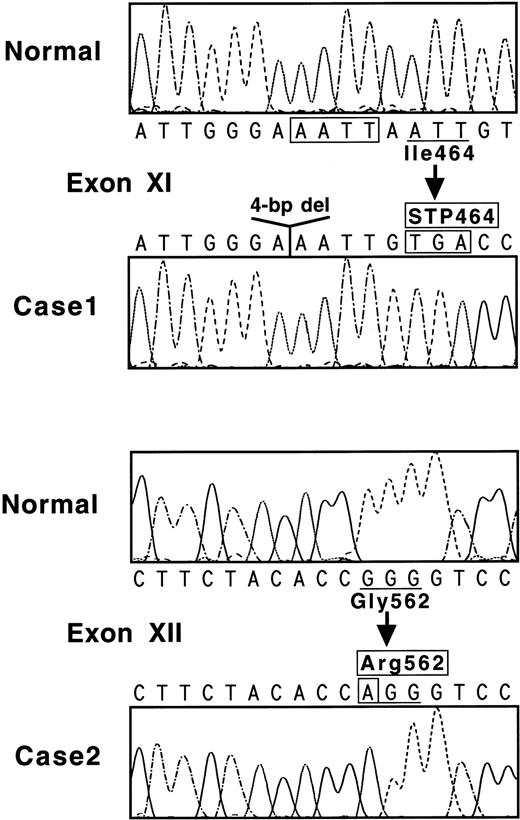
Nucleotide sequences of the 17 amplified fragments were then examined to search for differences from the normal sequence.7 When exon XI of case no. 1 was sequenced, all of 15 ssDNA samples showed AAATTGTGA (Fig 2, top), whereas the normal sequence is AAATTAATTGTGA. One obvious result of this 4-bp deletion is the creation of a new termination codon at amino acid position 464 (Fig 2, top).
(Top) Nucleotide sequence of a part of exon XI for normal and case no. 1. The 4-bp deletion is boxed. Codon 464 becomes a termination signal because of a reading frame-shift. (Bottom) Nucleotide sequence of a part of exon XII for normal and case no. 2. Mutated nucleotide A is indicated in a box, and a normal Gly562 residue is substituted with an abnormal Arg sequence.
(Top) Nucleotide sequence of a part of exon XI for normal and case no. 1. The 4-bp deletion is boxed. Codon 464 becomes a termination signal because of a reading frame-shift. (Bottom) Nucleotide sequence of a part of exon XII for normal and case no. 2. Mutated nucleotide A is indicated in a box, and a normal Gly562 residue is substituted with an abnormal Arg sequence.
Three additional changes in the nucleotide sequence were found in case no. 1 (Fig 1, top), but all of these were known genetic polymorphisms in the coding regions: Val34-Leu(GTG-TTG) in exon II, Pro331-Pro(CCA-CCC) in exon VIII, and Pro564-Leu(CCG-CTG) in exon XII.7 18 No other mutations were detected in and around the 15 exons or in a section of the 5′-flanking region. This region of about 300 bp, 1.3 kb upstream from the 5′-end of exon I, contains several TATA and CAAT boxes (A. Ichinose, unpublished data). Taken together, these results suggest that the 4-bp deletion is very likely causative for the A subunit deficiency in case no. 1.
When exon XII of case no. 2 was examined, all of 12 ssDNA samples demonstrated AGG for Arg in place of the normal sequence of GGG coding for Gly562 (Fig 2, bottom). Another nucleotide substitution, Val34-Leu (GTG-TTG), was identified in exon II; this is the same known polymorphism as found in case no. 1. No other mutation was detected in and around all exons or in a section of the 5′-flanking region. Accordingly, it was concluded that the Gly562-Arg mutation was responsible for the A subunit deficiency in case no. 2.
Detection of mutations by restriction enzyme digestion.
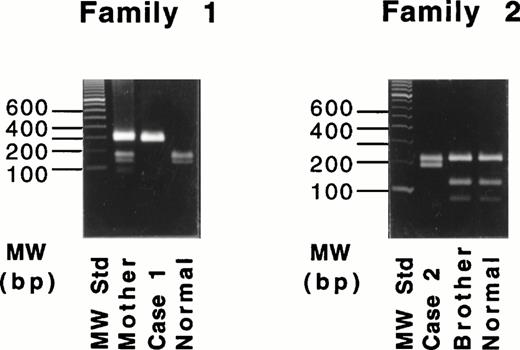
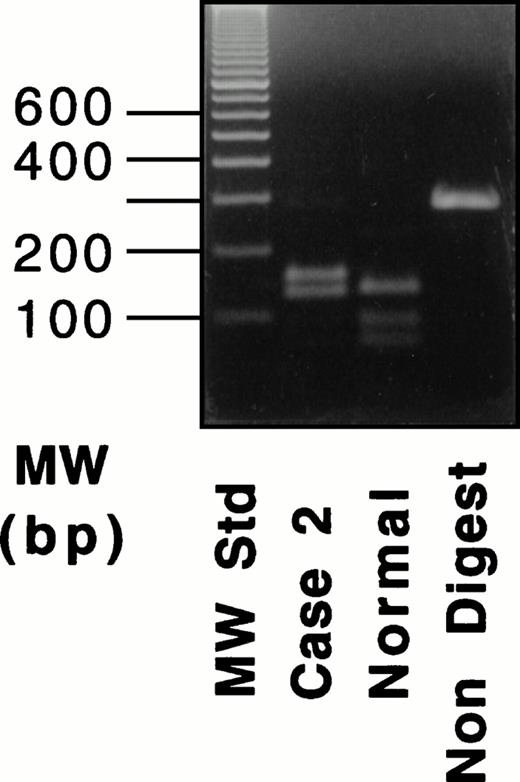
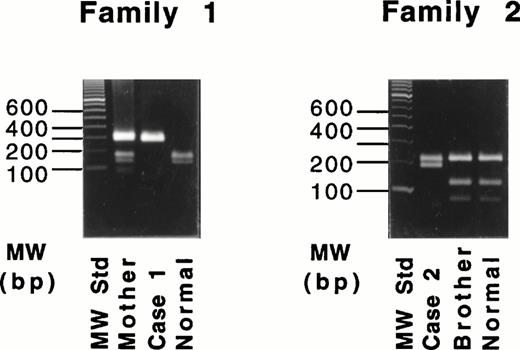
Because the deletion of 4-bp (AATT) in the DNA of case no. 1 destroys an intrinsic Tru9I site (TTAA), a fragment spanning exon XI was amplified and digested with Tru9I endonuclease. Digestion of the 291-bp fragment from a normal individual yielded two bands of 165 bp and 126 bp (Fig 3, left), whereas the 287-bp fragment of the proband remained unchanged. The proband's mother showed both cleaved and uncleaved bands. These results indicated that case no. 1 was a homozygote and that the mother was a heterozygote of this mutant allele. This same mutant allele was not found in 54 normal Italian individuals (data not shown).
Restriction digestion with Tru9I endonuclease of the amplified products of the exon XI for the 4-bp deletion (left) and with Hpa II endonuclease of the amplified products of exon XII for the Gly562-Arg mutation (right).
Restriction digestion with Tru9I endonuclease of the amplified products of the exon XI for the 4-bp deletion (left) and with Hpa II endonuclease of the amplified products of exon XII for the Gly562-Arg mutation (right).
Because the G-A substitution in case no. 2 destroyed an intrinsicHpa II site (CCGG), a fragment spanning exon XII was amplified and digested with Hpa II endonuclease. Digestion of the 389-bp fragments from a normal individual and the proband's brother (Fig 3, right) yielded three fragments of 208, 116, and 65 bp. In contrast, the digest of the 389-bp fragment from the proband yielded two fragments of 208 and 181 bp. Thus, the brother was normal, and case no. 1 was homozygous for the Gly562-Arg mutation. This mutant allele also was not found in 53 normal Italian individuals (data not shown).
Semiquantitation of mRNA for the A subunit.
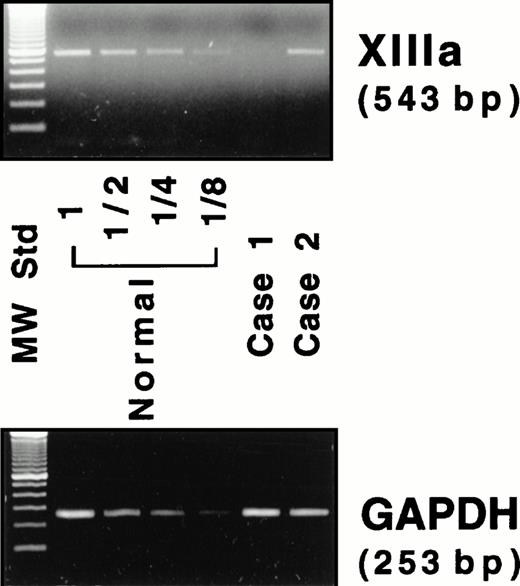
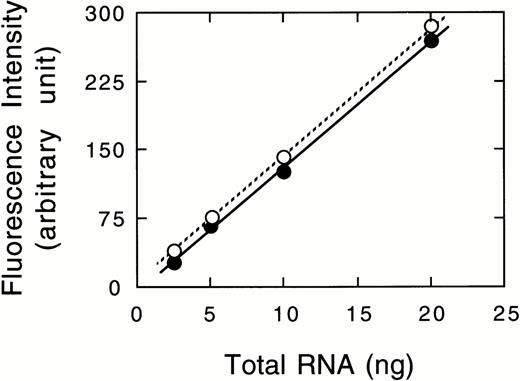
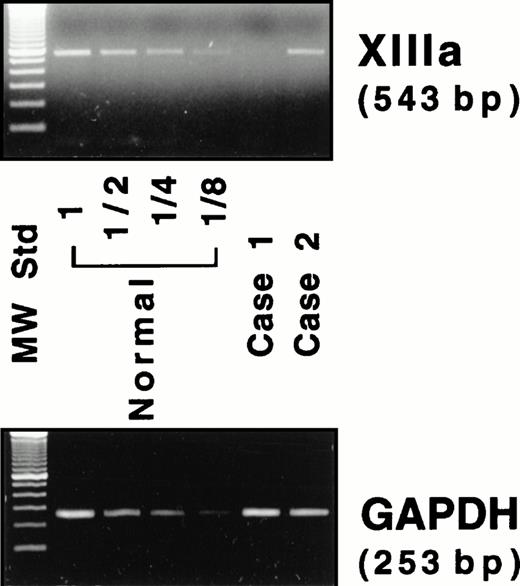
Because neither factor XIII activity in plasma28 nor the A subunit antigen in platelets11 was detected in either patient, the mRNA for the A subunit was examined. An RT-PCR assay was performed to estimate the amount of mRNA for the A subunit in peripheral blood cells, where the A subunit may be aberrantly expressed, although the site of biosynthesis for the plasma A subunit is still unknown. A 543-bp amplified fragment for the A subunit and a 253-bp fragment for GAPDH were obtained by RT-PCR from a normal individual (Fig 4). Under experimental conditions, a linear relationship was observed between the amounts of total RNA used and the fluorescence intensity of these bands (data not shown). A very faint band for the A subunit was detected in case no. 1, whereas a band was clearly observed in case no. 2 (Fig 4). The ratio of the A subunit mRNA/GAPDH mRNA was 1.0, 0.04, and 1.0 for a normal subject, case no. 1, and case no. 2, respectively. Thus, the amount of the mRNA in case no. 1 was markedly reduced, strongly suggesting that the mutant mRNA is not synthesized as stably as normal mRNA. In contrast, the amount of the mRNA of case no. 2 was comparable to that of the normal individual.
RT-PCR assay for the mRNAs of the A subunit (XIIIa) and GAPDH. Four lanes (normal) on the left of the panel contain amplified products of diluted (1 to 1/8) samples from a normal individual.
RT-PCR assay for the mRNAs of the A subunit (XIIIa) and GAPDH. Four lanes (normal) on the left of the panel contain amplified products of diluted (1 to 1/8) samples from a normal individual.
Detection of the mutant transcript for the A subunit.
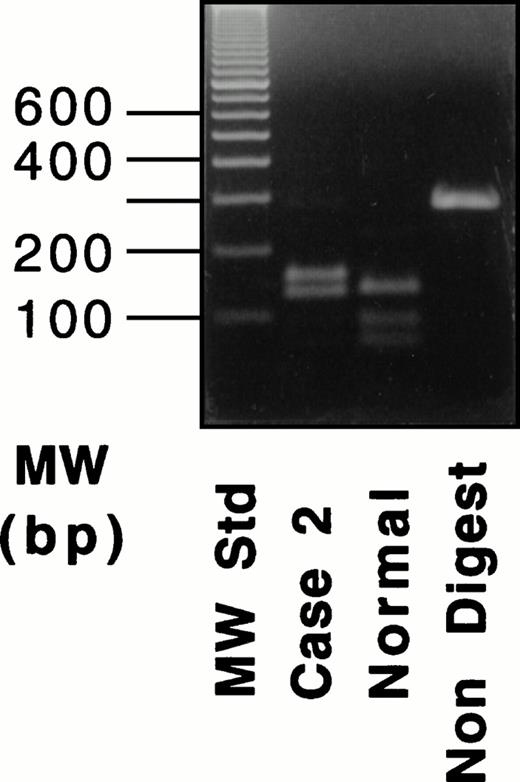
A band of normal size was obtained in the amplified sample from case no. 2 by using primers for exons XI-XIII. Digestion with Hpa II of the 290-bp fragment from case no. 2 yielded three bands of 155, 133, and 2 bp (the last fragment was unresolved), whereas a normal fragment produced four bands of 133, 90, 65, and 2 bp (Fig 5, right), indicating that the mRNA from case no. 2 contained the Gly562-Arg mutation. These results confirmed that the mRNA with the Gly562-Arg mutation was the sole transcript expressed in case no. 2.
RT-PCR and RFLP of a fragment containing exons XI-XIII for the Gly562-Arg mutation in case no. 2. An RT-PCR product was digested with Hpa II. For the results shown here, amplification was performed for 35 cycles.
RT-PCR and RFLP of a fragment containing exons XI-XIII for the Gly562-Arg mutation in case no. 2. An RT-PCR product was digested with Hpa II. For the results shown here, amplification was performed for 35 cycles.
No discrete band was obtained when the mRNA sample from case no. 1 was amplified, even by prolonged RT-PCR for 35 to 40 cycles using a pair of primers for exons X-XIII. Accordingly, PCR-restriction fragment length polymorphism (RFLP) analysis could not be performed for case no. 1.
Molecular modeling.
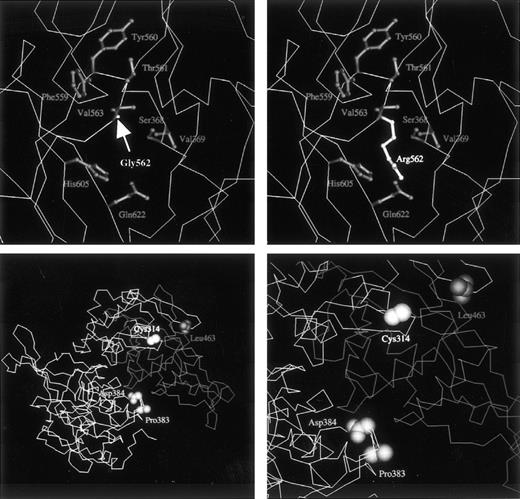
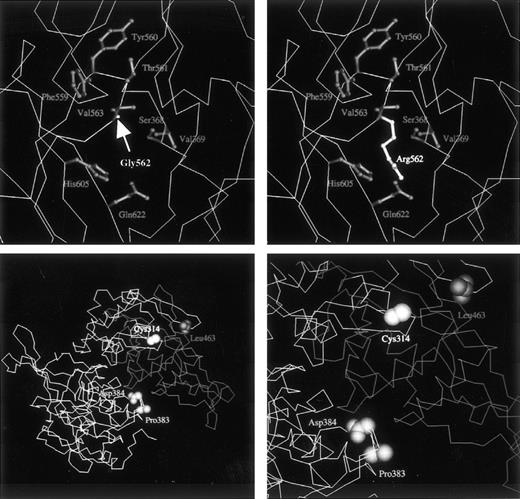
To calculate the effects of the Gly562-Arg mutation on the structure of the mutant A subunit, molecular modeling was performed. The solvent accessibility of the Arg562 side chain was 47% for domain IV alone and 6% for domains II, III, and IV, indicating that the Arg562 residue of the mutant was present on the surface of domain IV and on the interdomain surface between domains IV, II, and III of the molecule (refer to the coordinates for the mutant molecule presented athttp://prtds.pharm.kitasato-u.ac). Domain IV interacts with each of domains II and III, and Arg562 in domain IV interacts with several amino acid residues on the surface of domains II and III. Although the Gly562 residue is completely conserved among transglutaminases,13 the structure of the A subunit was partly compatible with that of the elongated side chain of the mutated Arg residue. However, there were atomic pairs that had distances shorter than van der Waals contacts between the Arg and other residues (Fig 6, top right); these included Phe559, Tyr560, Thr561, Val563, His605, and Gln622 in domain III, which were about 2.0 Å distant from the Arg residue; for domain IV, Ser368 and Val369 were at about 2.6 Å.
(Top) Close-up model view of the native (left, Gly562) or mutant (right, Arg562) A subunit. The tertiary structure of the normal A subunit is based on x-ray diffraction analysis.30 The main chain of the protein is drawn as an A carbon trace. Side chain groups of labeled residues are shown as ball-and-stick structures. Although the mutant Arg562 residue can be accommodated by the chemical structure, it generates unfavorable short contacts with neighboring residues. Accordingly, the substitution of a small residue by a large charged amino acid is expected to yield an unstable, misfolded structure. (Bottom left) Model view for the A subunit dimer including the normal domain II, the first half of domain III, the second half of domain III in a monomer, and the same domains II and III in the counterpart monomer. Domains IV and V are not shown. A total of 268 residues consisting of the second half of domain III and domains IV and V are removed from the molecule in the deletion mutant of case no. 1. The C-terminal Leu463 residue of the truncated molecule is shown. Pro383 and Asp384 residues contact with the second part of domain III, which is absent in the mutant. (Bottom right) Closer view of the environment around domains II and III. Because premature termination at position 464 would lead to the loss of a C-terminal part of the core domain and the entire domains IV and V (barrel 1 and 2), the protein is expected to misfold and/or be incapable of dimer formation.
(Top) Close-up model view of the native (left, Gly562) or mutant (right, Arg562) A subunit. The tertiary structure of the normal A subunit is based on x-ray diffraction analysis.30 The main chain of the protein is drawn as an A carbon trace. Side chain groups of labeled residues are shown as ball-and-stick structures. Although the mutant Arg562 residue can be accommodated by the chemical structure, it generates unfavorable short contacts with neighboring residues. Accordingly, the substitution of a small residue by a large charged amino acid is expected to yield an unstable, misfolded structure. (Bottom left) Model view for the A subunit dimer including the normal domain II, the first half of domain III, the second half of domain III in a monomer, and the same domains II and III in the counterpart monomer. Domains IV and V are not shown. A total of 268 residues consisting of the second half of domain III and domains IV and V are removed from the molecule in the deletion mutant of case no. 1. The C-terminal Leu463 residue of the truncated molecule is shown. Pro383 and Asp384 residues contact with the second part of domain III, which is absent in the mutant. (Bottom right) Closer view of the environment around domains II and III. Because premature termination at position 464 would lead to the loss of a C-terminal part of the core domain and the entire domains IV and V (barrel 1 and 2), the protein is expected to misfold and/or be incapable of dimer formation.
The molecular structure of the 464Stop mutant is truncated to 463 residues by the 4-bp deletion. In the native form, the first half of domain III (residues 320-463) makes contact with domain II and the second half of domain III (residues 464-513) over a wide area (Fig 6, bottom left). When the second half of domain III is absent, as in the deletion mutant of case no. 1, the surfaces of domain II and the first half of domain III become exposed extensively to solvents. Thus, it may be said that the surfaces of the mutant molecule will be susceptible to proteolytic degradation. The dimer formation of the mutant A subunit must also be impaired by the truncation of residues 464-731, because, in the native A subunit, this second part of domain III in one monomer contacts residues Pro383 and Asp384 directly in the counterpart monomer (Fig 6, bottom right).
Pulse-chase experiments in mammalian expression system.
To evaluate the intracellular stability of the newly synthesized A subunit, the wild-type and two mutants were examined by pulse-chase studies. RT-PCR analysis, using mRNA samples obtained from cells transfected with the expression vectors, showed that there were comparable amounts of the transcripts for both the wild-type and two mutants (data not shown).
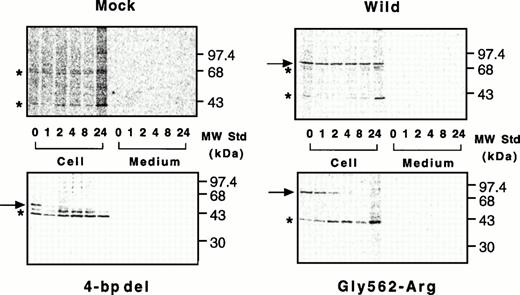
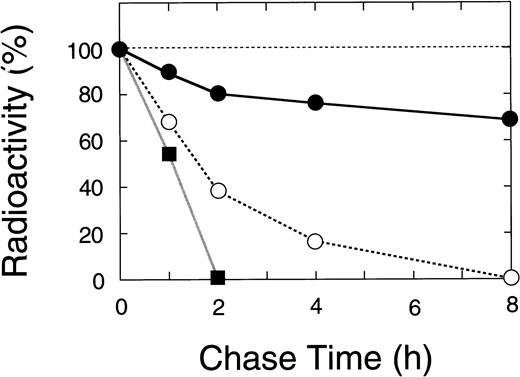
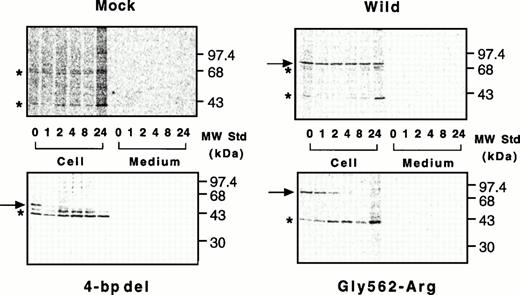
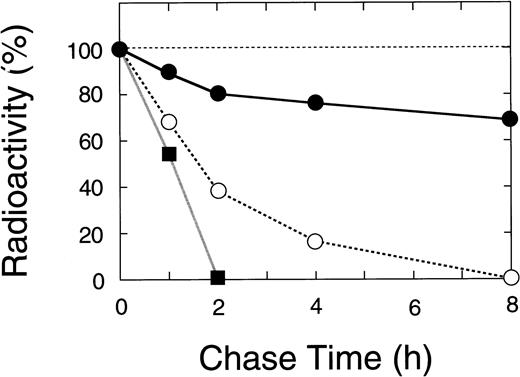
Nascent wild-type A subunit seen at 0 hours gradually decreased in the cells but did not appear in the culture medium at all (Fig 7, top right), as described previously.27 At 24 hours, the band of the wild-type A subunit was still intense (64% of 0 hours). In contrast, the mutant A subunits in the cell lysates rapidly decreased with time but, again, did not appear in the culture medium (Fig 7, bottom). The disappearance of a truncated 48-kD band in the 4-bp deletion mutant was much faster than that for the Gly562-Arg mutant (0% within 2 hours v 8 hours, Figs 7 and 8). The complete loss of both mutants in BHK cells indicated that these molecules degraded rapidly in an intracellular compartment(s).
Pulse-chase study of wild-type and mutant A subunits in BHK cells. Pulse-chase experiments were performed in a mammalian expression system as described in Materials and Methods. Radiolabeled cell lysates and culture media of cells were taken at various time intervals and immunoprecipitated with an antiserum against the A subunit, followed by electrophoresis on 10% SDS-polyacrylamide gels. Top left, mock; top right, wild-type; bottom left, the 4-bp deletion (4-bp del); bottom right, the Gly562-Arg mutant.
Pulse-chase study of wild-type and mutant A subunits in BHK cells. Pulse-chase experiments were performed in a mammalian expression system as described in Materials and Methods. Radiolabeled cell lysates and culture media of cells were taken at various time intervals and immunoprecipitated with an antiserum against the A subunit, followed by electrophoresis on 10% SDS-polyacrylamide gels. Top left, mock; top right, wild-type; bottom left, the 4-bp deletion (4-bp del); bottom right, the Gly562-Arg mutant.
Time course of the radiolabeled bands for wild-type and mutant A subunits in pulse-chase study. The radioactivity of the band for the recombinant A subunit was measured by a fluoroimage analyzer as described in Materials and Methods. (•) Wild-type, (▪) the 4-bp deletion (ie, C-terminal truncation), and (○) the Gly562-Arg mutation.
Time course of the radiolabeled bands for wild-type and mutant A subunits in pulse-chase study. The radioactivity of the band for the recombinant A subunit was measured by a fluoroimage analyzer as described in Materials and Methods. (•) Wild-type, (▪) the 4-bp deletion (ie, C-terminal truncation), and (○) the Gly562-Arg mutation.
These results strongly suggest that the mutant A subunits are unstable. This is in good agreement with the results obtained by molecular modeling as described above.
DISCUSSION
Factor XIII deficiency is caused by defects in either the A or B subunit of the gene.14 These can include a variety of missense and nonsense mutations, small deletions and insertions with or without out-of-frame shift/premature termination, splicing abnormalities, etc.11-26 Therefore, mutations causing factor XIII deficiency are highly heterogeneous. In the present study, nucleotide sequence analysis of amplified DNAs showed that cases no. 1 and 2 with severe factor XIII deficiency28 had a deletion of 4 bp in exon XI and a mutation of Gly562-Arg in exon XII of the gene for the A subunit, respectively.
The premature termination newly created by the 4-bp deletion leads to a marked reduction in the mRNA level of the A subunit in case no. 1. However, the mechanism by which a new in-frame termination codon results in a decrease in the concentration of steady-state cytoplasmic mRNA has not been understood to date. As discussed by Cooper,36 nonsense mutations would halt the pulling process of the pre-mRNA through the splicing apparatus and the nuclear pores, leaving the RNA molecule vulnerable to RNase digestion. Another model is proposed whereby detection of an in-frame termination codon by putative nuclear scanning machinery would result in a slowing down of mRNA splicing/translocation. Despite the fact that the steady-state mRNA of the mutant allele was not detectable in peripheral blood cells from case no. 1, the level of mutant mRNA was comparable to that of the wild-type in the transfected mammalian cells, strongly suggesting that this mutation impairs some steps involved in pre-mRNA processing before the completion of the mature mRNA. The marked reduction in the level of A subunit mRNA in case no. 1 would also result in a marked decrease in production of the A subunit protein.
The premature termination at codon 464, in turn, results in the deletion of the C-terminal third of the A subunit molecule (268 amino acids, residues 464-731) and in the creation of a C-terminal truncated protein. Because domains I and II (residues 1-319) are structurally independent of domains III, IV, and V,30 domains I and II of the mutant will fold like that of the native A subunit. It is generally accepted that a hydrophobic core in a domain plays essential roles in protein folding. Because residues 320-463 nearly complete the hydrophobic core in domain III, it is likely that the 464Stop mutant folds in a manner similar to the native A subunit. Because the missing C-terminal portion is essential for interdomain interactions and dimer formation of the A subunit, the truncated protein of 463 residues is predicted to be unstable. This hypothesis has been confirmed by pulse-chase experiments in a mammalian expression system in which the truncated protein of 48 kD was degraded rapidly inside BHK cells. The disappearance of the truncated mutant was even faster than that of the Gly562-Arg mutant. Accordingly, it may be said that the A subunit deficiency in case no. 1 was caused by two steps: first, by a severe reduction in its mRNA level; and, second, by the instability of its truncated molecule.
Alternatively, without residues 464-513, domain III may not be able to fold like the native domain III, in which case the result would also be the premature degradation of the entire mutant protein. However, this hypothesis cannot be substantiated at present.
Although this 4-bp deletion was previously identified at the DNA level in a compound heterozygote of French origin,23 it was not characterized further. A founder effect for this mutation is not likely, because none of the three polymorphisms found in our Italian homozygote was detected in the heterozygote mentioned above. This deletion mutation may be a recurring one, because the direct repeat of the AATT sequence7 at the deletion site in exon XI seems to be a hotspot for slippage of DNA polymerases.
The mRNA level of the Gly562-Arg mutation was not reduced at all in case no. 2. Gly562 is highly conserved throughout all 15 members of the transglutaminase family37,38; therefore, it may play an important role(s) in the enzymatic function and/or structural integration of transglutaminases. This amino acid is located on the interface of domains IV, II, and III, based on the three-dimensional structure of the molecule.30 It may be said that the Gly562-Arg mutant protein cannot fold into exactly the same structure as the wild-type, especially in the formation of the domain complex of II, III, and IV. Even if the gross structure consisting of domains II, III, and IV for the mutant were close to the native x-ray structure, molecular modelling showed a space somewhat smaller than required for the elongated side chain of Arg562. Furthermore, molecular mechanics predicted that the Gly562-Arg mutation increased the total potential energy of the A subunit molecule by +73.4 kcal/mole (Tsukamoto et al, unpublished data), suggesting that the mutation from Gly to Arg makes the molecule less stable than the native form. Therefore, the Gly562-Arg mutation is not adaptable for the A subunit molecule. This hypothesis is consistent with the results obtained by pulse-chase experiments that showed that the Gly562-Arg mutant was synthesized normally, but degraded rapidly in cells. Accordingly, the Gly562-Arg mutation leads to deficiency of the A subunit not only in plasma28 but in platelets as well.11
It may be that transport and secretion of the mutant A subunit are impaired in addition to its folding capabilities; however, these questions must remain unanswered until the mechanisms of its normal biosynthesis are better understood.
Eight additional missense mutations have been reported, including Asn60-Lys,19 Met242-Thr,18Arg252-Ile,22 Arg326-Gln,22Arg408-Gln,20 Leu498-Pro,22Gly501-Arg,24 and Leu667-Pro.19 To our knowledge, two of these mutants, Asn60-Lys and Gly501-Arg, have been expressed in yeast24 to date, but not in mammalian cells. It was concluded by Coggan et al24 that the Asn60-Lys mutant was very unstable and/or the subject of increased proteolytic degradation. However, this conclusion may not always be valid in vivo, because the quality control and proteolytic degradation mechanisms for abnormal molecules in mammalian cells may differ from those in other systems. For example, C-terminal truncated mutants (including one composed of 462 residues) were expressed inEscherichia coli at levels comparable to those of the wild-type,39 whereas our mutant of 463 residues was lost within 2 hours in mammalian cells, indicating that the effects of mutations on protein biosynthesis depend on the type of expression systems used. We have recently developed mammalian expression systems both for the A and B subunits9,13 and have confirmed that both recombinant A and B subunits were indistinguishable from the native A and B subunits of plasma factor XIII in terms of their physical and functional properties.27 Thus, in this study, we present for the first time the successful expression of novel mutant A subunits in mammalian cells and provide evidence indicating that rapid degradation of the A subunit mutants in the synthesizing cells leads to type II factor XIII deficiency.
Finally, this study has shown that molecular modeling is, at least to some extent, useful in predicting possible effects of missense mutations on the structure and stability of the A subunit, because an expression analysis of each mutant individually would require a great outlay of time and resources.
ACKNOWLEDGMENT
After completion of this manuscript, Mikkola et al40reported an expression of A subunit mutants in COS cells. The authors thank Drs V.C. Yee and D.C. Teller for providing the coordinates for the A subunit; Dr P. Bishop for the A subunit antibody; J. Harris for synthesis of the oligonucleotides; E. Espling for his excellent assistance; Drs T. Yamazaki, T. Izumi, and T. Saito for their helpful discussion; and L. Boba for her help in the preparation of the manuscript.
Supported in part by research grants from the Ministry of Education, Science and Culture, Japan (08457271), by the Ichiro Kanehara Foundation (Japan), and by the Japan Research Foundation for Clinical Pharmacology.
Address reprint requests to Akitada Ichinose, MD, PhD, Department of Molecular Pathological Biology, Yamagata University School of Medicine, Iida-Nishi 2-2-2, Yamagata, 990-9585 Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.