Abstract
Chromosomal abnormalities of band 8p11 are associated with a distinct subtype of acute myeloid leukemia with French-American-British M4/5 morphology and prominent erythrophagocytosis by the blast cells. This subtype is usually associated with the t(8;16)(p11;p13), a translocation that has recently been shown to result in a fusion between the MOZ and CBP genes. We have cloned the inv(8)(p11q13), an abnormality associated with the same leukemia phenotype, and found a novel fusion between MOZ and the nuclear receptor transcriptional coactivatorTIF2/GRIP-1/NCoA-2. This gene has not previously been implicated in the pathogenesis of leukemia or other malignancies. MOZ-TIF2 retains the histone acetyltransferase homology domains of both proteins and also the CBP binding domain of TIF2. We speculate that the apparently identical leukemia cell phenotype observed in cases with the t(8;16) and the inv(8) arises by recruitment of CBP by MOZ-TIF2, resulting in modulation of the transcriptional activity of target genes by a mechanism involving abnormal histone acetylation.
THE CHARACTERIZATION of recurrent chromosomal abnormalities in leukemia has provided a basis for disease classification and has helped to define prognosis in individual patients. Furthermore, the molecular cloning of the genes affected by these chromosomal changes has considerably advanced our understanding of the mechanisms of leukemogenesis and has enabled the design of molecular approaches to diagnosis and monitoring patient response to treatment.
Translocations involving chromosome band 8p11 are associated with a distinct subtype of acute myeloid leukemia (AML), with blast cells of a monocytoid phenotype that have pronounced erythrophagocytic activity. This subtype is found in approximately 2% of cases of AML with French-American-British M4 or M5 phenotype and typically found in association with the t(8;16)(p11;p13),1 but it is also seen in cases with the t(8;22)(p11;q13), t(8;19)(p11;q13), and inv(8)(p11q13).2-6 Recently the t(8;16) has been cloned and shown to fuse the MOZ gene at 8p11 to the CBP gene at 16p137; the molecular basis of the three other cytogenetic variants is unknown. Translocations of 8p11 are also observed in the 8p11 myeloproliferative syndrome, a rare disorder associated with the t(8;13)(p11;q12), t(8;9)(p11;q32), and t(6;8)(q27;p11).8However, the breakpoints in the t(8;13) are distinct from those associated with de novo AML and do not involve the MOZgene.5
Both MOZ and CBP have been implicated in histone acetylation,7,9,10 suggesting that the mechanism of leukemogenesis in patients with the t(8;16) involves specific alterations in gene expression by aberrant chromatin remodelling.CBP is also disrupted in the t(11;16), a translocation associated with therapy-related leukemia, by fusion to the MLLgene.11,12 In this paper we show that the inv(8)(p11q13) results in a fusion of MOZ to the nuclear receptor coactivatorTIF2/GRIP-1/NCoA-2 (transcriptional intermediary factor 2, hereafter referred to as TIF2),13-15 and propose a model to explain the similar leukemia phenotypes observed in patients with the t(8;16) and inv(8).
MATERIALS AND METHODS
Patient material/details.
Details of the patient's clinical course and leukemia morphology have been described elsewhere.5 6 Briefly, a bone marrow aspirate at presentation was hypercellular, with 98% blasts and prominent erythrophagocytosis; a diagnosis of AML-M5 was made. Cytogenetic analysis revealed an inv(8)(p11q13) in 70% of bone marrow metaphases.
Genomic cloning.
A genomic library was constructed to clone the 7-kb germlineMOZ and 5.1-kb BglII rearranged fragments detected by the MOZ 0.9 probe. Patient bone marrow genomic DNA (0.5 μg) was digested with BglII, ligated to λZAP Express arms and packaged with Gigapack III Gold Packaging Extract according to the manufacturer's instructions (Stratagene, Cambridge, UK). A total of 3.3 × 106plaque-forming units were screened with MOZ 0.9 and five positive clones were recovered. Germline and rearranged bands were distinguished by restriction mapping and one of each was sequenced (model ABI 373A; Applied Biosystems, Foster City, CA).
Fluorescence in situ hybridization (FISH).
A P1 artificial chromosome (PAC) clone (192D10) was isolated by screening the gridded human library RPCI1 (obtained from the Medical Research Council Human Genome Mapping Project (MRC HGMP) Resource Centre, Hinxton, UK) with the probe UN-1. FISH was performed on metaphases from phytohemagglutinin-stimulated peripheral blood lymphocytes from a normal individual. Digoxigenin-labeled probes were prepared by nick translation and detected with sheep antidigoxigenin (Boehringer Mannheim, Lewes, UK), rabbit anti-sheep fluorescein isothiocyanate (FITC; Vector, Bretton, Peterborough, UK), and finally swine anti-rabbit FITC (Dako, High Wycombe, UK). In addition, a biotinylated chromosome 8 centromere dye was included (Oncor, Durham, UK), and detected using Texas Red-Avidin (Vector). Chromosomes were counterstained with DAPI/antifade (Biovation, Aberdeen, UK). FISH-labeled metaphases were examined using an Olympus Vanox microscope, equipped with a fluorescence unit, a charge-coupled device camera, and images captured on SmartCapture Software (Vysis, Richmond, UK).
Rapid amplification of cDNA ends (RACE)–polymerase chain reaction (PCR).
One microgram of total patient bone marrow RNA was reverse-transcribed with primer RCT-A (5′-ATTGGTAGCTCTTGATCNNNNNN-3′) using standard conditions.16 A portion of cDNA was amplified with MOZ-RCO (5′-CCCTAGAGAATACTTCCGTC-3′) and a primer made up of the unique sequence from RCT-A (RC-B; 5′-ATTGGTAGCTCTTGATC-3′). Hemi-nested amplification was performed with an inner MOZ primer (MOZ-RCI: 5′-GGATGTACTCAGGTGTCAGT-3′) and RC-B. PCR products were shotgun cloned into pCR2.1 (Invitrogen, Leeu, The Netherlands), screened with a MOZ probe, and sequenced.
Yeast artificial chromosome (YAC) clones.
A gridded CEPH mega YAC library (obtained from the MRC HGMP Resource Centre) was screened with probes for the 5′ and 3′ ends of the published TIF2 cDNA sequence.13 Probes were amplified from human genomic DNA using the primers TIF2-A (5′-GGCACAGTTGCTGATATGTG-3′) and TIF2-B (5′-GTCCAAGTTGGTCAGGACAT-3′) for the 5′ end of TIF2, and TIF2-C (5′-GTGGCCTGCTTAGTAACATG-3′) and TIF2-D (5′-GGCTTGATACCAATCGAGCT-3′) for the 3′ end. The positions of positive clones were compared with existing chromosome 8 contig maps.17,18 Confirmation that these clones contained the TIF2 gene was obtained by PCR using the same primers combinations.
RESULTS
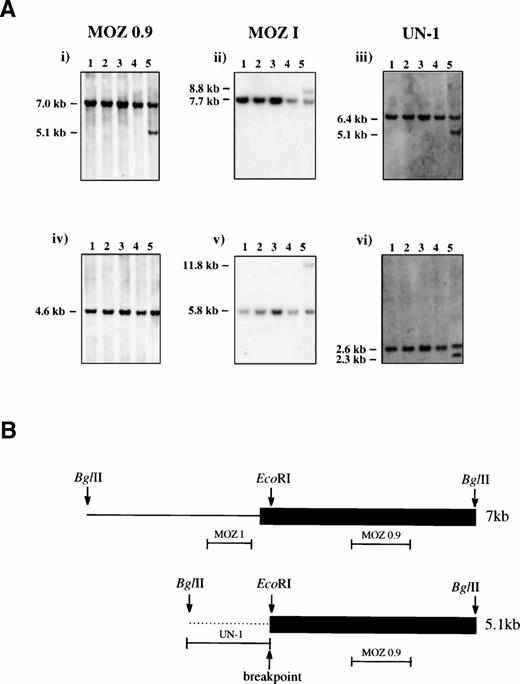
Southern blotting of patient bone marrow DNA with a MOZcDNA probe, MOZ 0.9, that spanned the published t(8;16) breakpoints revealed normal- and abnormal-sized bands after digestion withBglII5 (Fig 1A). A bacteriophage λ library was constructed from BglII-digested patient DNA and screened with MOZ 0.9. Two types of clone were recovered, corresponding to the normal 7-kb and rearranged 5.1-kb bands, respectively.
(A) Southern Blot analysis. Genomic DNA from four controls (lanes 1-4) and the inv(8) patient (lane 5) were digested withBglII (i-iii) or EcoRI (iv-vi) and probed with MOZ 0.9, MOZ I, and UN-1 as indicated. Germline bands were present in all samples and an additional band was seen for the patient for all enzyme/probe combinations except EcoRI/MOZ 0.9. (B) Schematic representation of the two sequenced bacteriophage λ clones. The 7-kb and 5.1-kb BglII fragments were derived from the germline and rearranged bands, respectively. The last MOZ exon is shown as a filled box and intron sequence as a line. The positions of the probes MOZ 0.9, MOZ I, and UN-1 are indicated. The breakpoint in the 5.1-kb clone coincides with the EcoRI site within the last exon of MOZ, 47 bp downstream of the MOZ intron-exon junction.
(A) Southern Blot analysis. Genomic DNA from four controls (lanes 1-4) and the inv(8) patient (lane 5) were digested withBglII (i-iii) or EcoRI (iv-vi) and probed with MOZ 0.9, MOZ I, and UN-1 as indicated. Germline bands were present in all samples and an additional band was seen for the patient for all enzyme/probe combinations except EcoRI/MOZ 0.9. (B) Schematic representation of the two sequenced bacteriophage λ clones. The 7-kb and 5.1-kb BglII fragments were derived from the germline and rearranged bands, respectively. The last MOZ exon is shown as a filled box and intron sequence as a line. The positions of the probes MOZ 0.9, MOZ I, and UN-1 are indicated. The breakpoint in the 5.1-kb clone coincides with the EcoRI site within the last exon of MOZ, 47 bp downstream of the MOZ intron-exon junction.
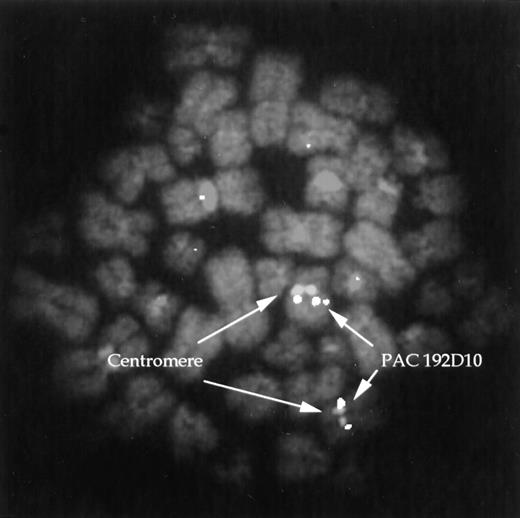
Sequence analysis of a 7-kb clone revealed a single large MOZexon of 4 kb from position 3746 of the cDNA (Genbank: U47742) to theBglII site at position 7742. Sequence of a 5.1-kb clone showed a break near the beginning of this exon within the coding sequence (position 3793), with 1.1 kb of unique sequence fused toMOZ (Fig 1B). The single published MOZ breakpoint in a patient with a t(8;16) also interrupted the same exon, but there was no sequence homology between that breakpoint and the one found here. A probe derived from the 1.1 kb of unique sequence (UN-1) was used to isolate a PAC clone which, as expected, mapped to 8q13 as determined by FISH (Fig 2).
FISH. PAC clone 192D10 was hybridized to normal lymphocyte metaphases along with a chromosome 8 centromere probe. Hybridization of the PAC is seen at 8q13.
FISH. PAC clone 192D10 was hybridized to normal lymphocyte metaphases along with a chromosome 8 centromere probe. Hybridization of the PAC is seen at 8q13.
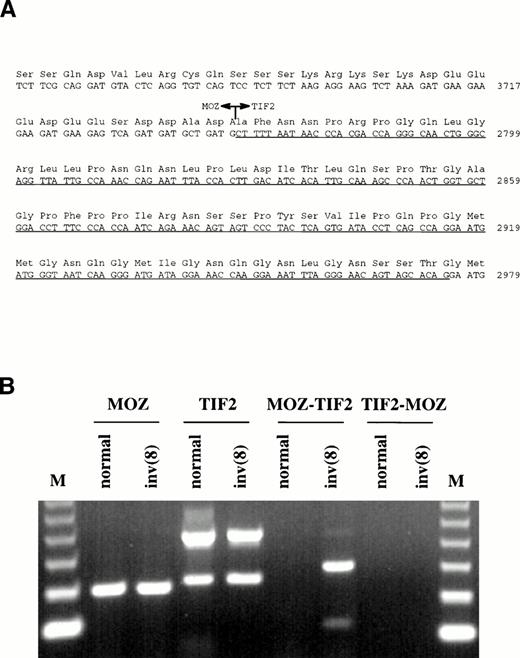
Primers were designed from the MOZ cDNA sequence upstream of the last exon and used for 3′ RACE-PCR with RNA extracted from the patient's bone marrow. In addition to normal MOZ products, clones were isolated in which the penultimate exon of this gene was fused to novel sequence. A search of the Genbank database showed that the sequence was derived from the TIF2 gene, a recently described member of the nuclear receptor transcriptional coactivator family of proteins.13-15 Nucleotide 3745 ofMOZ was joined to nucleotide 2768 of TIF2 (Genbank:X97674), resulting in an in-frame fusion (Fig3A).
(A) Sequence surrounding the MOZ-TIF2 fusion. Nucleotide 3745 of MOZ was joined to nucleotide 2768 ofTIF2. The underlined sequence indicates the region withinTIF2 that is alternatively spliced in both the normal and fusion genes. (B) RT-PCR analysis of bone marrow RNA from a normal individual and the patient with the inv(8). Normal MOZ andTIF2 transcripts were detected in both samples, the lowerTIF2 band resulting from alternative splicing. TheMOZ-TIF2 fusion was seen only in the patient and reciprocalTIF2-MOZ transcripts were not detected. The identity of all bands was confirmed by sequencing. M, 123-bp molecular weight marker.
(A) Sequence surrounding the MOZ-TIF2 fusion. Nucleotide 3745 of MOZ was joined to nucleotide 2768 ofTIF2. The underlined sequence indicates the region withinTIF2 that is alternatively spliced in both the normal and fusion genes. (B) RT-PCR analysis of bone marrow RNA from a normal individual and the patient with the inv(8). Normal MOZ andTIF2 transcripts were detected in both samples, the lowerTIF2 band resulting from alternative splicing. TheMOZ-TIF2 fusion was seen only in the patient and reciprocalTIF2-MOZ transcripts were not detected. The identity of all bands was confirmed by sequencing. M, 123-bp molecular weight marker.
Because the chromosomal localization of TIF2 had not previously been determined, we hybridized probes generated from the 5′ and 3′ ends of this gene to a gridded CEPH megaYAC library. Several positive clones were isolated that contained the entire TIF2 gene, including the YACs 798c3 and 815d1 which have been mapped previously to 8q13.2-3.17
The presence of a novel fusion transcript was confirmed by reverse transcriptase-PCR using cDNA primers either side of the junction. MOZ-TIF2 was specifically amplified from the patient's bone marrow RNA, but not from bone marrow RNA extracted from normal individuals (Fig 3B). Reciprocal TIF2-MOZ transcripts were not detected. Two MOZ-TIF2 bands were amplified, which, by sequencing, were found to result from alternative splicing within theTIF2 moiety (nucleotides 2767-2975). The same alternative splice was found for TIF2 amplified from both the patient and normal individual. Rearrangement of the TIF2 gene was confirmed by Southern blotting. After digestion with BglII, probe UN-1 detected the same-sized rearranged band as MOZ 0.9 (Fig 1A). Rearrangements were not seen in digests of DNA from normal individuals.
DISCUSSION
The MOZ gene was isolated as a consequence of its fusion toCBP by the t(8;16) in AML. Of its structural features, the most significant are a PHD/LAP domain, thought to be involved in protein-protein interactions,19,20 and a region that is homologous to a number of lower eukaryotic histone acetyltransferases (HATs).7 Although the precise function of MOZ is not known, a related gene in Drosophila, MOF, is required for dosage compensation, the mechanism which enables male flies with a single X chromosome to express the same level of X-linked products as females with two X chromosomes.21 The related yeast genesSAS2 and YBF2 influence transcriptional silencing and, taken together, it is likely that MOZ is a chromatin-bound HAT that modulates the transcription of specific target genes. CBP is a cointegrator/adaptor that is believed to coordinate the transcriptional effects of multiple signals from cell surface and nuclear receptors.22-24 Recently it has been shown that CBP also has HAT activity.9,10
Nuclear receptors are ligand-inducible transcription factors, which typically consist of three structural domains: an N-terminus containing an activation function, AF-1; a DNA-binding domain; and a ligand-binding C-terminus that contains a second activation function, AF-2.25 AF-2 mediates transcriptional activation through nuclear receptor coactivators (NRCoAs),26,27 which are thought to stimulate gene expression by facilitating the assembly of basal transcription factors into a stable preinitiation complex.28 Transcriptional interference/squelching experiments have indicated that NRCoAs are limiting factors in this process.13TIF2 was recently shown to be one such mediator of AF-2 function.13 This protein is homologous to other NRCoAs, specifically SRC-1 (also known as F-SRC-1 and NCoA-1),15,29,30 and pCIP (also known as RAC3, ACTR, and AIB1).15,31-33 Although the functional domains of TIF2 have not been determined, the related proteins SRC-1/F-SRC-1/NCoA-1 and ACTR/pCIP/RAC3/AIB1 have HAT activity and also interact directly with CBP.29,34-36 Therefore, it is likely that NRCoAs mediate transcriptional activation by a mechanism involving extensive chromatin remodeling. Acetylation of amino termini of core histones allows nucleosomes to unfold, thus increasing access to transcription factors.37 Conversely, histone deacetylation has been shown to repress transcription, at least in some situations.38
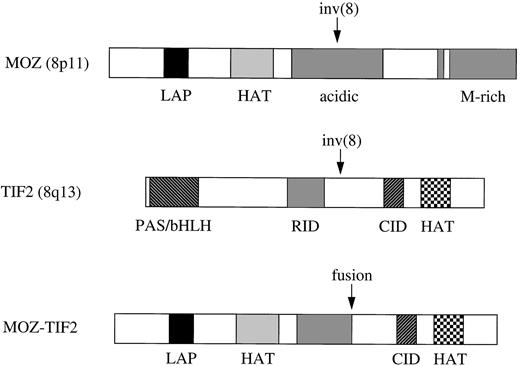
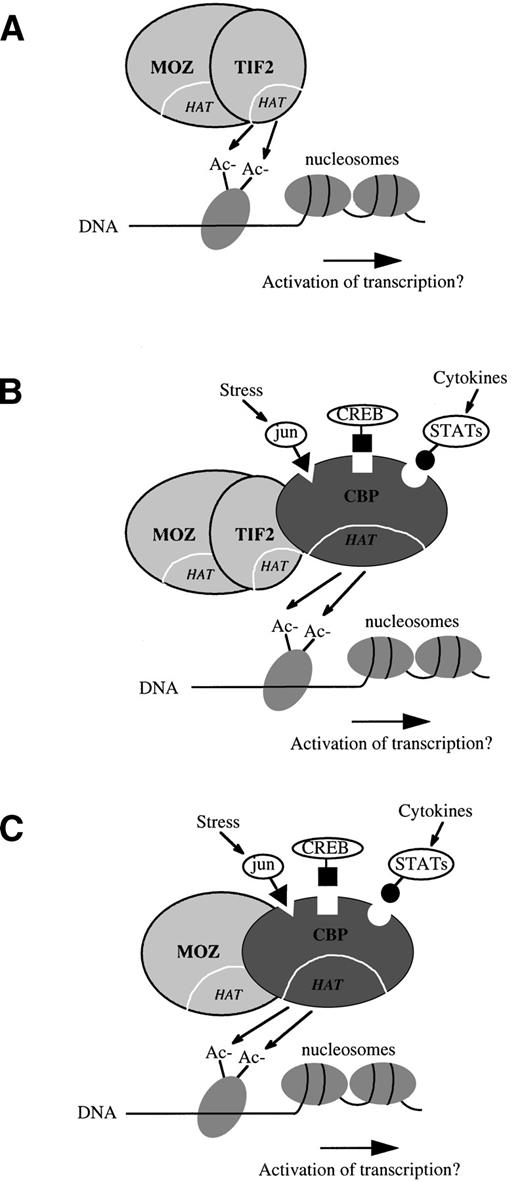
The MOZ-TIF2 fusion retains the LAP finger and HAT homology domains of MOZ, along with the putative CBP interacting domain (CID) and HAT domain of TIF2 (Fig 4). The fusion does not retain the TIF2 PAS/bHLH domain, believed to be involved in DNA binding and protein heterodimerization, or the receptor interacting domain (RID), which mediates the binding of transcriptional coactivators to nuclear receptors via conserved LXXLL motifs.15,29,39Therefore, it is unlikely that MOZ-TIF2 is able to interact with any of the upstream components that normally require TIF2 as a transcriptional intermediary. Instead, the HAT activity of TIF2 may directly modulate or augment the transcriptional activity of genes normally regulated by MOZ (Fig 5A). Alternatively, the TIF2 moiety may serve as a bridge between MOZ and CBP, and it is the HAT or other activities of CBP that are relevant to putative leukemogenic alterations in gene expression (Fig 5B). The latter model is particularly attractive in view of the striking similarity in cell phenotypes associated with the MOZ-CBP fusion in the t(8;16) (Fig 5C) and MOZ-TIF2 in the inv(8). The molecular basis of the two variant translocations t(8;19) and t(8;22) is unknown, but they may result in analogous mechanisms of leukemogenesis. For example, it has been postulated that the t(8;22) may involve p300 at 22q13, a gene that is structurally and functionally closely related toCBP.7 Indeed, it has been shown recently thatp300 is fused to the MLL gene in AML with the t(11;22)(q23;q13).40
Schematic representation of MOZ, TIF2, and MOZ-TIF2 fusion proteins. Domains are indicated as follows: LAP, leukemia-associated protein20; HAT, histone acetyltransferase; M-rich, methionine-rich; CID and RID, putative CBP and nuclear receptor interacting domains based on homology with SRC-1.29,30 The MOZ-TIF2 fusion retains the LAP finger and HAT homology domains of MOZ, along with the CID and HAT domains of TIF2.
Schematic representation of MOZ, TIF2, and MOZ-TIF2 fusion proteins. Domains are indicated as follows: LAP, leukemia-associated protein20; HAT, histone acetyltransferase; M-rich, methionine-rich; CID and RID, putative CBP and nuclear receptor interacting domains based on homology with SRC-1.29,30 The MOZ-TIF2 fusion retains the LAP finger and HAT homology domains of MOZ, along with the CID and HAT domains of TIF2.
Hypothetical models of the mode of action of the MOZ-TIF2 fusion protein. (A) TIF2 may directly modulate the transcriptional activity of genes normally regulated by MOZ through the addition or removal of histone acetyl (Ac−) groups by its histone acetyltransferase (HAT) domain. (B) The TIF2 moiety may serve as a bridge between MOZ and CBP, and it is the HAT or other activities of CBP that leads to leukemogenic alterations in gene expression. Chromatin-associated CBP may be responsive to other cellular signals such as those mediated by jun, CREB, or STAT proteins.22-24(C) The MOZ-CBP fusion in the t(8;16),7 which is associated with a strikingly similar leukemia cell phenotype to that seen in cases with the inv(8).
Hypothetical models of the mode of action of the MOZ-TIF2 fusion protein. (A) TIF2 may directly modulate the transcriptional activity of genes normally regulated by MOZ through the addition or removal of histone acetyl (Ac−) groups by its histone acetyltransferase (HAT) domain. (B) The TIF2 moiety may serve as a bridge between MOZ and CBP, and it is the HAT or other activities of CBP that leads to leukemogenic alterations in gene expression. Chromatin-associated CBP may be responsive to other cellular signals such as those mediated by jun, CREB, or STAT proteins.22-24(C) The MOZ-CBP fusion in the t(8;16),7 which is associated with a strikingly similar leukemia cell phenotype to that seen in cases with the inv(8).
Although TIF2 has not previously been implicated in malignancy, several sporadic leukemias have been described with translocations or other structural rearrangements that involve chromosome band 8q13.41 Furthermore, the related geneAIB1/pCIP/RAC3/ACTR at 20q12 is amplified in breast and ovarian cancers,33 and, in addition, the ARA70gene was fused to RET in a case of human thyroid papillary carcinoma42,43 and TIF1 fused to B-RAF in the mouse hepatoma-derived oncogene T18.44 Although ARA70 and TIF1 do not share sequence homology with the TIF2 family of NRCoAs, they are also involved in the ligand-dependent activation function of nuclear receptors.43,44 These observations suggest that subversion of the mechanisms by which nuclear receptors modulate gene transcription, including specific acetylation of histones, is widely involved in malignancy.
ACKNOWLEDGMENT
We thank the MRC HGMP Resource Centre for providing the PAC clones and CEPH megaYAC clones.
Supported by the Leukaemia Research Fund. R.C.T.A. was supported by CNPq (Conselho Nacional de Desenvolvimento Cientı́fico e Tecnológico-No 200995/94-4), Brazil.
Address reprint requests to Nicholas C.P. Cross, PhD, Dept of Haematology, Imperial College School of Medicine, Hammersmith Hospital, Du Cane Rd, London, W12 ONN, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.