Using the polymerase chain reaction (PCR) technique and total DNA extracts of Hodgkin's disease (HD)-involved lymph nodes, the t(14;18)(q32;q21) translocation was detected in 37 of 115 (32.2%) cases studied. No correlation was found between the presence of this translocation and bcl-2 protein expression in Hodgkin and Reed-Sternberg (HRS) cells detected by immunohistochemistry in 58 of 96 (60.4%) cases. To identify the cells carrying the t(14;18) translocation, single-cell DNA from HRS cells isolated by micromanipulation from frozen tissue sections of lymph nodes was investigated by PCR amplification. Eleven cases showing a positive band of the same size in at least two of five PCR experiments performed on the same total DNA extract were selected for single-cell PCR. We postulated that this repeated successful amplification could be indicative of the presence of the t(14;18) translocation in the neoplastic HRS cells. Single cells from frozen tumor sections of the t(14;18)-positive OCI LY8 cell line grafted into nude mice served as a positive control. The bcl-2/JH rearrangement, involved in this translocation, could be amplified from single-cell DNA of the latter tumor, whereas, in all of the HD cases, HRS cells were found to be negative. We conclude that the t(14;18) translocation is not localized in HRS cells, but in nonmalignant B bystander lymphocytes, admixed with these neoplastic cells.
CYTOGENETIC ANALYSIS of Hodgkin's disease (HD) has yielded conflicting results on account of the few number of the neoplastic Hodgkin and Reed-Sternberg (HRS) cells that present an infrequent mitotic rate against a large background of nonneoplastic lymphocytes and reactive cells.1,2 For instance, the t(14;18)(q32;q21) translocation, usually found in follicular lymphomas, has been reported in only two cases of HD by cytogenetic analysis,3,4 whereas polymerase chain reaction (PCR) amplification has yielded controversial results. Using this technique, some groups have reported detection rates ranging from 10% to 40% of cases,4-7 whereas some others failed to detect this chromosomal abnormality in any of their cases.8-10This translocation places the bcl-2 gene of the 18q21 chromosomal region under the transcriptional control of the Ig heavy chain gene (IgH) region inducing an overexpression of the bcl-2 protein.11 There are also several reports about the expression of bcl-2 gene product by HRS cells in 20% to 60% of HD cases.12-14 The bcl-2 protein has an established role in the protection of cells from apoptosis.15 The drawbacks of the aforementioned investigations are that these studies have been performed by PCR on total DNA extracts of lymph nodes involved by HD, thus precluding information on the relation between the true nature of the cells carrying the translocation and expression of the bcl-2 gene product. In this study, we report the results obtained using the recently described single-cell PCR technique on HRS isolated cells of 11 cases of HD that were positive for the t(14;18) translocation by total DNA extracts of lymph nodes.
MATERIALS AND METHODS
Tissue Sources
Lymph nodes from 115 cases of HD were retrieved from our tissue bank. The phenotype, genotype, and Epstein-Barr virus (EBV) status of most of these cases were reported previously.16 The diagnosis of HD subtypes was made by established immunomorphologic criteria.17,18 The distribution of cases according to immunomorphologic subtypes was as follows: 5 cases of lymphocyte predominance (LPHD), 54 cases of nodular sclerosis (NSHD), and 56 cases of mixed cellularity (MCHD). Fifteen hyperplastic lymph nodes, the t(14;18)-negative HSB-2 cell line (ATTC CCL 120.1), the OCI LY8 cell line with the t(14;18) translocation involving the major breakpoint region (MBR),19 and a case of follicular lymphoma carrying the t(14;18) translocation involving the minor cluster region (mcr) were used as negative and positive controls, respectively. Standard precautions were taken to avoid cross-contamination of amplified material, which was physically separated from unamplified ones at all times.20 In each experiment, a negative control (sterile water [blank]) was systematically included.
Detection of Bcl-2 Onco-Protein Expression
In 96 cases of HD, expression of bcl-2 gene product was visualized on paraffin sections with anti–bcl-2 onco-protein monoclonal antibody (MoAb; Dako A/S, Copenhagen, Denmark) and the antigen retrieval method.21 Staining was performed by the streptavidine-biotin-peroxidase complex (ABC) method using the streptABComplex/HRP duet (mouse/rabbit) kit (no. K492; Dako).
DNA Extraction of HD Tissue Samples
Total DNA extraction.
In the first step, total cellular DNA from lymph node samples of all cases was extracted from 10 frozen sections (10-μm thick) using proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation.22 Amplification of a fragment of the housekeeping genes β-actin (606-bp fragment)23 orc-raf-1 (258-bp fragment)24 25 was used as a positive control for successful amplification of the extracted DNA. The sequences of primers used for the PCR amplifications were as follows: β1-actin, 5′-TCA TGT TTG AGA CCT TCA A-3′; β2-actin, 5′-GTC TTT GCG GAT GTC CAC G-3′; Raf 8+, 5′-GAT GCA ATT CGA AGT CAC AGC G-3′; and Raf 9−, 5′-TTT TCT CCT GGG TCC CAG ATA-3′.
Single-cell DNA extraction.
After identification of 11 cases that showed t(14;18) translocation by PCR on total DNA extracts, we used 8-μm–thick frozen sections for studies on single HRS cells. Two negative cases were also studied as negative controls. To visualize the HRS cells in the EBV+cases, tissue sections were immunostained using the APAAP technique26 with MoAb directed against the latent membrane protein (LMP-1) of EBV (anti-LMP1 antibody [CS1-4]; Dako). In EBV− HD, CD30/HSR4 MoAb (Immunotech, Marseilles, France) was used (Fig 1). Because of the expression of the CD30 antigen by both HRS cells and reactive immunoblasts, only clearly atypical CD30+ cells were selected for further experiments. The sampling technique and the single-cell PCR amplification were validated by using frozen sections of the t(14;18)-positive OCI LY8 and t(14;18)-negative HSB-2 tumors (generated by the xenograft into nude mice of OCI LY8 [Fig 2] or HSB-2 cells27). In addition, two follicular lymphomas t(14;18)/MBR-positive from our tissue bank were also studied to confirm the reproducibility of our methodology.
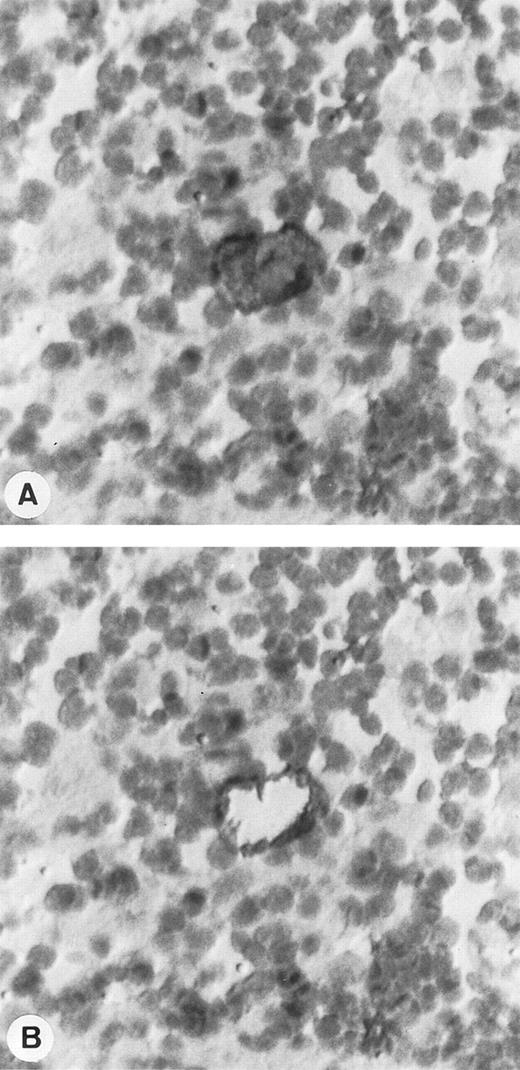
Single-cell isolation of one HRS cell from a frozen section of a lymph node involved by HD and immunostained with CD30/HSR4 MoAb. (A) Tissue section before single-cell picking: HRS cell is strongly positive with the anti-CD30 antibody, whereas the surrounding lymphocytes are negative. (B) Tissue section after the micromanipulation of the nucleus of HRS cell (APAAP technique with nuclear counterstaining; original magnification × 400).
Single-cell isolation of one HRS cell from a frozen section of a lymph node involved by HD and immunostained with CD30/HSR4 MoAb. (A) Tissue section before single-cell picking: HRS cell is strongly positive with the anti-CD30 antibody, whereas the surrounding lymphocytes are negative. (B) Tissue section after the micromanipulation of the nucleus of HRS cell (APAAP technique with nuclear counterstaining; original magnification × 400).
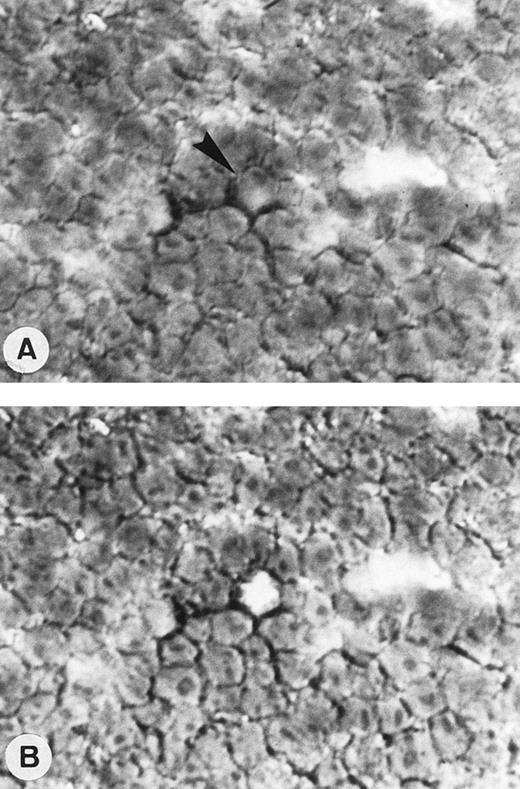
Single-cell isolation of one OCI LY8 cell from a frozen section of OCI LY8 tumor generated into nude mice and immunostained with CD20/L26 MoAb. Tissue section before (A) and after a single-cell sampling by micromanipulation (arrow; APAAP technique with nuclear counterstaining; original magnification × 400).
Single-cell isolation of one OCI LY8 cell from a frozen section of OCI LY8 tumor generated into nude mice and immunostained with CD20/L26 MoAb. Tissue section before (A) and after a single-cell sampling by micromanipulation (arrow; APAAP technique with nuclear counterstaining; original magnification × 400).
Tissue sections from these tumors were labeled with anti-CD20/L26, anti-CD3 (Dako) and anti-CD10 (Immunotech) MoAbs, respectively. For single-cell studies, stained sections were overlaid with phosphate-buffered saline (PBS). Single cells were picked up under the microscope by a closed glass capillary using an hydraulic micromanipulator and then transferred by aspiration to an open glass capillary with another micromanipulator, as described by Küppers et al28 (Fig 1). Each isolated cell was placed in a tube containing 12 μL extraction solution (50 mmol/L Tris, 10 mmol/L EDTA, 100 mmol/L NaCl, pH 8, 200 μg/mL proteinase-K). After an overnight proteinase K digestion at 37°C followed by 10 minutes of inactivation of the proteolytic enzyme at 95°C, the extracted DNA was either stored at −20°C or used immediately for PCR amplification.
Selection of HD Cases for the Single PCR Assay
For the selection of HD cases for the single-cell study, we performed for every t(14;18)/MBR-positive HD case five seminested bcl-2/JH PCR amplifications. Only the cases showing a positive band of the same size, as detected by gel electrophoresis, in at least two of five PCR runs performed on the same HD-involved DNA sample were selected for the single-cell study. We postulated that repeated successful amplification of the same band could be indicative of the presence of the t(14;18) translocation in neoplastic HRS cells. Two negative cases were also studied as negative controls.
PCR Analysis of HD Tissue Samples
Primers design.
Both the MBR and mcr of the t(14;18) chromosomal translocation were analyzed by PCR.29,30 PCR was performed either by a one-round PCR amplification followed by a Southern blotting hybridization with an internal oligonucleotide probe or by a two-round PCR amplification using a seminested PCR protocol. The amplification of the t(14;18) translocation was performed using the t(14;18) MBR and mcr specific primers. Mbr3+ oligonucleotide primer localized at MBR (5′-TTT GAC CTT TAG AGA GTT GCT TTA CG-3′) or MC8 primer (5′-GAC TCC TTT ACG TGC TGG TAC C-3′) at the mcr were used in conjunction with JH- consensus primer (5′-ACC TGA GGA GAC GGT GAC C-3′) directed to the JHregion.25,31,32 For reamplification, a second round of PCR was performed with 18q21(+)II (5′-CAC AGA CCC ACC CAG AGC CC-3′)25,31 or MC12 (5′-GAT GGC TTT GCT GAG AGG TAT-3′)32 primers, with the JH- primer for the t(14;18)/MBR and t(14;18)/mcr, respectively. For the housekeeping genec-raf-1, the first round of amplification was performed with Raf 8+ and Raf 9- primers, whereas the reamplification of the PCR amplification product was realized using the Raf 8+ primer with an internal Raf-SN (5′-CAG ATT GTT GGG GCT ACT GGA C-3′) primer.24
PCR conditions.
For each initial amplification, 1 μg of total DNA extracted from the studied lymph node in 100 μL PCR buffer volume or the total amount of the DNA extracted from the single-cell procedure in 50 μL was subjected to 34 cycles of PCR amplification as described previously using conditions adapted for each set of primers.33 The PCR cycle consisted of denaturation at 95°C, and annealing at 55°C for bcl-2/JH and c-raf-1 and at 50°C for β-actin, followed by extension at 72°C. The length of each step of amplification cycle was 1 minute, except for β-actin, for which the amplification cycle was 2 minutes for each step and the period of extension for bcl-2-mcr/JH was 2 minutes. For all PCR amplifications, a final period of 10 minutes at 72°C was performed to complete the reaction. For the reamplification step of the PCR products, 1% or 10% of the first round was used as template and subjected to 26 cycles of amplification for the total DNA and single-cell DNA extracts, respectively.
A volume of 20 μL of each amplified product was applied to a 2% agarose/NuSieve (1:1; FMC, Rockland, ME) gel electrophoresis and either visualized by staining with ethidium bromide and photographed directly after the seminested PCR procedure or proceeded for Southern blotting hybridization with an internal oligonucleotide probe after the one-round PCR amplification.
Southern Blotting Hybridization
Amplification products of the one-round PCR amplifications were transferred22 to a nylon membrane (Hybond N+; Amersham, Les Ulis, France) in 0.4 mol/L NaOH, after a 2% agarose/NuSieve (1:1) gel electrophoresis. Hybridization22 was performed with T4-polynucleotide kinase 32P-labeled oligonucleotide probes at 63°C for MBR-P+ (bcl-2-MBR internal probe, 5′-GCC TGT TTC AAC ACA GAC CCA C-3′)33,34 and Raf-P+ (c-raf-1 gene internal probe, 5′-GTC CAG TAG CCC CAA CAA TCT G-3′)24 and at 59°C for MC12 (bcl-2-mcr internal probe).32
Direct Sequencing of PCR Products
The bcl-2(MBR)/JH junctional region was sequenced in 2 cases of HD and in OCI LY8 tumor. In the first HD case, we analyzed the bcl-2/JH rearrangement obtained from total DNA extract of the lymph node to confirm the clonal relationship of the bands of the same size obtained from the five repeated PCR amplifications. In the second case of HD, sequencing was performed to investigate the different bcl-2/JH rearranged bands obtained in the 5 PCR runs. For OCI LY8 tumor, we sequenced each rearranged bcl-2/JH band obtained from the total DNA extracted from OCI LY8 tumor and from two DNA products of single-cell PCR to ascertain that we were able to amplify a single bcl-2/JH rearranged gene.
For direct sequencing, the amplified products obtained by seminested PCR amplification were separated on a 2% low melting point agarose (Appligene Oncor, Illkirch, France) gel electrophoresis and the appropriate fragments were cut from the gel and purified using the JETsorb gel extraction kit (GENOMED Inc, NC). An aliquot of the isolated DNA was sequenced with the dsDNA Cycle Sequencing System (GIBCO BRL, Cergy Pontoise, France), as recommended by the supplier. Both strands of the PCR products were sequenced with the primers used in the second round of amplification. The resulting sequences were compared with published germline sequences35-37 to determine the bcl-2/JHjunctional region sequence, which is considered as a specific marker of cell clonality.38
RESULTS
Detection of bcl-2 Expression in HRS of HD
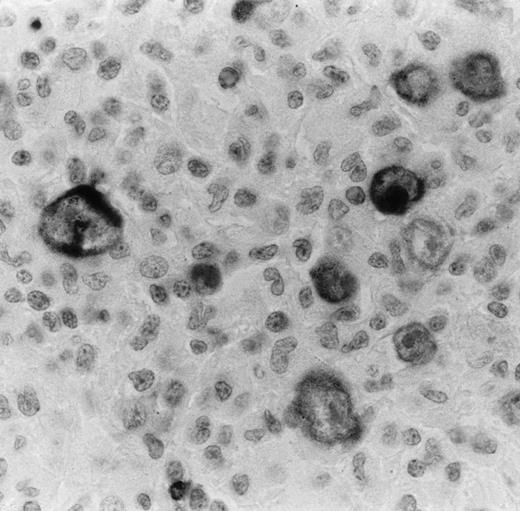
Bcl-2 oncoprotein expression in HRS cells was found in 58 of 96 cases (60.4%; 1/3 LPHD, 32/47 NSHD, and 25/46 MCHD). The number of positive cells varied from case to case. Some cases showed only some positive HRS cells, whereas in other cases almost all HRS cells were strongly labeled (Fig 3). In all cases, bcl-2–positive small lymphocytes were also detected and used as an internal control of the immunostaining (Fig 3). Because of the possible induction of bcl-2 expression by EBV infection in HD,39 40we correlated the EBV findings with bcl-2 expression by HRS cells. EBV was detected in HRS cells by in situ hybridization and LMP1 expression in 50 of the 115 cases (43.5%). It appeared that only 22 of the 43 EBV+ cases (51.1%) expressed the bcl-2 protein. Twenty-one cases containing the EBV were found to be negative for bcl-2.
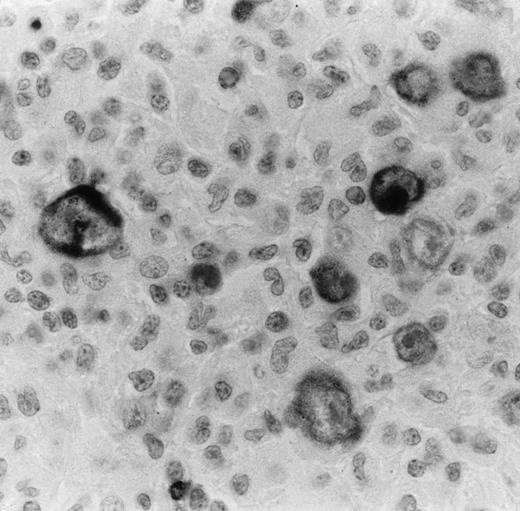
Bcl-2 onco-protein expression in HRS in a case of nodular sclerosis HD as detected by anti–bcl-2 MoAb on paraffin section. Virtually all HRS cells are immunostained with this antibody. The staining is mainly cytoplasmic and finely granular. Bcl-2–positive small lymphocytes are also seen and used as an inner control of the immunostaining (immunoperoxidase technique with nuclear counterstaining; original magnification × 400).
Bcl-2 onco-protein expression in HRS in a case of nodular sclerosis HD as detected by anti–bcl-2 MoAb on paraffin section. Virtually all HRS cells are immunostained with this antibody. The staining is mainly cytoplasmic and finely granular. Bcl-2–positive small lymphocytes are also seen and used as an inner control of the immunostaining (immunoperoxidase technique with nuclear counterstaining; original magnification × 400).
Detection of the t(14;18) Translocation in Total DNA Extracts From Lymph Node Involved by HD and From Reactive Lymph Nodes
In these experiments, PCR products were expected to range from approximately 150 to 400 bp for t(14;18)/MBR25 and 500 to 1,000 bp for t(14;18)/mcr.32 The t(14;18)/MBR translocation was detected in 29 of the 115 (25.2%) HD cases studied (12/54 NSHD and 17/56 MCHD) and in 5 of 15 (33.3%) reactive lymph nodes. The t(14;18)/mcr was demonstrated in 5 of these 115 HD cases (4.3%; 2/54 NSHD and 3/56 MCHD) and in 1 reactive lymph node case (1/15; 6.7%). Both t(14;18)/MBR and /mcr was found in 3 additional HD cases (2.6%; 2/54 NSHD and 1/56 MCHD). No correlation was found between the detection of the translocation and bcl-2 protein expression as detected by PCR and immunohistochemistry, because only 22 of 32 cases positive for the t(14;18) translocation showed the bcl-2 protein in HRS cells.
The 32 t(14;18)/MBR-positive HD cases were investigated by five PCR runs from the same DNA sample. In 11 cases, at least two of the five PCR runs showed one rearranged band of the same size (Fig 4, cases a, b, and c). DNA sequencing analysis of the bcl-2/JH junctional region was performed in 1 case and confirmed that these bands were clonally related (Table 1). As shown in Table 1, in every case bcl-2/JH junctional sequence was unique and different sequences were observed in each of the 2 cases, excluding contamination as a source of error. In addition, none of these bcl-2/JH junctional sequences conformed to t(14;18) breakpoint sequences previously characterized in our laboratory. The five repeated PCR strategy was also applied to the 5 cases of reactive lymph node samples positive for the t(14;18)/MBR. A rearranged band of the same size was detected in the five PCR runs from 1 case (Fig 5, case a). DNA sequencing study of the bcl-2/JH junctional region of the t(14;18)/MBR PCR amplification products demonstrated that only two of the five bands were clonally related (Table 1).
Multiple PCR analysis of bcl-2/JH junction region using primers specific for the major translocation breakpoint region (MBR). Total DNA extracts of lymph node from 3 cases of HD (cases a, b, and c) were amplified, separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor DNA]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in five (cases a and b) or four (case c) of the five PCR runs performed on the same DNA sample in each case. Note also that the sizes of these bands vary from one case to another.
Multiple PCR analysis of bcl-2/JH junction region using primers specific for the major translocation breakpoint region (MBR). Total DNA extracts of lymph node from 3 cases of HD (cases a, b, and c) were amplified, separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor DNA]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in five (cases a and b) or four (case c) of the five PCR runs performed on the same DNA sample in each case. Note also that the sizes of these bands vary from one case to another.
Multiple PCR analysis of the t(14;18)-MBR translocation of total DNA samples extracted from 3 reactive lymph nodes (RLN; cases a, b, and c). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor)]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in the five PCR runs performed on the same DNA sample (case a), whereas cases b and c present two bands of different size in two of the five runs performed from each sample. Note also that the sizes of the bands vary from one case to another.
Multiple PCR analysis of the t(14;18)-MBR translocation of total DNA samples extracted from 3 reactive lymph nodes (RLN; cases a, b, and c). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor)]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in the five PCR runs performed on the same DNA sample (case a), whereas cases b and c present two bands of different size in two of the five runs performed from each sample. Note also that the sizes of the bands vary from one case to another.
Single-Cell PCR Study of the t(14;18) Translocation in OCI LY8 and HSB-2 Cell Line Tumors
C-raf-1 gene DNA from single cells of the OCI LY8 and HSB-2 tumors, isolated by micromanipulation, was amplified in 22 of 30 reaction tubes using either the seminested PCR procedure (Fig 6A) or one-round PCR amplification followed by 32P-internal oligonucleotide probe hybridization. As expected, bcl-2/JH rearranged PCR amplification products were only obtained from OCI LY8 tumor sections (42+/59 cells sampled; Fig 6B), whereas HSB-2 cells were always negative (0+/52 cells; Fig 6B). Sequencing analysis of the t(14;18)/MBR rearranged bands of OCI LY8 tumor (Table 1), obtained by PCR on total DNA extracted from OCI LY8 tumor and by single-cell PCR, showed the same bcl-2/JH junctional sequence as that previously found using total DNA extract from this cell line.33 These results demonstrated the sensitivity and the reproducibility of our approach for the amplification of bcl-2/JH rearranged genes in a single t(14;18)-carrying cell. In addition, the 2 t(14;18)/MBR-positive follicular lymphomas investigated by single PCR analysis and bcl-2/JH sequencing gave identical results to those obtained from whole lymph node DNA (Fig 7). This further validated the reproducibility of our method for the t(14;18) translocation detection at the single-cell level.
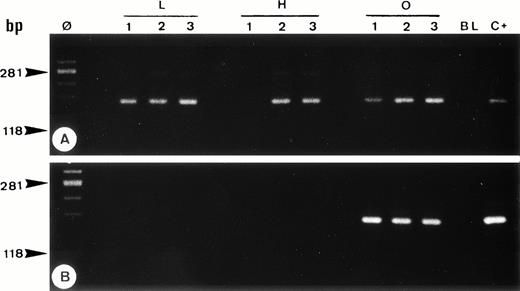
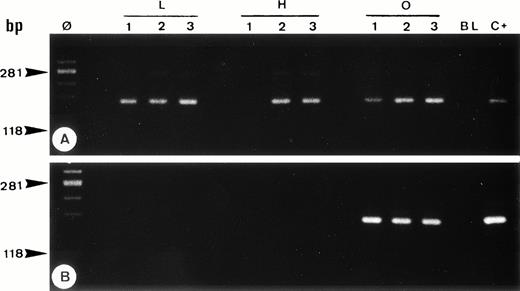
Single-cell PCR analysis of the housekeeping genec-raf-1 (A) and the bcl-2/JH rearrangement (B) by seminested PCR analysis of cells isolated from frozen sections of OCI LY8 (lanes O: 1 through 3) and HSB-2 (lanes H: 1 through 3) tumor cells and of small lymphocytes from lymph node involved by HD (lanes L: 1 through 3). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular weight markers; BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control (OCI LY8 tumor DNA). C-raf-1 gene has been successfully amplified from 8 of 9 isolated single cells (A) as demonstrated by the presence of a band of 180 bp in 8 of 9 micromanipulated single-cell DNA, which indicates the validity of our methodology for the single-cell PCR amplification. As expected, only OCI LY8 tumor cells showed rearranged bcl-2/JH bands within the expected range of size (110 to 360 bp; lanes O: 1 through 3) (B).
Single-cell PCR analysis of the housekeeping genec-raf-1 (A) and the bcl-2/JH rearrangement (B) by seminested PCR analysis of cells isolated from frozen sections of OCI LY8 (lanes O: 1 through 3) and HSB-2 (lanes H: 1 through 3) tumor cells and of small lymphocytes from lymph node involved by HD (lanes L: 1 through 3). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular weight markers; BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control (OCI LY8 tumor DNA). C-raf-1 gene has been successfully amplified from 8 of 9 isolated single cells (A) as demonstrated by the presence of a band of 180 bp in 8 of 9 micromanipulated single-cell DNA, which indicates the validity of our methodology for the single-cell PCR amplification. As expected, only OCI LY8 tumor cells showed rearranged bcl-2/JH bands within the expected range of size (110 to 360 bp; lanes O: 1 through 3) (B).
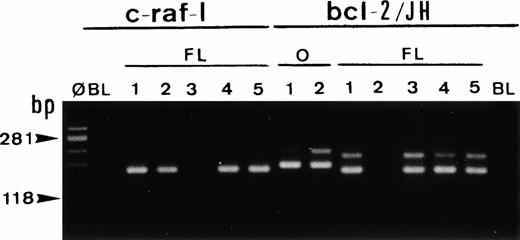
Single-cell PCR analysis of the housekeeping genec-raf-1 and the bcl-2/JH rearrangement by seminested PCR analysis of cells isolated from frozen sections of a case of t(14;18)/MBR-positive follicular lymphoma (FL). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular weight markers; BL (blank), PCR amplification without DNA template to rule out contamination; O, single-cell positive control: t(14;18)/MBR-positive OCI LY8 tumor (lanes O, bcl-2/JH: 1 and 2). Single-cell DNA has been successfully amplified using c-Raf-1 primers (lanes FL: c-raf-1: 1 through 5): 4 positive cells (lanes 1, 2, 4, and 5) and bcl-2/JH primers (lanes FL, bcl-2/JH: 1 through 5): 4 positive cells (lanes 1 and 3 through 5). As expected, the amplification of the c-Raf-1 gene shows a band of 180 bp and the amplification of the t(14;18)/MBR shows a rearranged bcl-2/JH band within the expected range of size (110 to 360 bp).
Single-cell PCR analysis of the housekeeping genec-raf-1 and the bcl-2/JH rearrangement by seminested PCR analysis of cells isolated from frozen sections of a case of t(14;18)/MBR-positive follicular lymphoma (FL). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular weight markers; BL (blank), PCR amplification without DNA template to rule out contamination; O, single-cell positive control: t(14;18)/MBR-positive OCI LY8 tumor (lanes O, bcl-2/JH: 1 and 2). Single-cell DNA has been successfully amplified using c-Raf-1 primers (lanes FL: c-raf-1: 1 through 5): 4 positive cells (lanes 1, 2, 4, and 5) and bcl-2/JH primers (lanes FL, bcl-2/JH: 1 through 5): 4 positive cells (lanes 1 and 3 through 5). As expected, the amplification of the c-Raf-1 gene shows a band of 180 bp and the amplification of the t(14;18)/MBR shows a rearranged bcl-2/JH band within the expected range of size (110 to 360 bp).
Single-Cell PCR Study of the t(14;18) Translocation in HRS of HD-Involved Lymph Nodes
After establishing PCR conditions suitable for single-cell analysis using the OCI LY8 tumor, HRS cells were analyzed at the single-cell level for the t(14;18)/MBR translocation in the 13 selected HD cases. In each case, 20 isolated HRS cells were analyzed and each experiment was repeated twice. Bcl-2/JH gene rearrangement was not detected in HRS cells in the 13 investigated cases (Fig 8), whereas c-raf-1 DNA could be amplified from HRS cells micromanipulated in parallel reactions (Fig 9). In each PCR amplification experiment, four OCI LY8 isolated cells were used as a positive control to ascertain the successful amplification of bcl-2/JH gene rearrangement at the single-cell level.
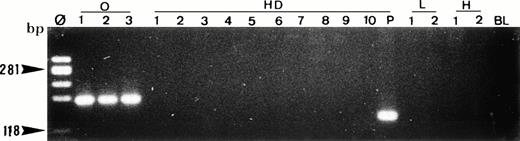
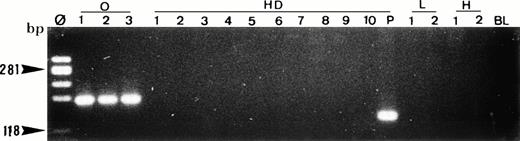
Single-cell PCR analysis of the t(14;18)-MBR translocation by seminested PCR analysis of isolated OCI LY8 tumor cells (lane O: 1 through 3), HRS cells from a lymph node involved by HD (lanes HD: 1 through 10), small lymphocytes from the same lymph node (lanes L: 1 and 2), and t(14;18)-negative HSB-2 tumor cells (lanes H: 1 and 2). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular weight markers; BL (blank), PCR amplification without DNA template to rule out contamination; P, total DNA extracted from the same lymph node involved by HD. Only OCI LY8 tumor cells show rearranged bcl-2/JH bands within the expected range of size (110 to 360 bp; lanes O: 1 through 3). All single HD cells are negative for this translocation, whereas total DNA extracted from the same patient (P) is positive.
Single-cell PCR analysis of the t(14;18)-MBR translocation by seminested PCR analysis of isolated OCI LY8 tumor cells (lane O: 1 through 3), HRS cells from a lymph node involved by HD (lanes HD: 1 through 10), small lymphocytes from the same lymph node (lanes L: 1 and 2), and t(14;18)-negative HSB-2 tumor cells (lanes H: 1 and 2). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular weight markers; BL (blank), PCR amplification without DNA template to rule out contamination; P, total DNA extracted from the same lymph node involved by HD. Only OCI LY8 tumor cells show rearranged bcl-2/JH bands within the expected range of size (110 to 360 bp; lanes O: 1 through 3). All single HD cells are negative for this translocation, whereas total DNA extracted from the same patient (P) is positive.
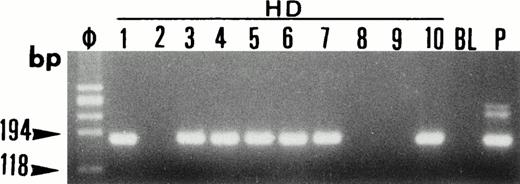
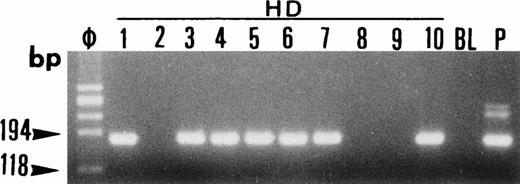
Single-cell PCR analysis of the housekeeping genec-raf-1 by seminested PCR analysis of HRS cells from lymph node involved by HD (lanes HD: 1 through 10). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular size markers; BL (blank), PCR amplification without DNA template to rule out contamination; P, total DNA extracted from the same lymph node involved by HD. C-raf-1 has been successfully amplified from 7 of 10 isolated single cells as demonstrated by the presence of a band of 180 bp in 7 of 10 micromanipulated single-cell DNA. This indicates the validity of our methodology for the single-cell isolation and the PCR amplification of the extracted DNA from the majority of HRS cells isolated from tissue sections of lymph node involved by HD.
Single-cell PCR analysis of the housekeeping genec-raf-1 by seminested PCR analysis of HRS cells from lymph node involved by HD (lanes HD: 1 through 10). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis and visualized by staining with ethidium bromide. Ø, molecular size markers; BL (blank), PCR amplification without DNA template to rule out contamination; P, total DNA extracted from the same lymph node involved by HD. C-raf-1 has been successfully amplified from 7 of 10 isolated single cells as demonstrated by the presence of a band of 180 bp in 7 of 10 micromanipulated single-cell DNA. This indicates the validity of our methodology for the single-cell isolation and the PCR amplification of the extracted DNA from the majority of HRS cells isolated from tissue sections of lymph node involved by HD.
In every lymph node studied, four B and four T lymphocytes isolated from the same tissue section were analyzed together with HRS cells using the same PCR conditions and were also found to be negative for the t(14;18) translocation (Fig 6B) despite the successful amplification of the c-raf-1 gene DNA at the single-cell level (Fig 6A).
DISCUSSION
Recurrent chromosomal translocations play an important role in many human lymphoid tumors and are often responsible for the deregulation of genes involved in the control of normal cell survival and proliferation.41 The best example is provided by follicular lymphoma in which the t(14;18)(q32;q21) translocation is associated with the overexpression of the bcl-2 oncoprotein.15,42Little information is available about the molecular events that lead to the development of HRS cells and studies dealing with the status of oncogenes have yielded conflicting results in HD.43-46 The demonstration by some investigators of the bcl-2/JH gene rearrangement in 10% to 40% of cases using total DNA extracts could suggest the implication of the t(14;18) translocation in HD.4-7 However, the presence of rare t(14;18)-bearing nonmalignant B cells has been demonstrated in hyperplastic lymph nodes of healthy individuals.47,48 As reported previously,5,10 our finding of bcl-2/JHrearrangement in one third of reactive lymph nodes led to the postulate that the t(14;18) translocation attributed to HRS cells could originate from reactive B lymphocytes in tissues involved by HD. Single-cell PCR study was thus needed to verify the postulate. This method has been effectively used recently to demonstrate IgH gene rearrangements in HRS cells,28,49-51 and the results suggest that HRS cells might originate from germinal center B cells. The later studies have demonstrated that rearranged VH genes from HRS cells carried a high load of somatic mutations with stop codons that prevent antigenic selection. However, the HRS precursors probably escape apoptosis through some transforming events, resulting in premature upregulation of genes known to protect germinal center cells from apoptosis, particularly bcl-2.51,52 This hypothesis is in agreement with the expression of bcl-2 protein by HRS found in the majority of our cases. However, in contrast to follicular lymphoma, the expression of bcl-2 is not associated with the t(14;18) translocation. We and others have previously shown that the current PCR methodology to detect the bcl-2/JH rearranged genes is highly sensitive and capable of detecting one copy of a rearranged gene.33,53 Using single-cell PCR on the t(14;18)-positive OCI LY8 cell line, we showed that it was also possible to amplify and sequence bcl-2/JH rearrangement. Thus, we selected for single-cell PCR analysis only cases of HD that yielded the same bcl-2/JH rearrangement in at least two of five PCR amplifications performed on the same DNA sample. We postulated that the detection of a rearranged band of the same size and sequence, in more than one PCR run performed on the same sample, was indicative of the presence of this rearrangement in neoplastic HRS cells.54All selected cases of HD that were found to be positive for t(14;18) translocation by total DNA extracts proved to be negative at the single-cell level, whereas the housekeeping gene c-raf-1 could be amplified from the extracted DNA in parallel reactions. These results suggest that the t(14;18) translocation is not present in HRS cells of HD. These findings are in agreement with reported cytogenetic data indicating that detection of the t(14;18) in HD is either absent or exceedingly rare.4 Also, even if the bcl-2 protein can be demonstrated in HRS cells of 20% to 60% of cases12-14(present study), the bcl-2 expression is not due to the t(14;18) translocation. This finding is in agreement with previous studies pointing out that the t(14;18) translocation is one of many possible mechanisms that lead to overexpression of bcl-2 protein55,56 and that the translocation is not always necessary for the production of this protein.57 Moreover, the results of the present study and other studies14,58,59indicate a lack of correlation between bcl-2 expression and EBV status. Thus, in contrast to data obtained from in vitro cell culture studies on lymphoid cells,39,40 LMP-1 does not upregulate bcl-2 expression in EBV+ HRS cells. However, a posttranscriptional regulation of the bcl-2 expression in HRS cells, comparable to that previously demonstrated in germinal center cells, cannot be excluded.59 60
It is noteworthy that the finding of clonally related t(14;18)-carrying cells in reactive lymphoid cell populations in HD-involved tissue and hyperplastic lymph node samples could be explained by an oligoclonal expansion, after different antigenic stimulation, of the long-lived t(14;18)-bearing B-cell clones.47,48 Indeed, due to the relative stability of the t(14;18) junction region sequence,33,61,62 these clonal related t(14;18)-positive cell populations are likely generated by the outgrowth of clones of the lymph node germinal founder B cells.52 63
Finally, the result of this study showed clearly that t(14;18) translocation does not play a role in the pathogenesis of HD. Further studies are necessary to determine if and to what extent other oncogenes are involved in the pathogenesis of this enigmatic disease. Single-cell PCR with direct sequencing of amplification products will be a powerful tool to examine these questions.
ACKNOWLEDGMENT
The authors thank Dr N.L. Berinstein (Regional Cancer Center, Toronto, Ontario, Canada) for kindly providing the OCI LY8 lymphoma cell line. We are grateful to the staff of Pathological Anatomy Laboratory and CIGH/CNRS, in particular Marie Andrée Dussion, Daniel Roda, Reine Claude Zenou, and Michel March, for their excellent technical assistance.
Supported by grants from Association pour la Recherche Contre le Cancer (ARC) and Délégation à la Recherche Clinique, CHU-Purpan.
Address reprint requests to Talal Al Saati, MD, PhD, Laboratoire d'Anatomie Pathologique, CHU Purpan, Place du Dr. Baylac, 31059 Toulouse Cedex, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 4. Multiple PCR analysis of bcl-2/JH junction region using primers specific for the major translocation breakpoint region (MBR). Total DNA extracts of lymph node from 3 cases of HD (cases a, b, and c) were amplified, separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor DNA]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in five (cases a and b) or four (case c) of the five PCR runs performed on the same DNA sample in each case. Note also that the sizes of these bands vary from one case to another.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/8/10.1182_blood.v91.8.2866.2866_2866_2874/5/m_blod4083404.jpeg?Expires=1768126070&Signature=VJVzvJ17K9g8b7zvfHSjU4NDR92nd41VI1gB~L~jfdkHRA91bls~zKr~z54KhMVNVPjTpo-53FVBeXpV4QCJ1kuXXVCwGreL5-3Y6Z6~q2dWUibzulk9pUNuX0iM6SuG1xHf62rRLJ-IQRE8nDZawYH~1R8rRMHU0uRizoKeUdvzxZbpLPX0KI7-6FmiFNxdKn0UEOa7~zcB4Nvmzd2BomxBt6LqENHWwGz4RQFGkW07eej6NW8twWVB1u07QPOdZfvaRYYiG7xmo5RiM12F76y9RzGhVrvG229I6w~fiakUtL02LEZeTlaEJcVRAiwCO7tOWQxHnKzmnK6vl8SKSg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Multiple PCR analysis of the t(14;18)-MBR translocation of total DNA samples extracted from 3 reactive lymph nodes (RLN; cases a, b, and c). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor)]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in the five PCR runs performed on the same DNA sample (case a), whereas cases b and c present two bands of different size in two of the five runs performed from each sample. Note also that the sizes of the bands vary from one case to another.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/8/10.1182_blood.v91.8.2866.2866_2866_2874/5/m_blod4083405.jpeg?Expires=1768126070&Signature=rv48qShdLz05PpLddfZH6MR8tk42yuppYX3WOw13IJUEAxRb8ps19La4i-jvs2PXKzNO00Q46hLsckOJrVEAdXk7VgW0Ysla89dnVNIs50PypPoRj9pwcbBcXoTP4-Wmmolw5jsfn7Ooiqrz08~C2ISQtgj9fZOUV7sj92lZIExZqdJB993qM4ej5iBFdjbuPOvjDTRBC~wXzzGLokJm9nTy7OUq0wVzptj1zqzpWcPtC0tBICnIvJn9siIMfXqS9TrWjuN21IYNBqbtEKdkaN4~WbQSIpTG5F5op1kYdqF0pktVHPjqLrbj6xQHYQKBxtH1g4Ad5XuLj3PXUPhdig__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Fig. 4. Multiple PCR analysis of bcl-2/JH junction region using primers specific for the major translocation breakpoint region (MBR). Total DNA extracts of lymph node from 3 cases of HD (cases a, b, and c) were amplified, separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor DNA]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in five (cases a and b) or four (case c) of the five PCR runs performed on the same DNA sample in each case. Note also that the sizes of these bands vary from one case to another.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/8/10.1182_blood.v91.8.2866.2866_2866_2874/5/m_blod4083404.jpeg?Expires=1768794897&Signature=XuMrqSu-uZqR~rAgvLcHwPhDDK8poGByMMAZ4ux19uL43I7C5ejYz7P-ZGcS~7YaCkJFj4j6mPKFLgzqPMwHiKC8fG-jZsaLSMjRUFOCnecAmn6-RT~2kH8wo0SzgIvvut8HlYZOYAcwIHoT-ovYN1cNPPu8t4MlJHJhqOA0Svttl3hIKFqd97mbuJM5co0g2uJA3sd7NtuOb0uJogVZbDZNkWct0sn5WToUMF2wJsUsXVf8FUfTzWo0toU7Gp4bMSII52ORlChBF7SDGHRwA3KA6JwSx0zxLZ9yhYIGPakTqj3VXZACdfz9ExnbywQY6uRjm3-TuwAVhJij-ELFEA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Multiple PCR analysis of the t(14;18)-MBR translocation of total DNA samples extracted from 3 reactive lymph nodes (RLN; cases a, b, and c). The amplified DNA was separated on 2% agarose/NuSieve gel electrophoresis, transferred to nylon membrane, and hybridized with32P-endlabeled MBR-P+ oligonucleotide probe specific for the bcl-2/MBR region. BL (blank), PCR amplification without DNA template to rule out contamination; C+, positive control [t(14;18)-positive tumor)]. Analysis of the different samples shows rearranged bcl-2/JH bands within the expected range of size (150 to 400 bp). Note the presence of rearranged bands of the same size in the five PCR runs performed on the same DNA sample (case a), whereas cases b and c present two bands of different size in two of the five runs performed from each sample. Note also that the sizes of the bands vary from one case to another.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/8/10.1182_blood.v91.8.2866.2866_2866_2874/5/m_blod4083405.jpeg?Expires=1768794897&Signature=MrKTuRjjxncjYxZBPEuLP5E5TllZgEsMgunrT6rqykSBnfo5tNREbzC1sWx0X1iX6s-t8LNx9cw3evprWfs3tJy3s~iH9yqRKaXhX6SlGwsXRhYCe3jsUctqcHNW9o7eqraHKlE4KItCgW0lXc2~fnviSzjaXPt736LsXCEBSvEe8zjiPtyQ2TX9ZgbDU3wj03MB1ZJmC85cWshGHvln3zCdGEvfsi1qgBdSLwjKhD498VVyVbsEOf3dLGmsCQFLlacJo4JsJ8P9oEtKj0sctnXkrQIHkl5eWSGCsjR4p4oxamok70iqtqQn0FAgItPTErLfV9RDMLyBAPYoJrEycw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)