Factor VIII (fVIII) functions as a cofactor of factor IXa in the intrinsic pathway of blood coagulation. Its absence or abnormality causes the bleeding disorder hemophilia A. About 23% of hemophiliacs who receive therapeutic fVIII infusions develop antibodies that inhibit its activity. We previously showed by inhibitor neutralization assays that the fVIII A2 and C2 domain polypeptides contain common inhibitor epitopes. Often hemophilic inhibitor plasmas were partially neutralized by C2 and more completely neutralized by fVIII light chain (A3-C1-C2), suggesting the presence of an additional major inhibitor epitope(s) within the A3-C1 domains. In immunoprecipitation assays, 17 of 18 inhibitor IgGs bound to recombinant 35S-A3-C1. Amino acids 1811-1818 of the A3 domain comprise a binding site for factors IX and IXa. Three inhibitor IgGs prevented binding of factor IXa to fVIII light chain, and the binding of each IgG to light chain was competed by A3 peptide 1804-1819. The generation of factor Xa by the fVIIIa/fIXa complex in a chromogenic assay was prevented by these inhibitors. Therefore, we propose that another important mechanism of fVIII inactivation by human inhibitors is the prevention of fVIIIa/fIXa association.
COAGULATION FACTOR VIII (fVIII) functions as a cofactor of the protease factor IXa (fIXa) in the intrinsic pathway of blood coagulation. Activated fVIII (fVIIIa) forms a complex with fIXa and factor X in the presence of CaCl2 on a cell surface containing negatively charged phospholipid where it acts to increase the Vmax for the conversion of factor X to Xa.1
FVIII is a high-molecular-weight glycoprotein composed of three different types of domains arranged in the order NH2-A1-A2-B-A3-C1-C2-COOH, as deduced from the cDNA sequence.2,3 It circulates in the plasma as a heterodimer of a light chain (A3-C1-C2) and a heavy chain (A1-A2-B)4linked by a metal ion bridge. Lack of or defective fVIII causes the disease hemophilia A, and plasma-derived or recombinant fVIII is used to correct this bleeding disorder. In eight studies, 23% (6.2% to 33%) of hemophiliacs developed antibodies that inhibit fVIII activity,5 and they present a serious clinical complication.
Epitopes of common inhibitor antibodies were previously localized to the A2 and C2 domains by assays in which recombinant A2 and C2 polypeptides neutralized the inhibitor activity.6 Anti-A2 antibodies prevent the function of the fIXa/fVIIIa enzyme complex on a phospholipid surface but not its formation,7 whereas anti-C2 antibodies prevent the interaction of fVIII with phospholipid and von Willebrand factor.8 The greater neutralization of many inhibitor plasmas by fVIII light chain than by recombinant C2 suggested the presence of another common inhibitor epitope(s) within the A3 and/or C1 domains.6
FVIII binding sites for fIXa were localized to amino acid residues 558-565 of the A2 domain9 and residues 1811-1818 of the A3 domain.10 Synthetic peptides corresponding to each region inhibited the factor Xase activity of fIXa, which suggested that both sites are required for maximal affinity of fVIII for fIXa. The monoclonal antibody (MoAb) CLB-CAg A with an epitope in A3 domain residues 1804-1818 prevented fVIII light chain binding to fIXa and inhibited fVIII cofactor activity.10 Because of the characteristics of the MoAb, we investigated the possibility that the anti–A3-C1 inhibitors suggested in our previous experiments6 may also prevent fVIII/fIX binding.
MATERIALS AND METHODS
Antibodies.
MoAbs CLB-CAg A and NMC-VIII/5 were kindly provided by Drs Jan van Mourik (Central Laboratory, Netherlands Red Cross) and Midori Shima (Nara Medical College, Japan), respectively. The preparation of IgG from plasmas of severe hemophilia A patients with inhibitors was previously described.6 Inhibitor titers were determined in the Bethesda assay.11
Construction of recombinant A3-C1 and (His)5-C2 polypeptide expression vectors.
A cDNA construct encoding the signal peptide of interferon-β in frame with the A3 and C1 domains (amino acids 1690-2172) and two stop codons was assembled by overlap extension12 and cloned into plasmid pMT2,13 which also encodes dihydrofolate reductase (dhfr). The pMT2 A3-C1 construct was used to transfect dhfr− Chinese hamster ovary (CHO) cells, a stable dhfr+ cell line expressing A3-C1 was isolated, and CHO cells containing amplified copies of A3-C1, verified by Southern blotting, were selected in increasing concentrations of methotrexate13 to 0.32 μmol/L. The C2 cDNA including the stop codon was attached to a 5′ sequence encoding the prepro polypeptide of tissue plasminogen activator,14 five histidines, and a thrombin cleavage site by overlap extension,12 and the resulting DNA was cloned into baculovirus vector pVL1393 (Invitrogen, Carlsbad, CA). Using the Bac to Bac expression system (Life Technologies, Gaithersburg, MD), the C2 cDNA was transferred to a baculovirus plasmid by in vivo site specific transposition in Escherichia coli. The C2-baculovirus plasmid was purified and used to transfect Sf9 insect cells for generation of baculoviruses expressing C2. Secreted C2 was quantitated by enzyme-linked immunosorbent assay (ELISA)14 as 27 μg/mL. All cDNA constructs were sequenced to verify the absence of DNA errors.
The amplified A3-C1 cell line, 2 × 106 in 100-mm tissue culture dishes, was grown in Dulbecco's modified Eagle medium (DMEM) α-medium (Life Technologies) with 10% fetal calf serum for 48 to 72 hours to 90% confluency. The cells were washed two times with methionine-free DMEM and labeled in 4 mL of this medium with 0.5 mCi Tran35S-Label (ICN Biomedicals, Costa Mesa, CA) for 4 hours at 37°C. The culture medium was collected, and 1% NP40 (Sigma, St Louis, MO) was added. Cell lysates were suspended in 50 mmol/L Tris, pH 7.4, 150 mmol/L NaCl (TBS), 1% NP40. The growth medium and cellular fractions were centrifuged at 10,000 rpm for 10 minutes at 4°C. Aliquots of the supernatants were stored at −80°C.
Preparation and radiolabeling of fVIII fragments.
Plasma derived fVIII was purified from fVIII concentrate Method M (American Red Cross),15 and heavy and light chains were further purified as described.15,16 The A1 and A2 domains were purified from plasma fVIII after thrombin cleavage17, and recombinant C2 polypeptide was purified from Sf9 insect cell growth medium.14 Ammonium sulfate (38%) was added to the Sf9 medium containing 0.7 mg (His)5-C2, and the C2 precipitate was dialyzed into 50 mmol/L phosphate, 0.3 mol/L NaCl, 0.05% Tween 20, pH 8.0, and passed over a 2-mL Ni2+-NTA agarose column (Qiagen, Valencia, CA). The column was washed with the above buffer adjusted to pH 5.8 followed by elution of (His)5-C2 with a pH gradient of 5.6 to 4.0 in the same buffer. Recovery of (His)5-C2 was 65%. Recombinant fVIII and B domain were generously provided by Baxter/Hyland (Glendale, CA).
Polypeptides were radiolabeled with 5 μL Na 125I (100 mCi/mL; Amersham, Arlington Heights, IL) by immobilized lactoperoxidase (Worthington, Freehold, NJ) as described.17 Specific radioactivities ranged from 5 to 13 μCi/μg for all polypeptides. They were aliquoted and stored at −80°C for up to 1 month.
Immunoprecipitation of radiolabeled polypeptides by inhibitor IgG.
Details were previously described.17 Briefly, each125I-polypeptide (0.75 nmol/L) was incubated with inhibitor IgG or plasma dilutions at 4°C overnight. Immune complexes were precipitated with Protein G Sepharose (Pharmacia Biotech, Piscataway, NJ) and washed to remove unbound 125I-polypeptide. The negative control contained all components except antibody. Dose-response binding curves were done for each IgG, and data points from the linear portion of each curve were used to calculate immunoprecipitation (IP) units per milliliter: [(Bound/Total125I-fVIII Polypeptide − Background) × Plasma Dilution × 20 (to Convert to IP Units/mL)]. IP of35S-labeled A3-C1 polypeptide in cell culture medium with inhibitor was performed as described above, except that bound35S-labeled A3-C1 was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide) and autoradiography.
To determine the concentration of A3-C1, aliquots of the CHO cell culture medium containing A3-C1 were used to compete the binding of125I-recombinant fVIII to MoAb CLB-CAg A IgG in competitive IP assays, a modification of the above IP method. Recombinant fVIII was used to generate a standard curve.
Depletion of anti-C2 antibodies from inhibitor IgG.
Purified recombinant (His)5-C2 was immobilized on Ni+2-NTA agarose at 250 μg per mL of resin via the (His)5 tag. Inhibitor IgGs were diluted to 2 mg/mL in HBS (20 mmol/L HEPES, 0.15 mol/L NaCl, pH 7.4, 0.01% Tween 80), mixed with resin at a 2:1 vol/vol ratio, and incubated for 18 hours at 4°C. Upon removal of the resin by centrifugation, the supernatants were tested for the presence of anti-C2 antibodies by IP assay using125I-labeled C2 polypeptide.
Chromogenic factor Xase assay.
Details were previously described.16 Briefly, plasma fVIII (2 nmol/L) was incubated with increasing concentrations of MoAb CLB-CAg A or inhibitor IgG for 30 minutes at 37°C. Aliquots were diluted 10-fold into HBS (above) with 5 mmol/L CaCl2, 2 nmol/L fIXa (Enzyme Research Laboratories, South Bend, IN), and 20 μmol/L phosphatidylserine/phosphatidylcholine vesicles (75/25, wt/wt). fVIII was actived with 40 nmol/L thrombin for 30 seconds, and 300 nmol/L factor X was added. Aliquots were withdrawn after 15, 30, 45, and 60 seconds, and factor X activation was stopped with 0.05 mol/L EDTA. Factor Xa generation was measured by cleavage of synthetic substrate S-2765, 0.3 mmol/L (Pharmacia Hepar, Franklin, OH) using a Vmax microplate reader (Molecular Devices, Menlo Park, CA). A purified factor Xa standard (Enzyme Research Laboratories) was used to convert absorbance (410 nm) into factor Xa concentration. The molar concentrations of fIXa, factor X, and factor Xa were calculated using molecular masses of 45 kD, 57 kD, and 45.3 kD, respectively.
FVIII or light-chain binding to a ligand using biosensor technology.
The kinetics of light-chain binding to ligands was determined by surface plasmon resonance using the IAsys biosensor (Fisons, Cambridge, UK). Anti-C2 MoAb NMC-VIII/5 (50 μg/mL) in 10 mmol/L sodium acetate, pH 5.0, was covalently coupled to an activated carboxymethyldextran coated biosensor cuvette (Affinity Sensors, Paramus, NJ) via amino groups using succinimide ester chemistry.18 fIXa was inactivated for use in the binding studies by reaction with the active site specific reagent DEGR-CK (Calbiochem, La Jolla, CA) as described.19 All measurements of fIXa-DEGR binding to fVIII light chain were performed in 200 μL HBS, 5 mmol/L CaCl2at 37°C. Dissociation of bound fIXa-DEGR was initiated by substitution with 200 μL buffer lacking fIXa-DEGR. Binding of MoAb or inhibitor IgG to immobilized light chain in the presence or absence of the synthetic peptide was performed under the above conditions. The cuvette was regenerated by addition of 0.1 mol/L glycine, pH 3.0, for 3 minutes, resulting in complete dissociation of light chain from MoAb.
RESULTS
Expression of recombinant A3-C1 polypeptide.
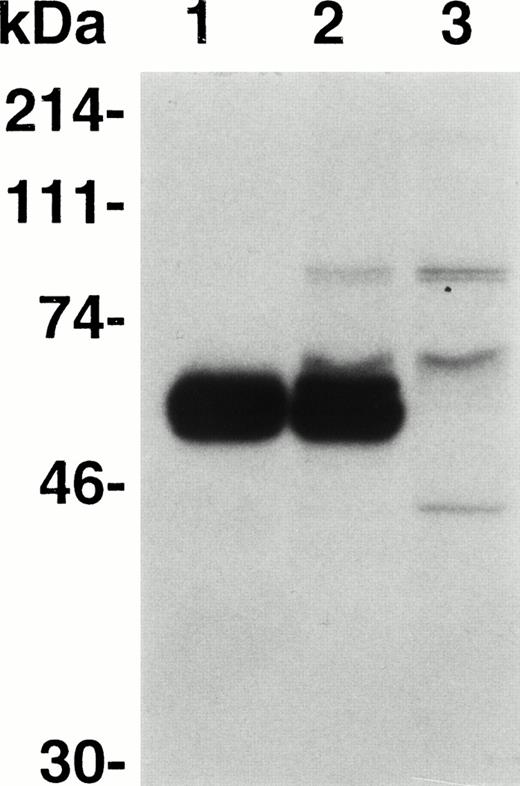
The previously observed greater neutralization of inhibitors by light chain than by C26 suggested the presence of one or more additional inhibitor epitopes in the A3-C1 domains of the light chain. To determine if antibodies from inhibitor plasmas bind to this region, we constructed an expression plasmid, pMT2,13 encoding the A3 and C1 domains of the light chain but not its amino terminal acidic region, residues 1649-1689. The interferon-β (IFN-β) signal sequence cDNA was linked in frame to the 5′ end of the A3-C1 cDNA for potential secretion of the recombinant polypeptide. A stable CHO cell line containing multiple copies of the A3-C1 cDNA was established (Materials and Methods). Expression of recombinant A3-C1 was analyzed by IP of the growth medium and the cellular fraction of the35S-Met labeled CHO cell line (Fig 1) with MoAb CLB-CAg A IgG (epitope A3 residues 1804-182010) followed by SDS-PAGE. Similar concentrations of 35S-A3-C1 with the expected molecular weight (∼60 kD) were seen in the extracellular (lane 1) and the intracellular (lane 2) fractions, demonstrating approximately 50% secretion. Additional bands seen in the cell fraction were not fVIII specific because they were also present when MoAb CLB-CAg A was omitted (Fig 1, lane 3). The A3-C1 concentration in the growth medium was determined to be 0.76 μg/mL in a competitive IP assay (Materials and Methods).
Expression and secretion of A3-C1 polypeptide. Immunoprecipitation of 35 S-A3-C1 by MoAb CLB-CAg A IgG (30 μg/mL) was analyzed by 10% SDS-PAGE and autoradiography. The cellular and growth medium fractions were adjusted to equal volumes. Lane 1, culture medium; lane 2, cell lysate; lane 3, cell lysate without CLB-CAg A. Molecular weight standards are shown in kilodaltons (kDa) at the left.
Expression and secretion of A3-C1 polypeptide. Immunoprecipitation of 35 S-A3-C1 by MoAb CLB-CAg A IgG (30 μg/mL) was analyzed by 10% SDS-PAGE and autoradiography. The cellular and growth medium fractions were adjusted to equal volumes. Lane 1, culture medium; lane 2, cell lysate; lane 3, cell lysate without CLB-CAg A. Molecular weight standards are shown in kilodaltons (kDa) at the left.
Binding of human antibodies to the A3-C1 polypeptide.
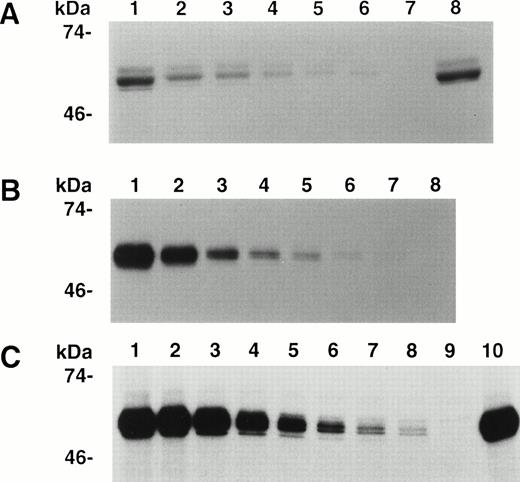
The binding of 35S-A3-C1 to decreasing concentrations of IgG purified from each of 10 inhibitor plasmas was measured by IP followed by SDS-PAGE (Materials and Methods), and representative results are shown in Fig 2. The binding of the positive control, anti-A3 MoAb CLB-CAg A IgG10 (Fig2B), and human inhibitors RI (Fig 2A) and MU (Fig 2C) was dose dependent, and controls without antibody were negative. As both complex antibodies and impure A3-C1 were used, the results in Fig 2 cannot be used for quantitative comparisons among the IgGs. The binding of the eight other human IgGs was also dose dependent in this assay (not shown). For a more extensive analysis of the frequency of A3-C1 binding by IgG in inhibitor plasmas, eight were diluted 1:5 and tested in single-point IP assays. Binding was positive in seven and negative in one (not shown). A3-C1 binding by 17 of 18 human inhibitors from both experiments demonstrated that specificity for A3-C1 is common.
Immunoprecipitation of A3-C1 polypeptide by MoAb and hemophilic inhibitor IgGs. Cell culture medium containing35S-methionine–labeled rA3-C1 was incubated with increasing concentrations of IgG from human inhibitors RI or MU and from MoAb CLB-CAg A. Immunoprecipitated 35S-A3-C1 was analyzed as in Fig 1. (A) RI, lanes 1 through 6: 20, 10, 5, 2.5, 1.25, and 0.625 μg/mL; lane 7: no antibody; lane 8: 3 μg/mL of CLB-CAg A IgG. (B) MoAb CLB-CAg A, lanes 1 through 7: 2, 1, 0.5, 0.25, 0.12, 0.06, and 0.03 μg/mL; lane 8: no antibody. (C) MU, lanes 1 through 8: 20, 10, 5, 2.5, 1.25, 0.62, 0.31, and 0.15 μg/mL; lane 9: no antibody; lane 10: 3 μg/mL of CLB-CAg A IgG.
Immunoprecipitation of A3-C1 polypeptide by MoAb and hemophilic inhibitor IgGs. Cell culture medium containing35S-methionine–labeled rA3-C1 was incubated with increasing concentrations of IgG from human inhibitors RI or MU and from MoAb CLB-CAg A. Immunoprecipitated 35S-A3-C1 was analyzed as in Fig 1. (A) RI, lanes 1 through 6: 20, 10, 5, 2.5, 1.25, and 0.625 μg/mL; lane 7: no antibody; lane 8: 3 μg/mL of CLB-CAg A IgG. (B) MoAb CLB-CAg A, lanes 1 through 7: 2, 1, 0.5, 0.25, 0.12, 0.06, and 0.03 μg/mL; lane 8: no antibody. (C) MU, lanes 1 through 8: 20, 10, 5, 2.5, 1.25, 0.62, 0.31, and 0.15 μg/mL; lane 9: no antibody; lane 10: 3 μg/mL of CLB-CAg A IgG.
Characteristics of IgGs used in determination of inhibitor mechanism.
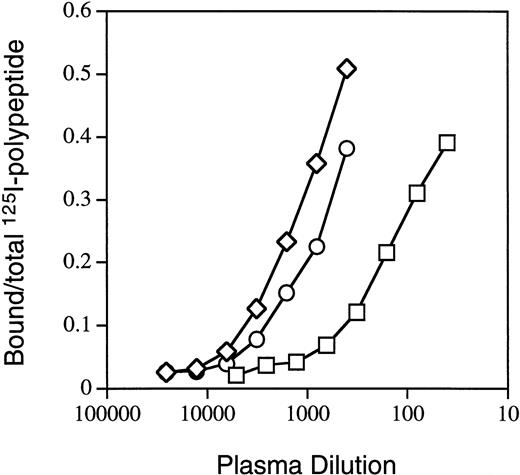
For further characterization of antibodies directed against A3-C1, we selected three IgGs from hemophilia A patients with high Bethesda titers. In previous inhibitor neutralization assays6 the MU, RI, and MS IgGs were neutralized to a greater extent by light chain than by C2 (Table 1), suggesting that they contained 44%, 48%, and 79% anti–A3-C1 and 15%, 26%, and 30% anti-C2 inhibitors, respectively. MU and RI were not detectably neutralized by the A2 domain (≤10%) but MS was 27% neutralized. In addition, binding of each IgG to the individual fVIII heavy-chain domains, the intact light chain, and the isolated C2 domain by semi-quantitative IP assays (Materials and Methods) was also tested. Figure 3 contains representative binding curves for RI binding to the isolated fVIII fragments, and the total results are summarized in Table 1. The combined anti–heavy-chain titers were ≤0.1 of the anti–light-chain titers. The percent anti–A3-C1 antibodies (LCh-C2) was 61%, 37%, and 91% and anti-C2 antibodies was 39%, 64%, and 9% for MU, RI, and MS, respectively. These percentages are not the same as those determined by the inhibitor neutralization assay due to the presence in plasmas of both inhibitory and noninhibitory antibodies.20
Binding of inhibitor RI IgG to fVIII-derived polypeptides. IP assays with radiolabeled fVIII domains A2 (□), C2 (○), and light chain (◊) were performed as described in Materials and Methods.
Binding of inhibitor RI IgG to fVIII-derived polypeptides. IP assays with radiolabeled fVIII domains A2 (□), C2 (○), and light chain (◊) were performed as described in Materials and Methods.
Because the neutralization assays indicated that 15% to 30% of the inhibitor titers were caused by anti-C2 antibodies, each IgG was depleted of these antibodies (Materials and Methods) to measure only the effects of anti–A3-C1 antibodies in the following experiments. IP assays of 125I-C2 binding by each IgG before and after the depletion showed the >300-fold removal of anti-C2 antibodies in all three IgGs (Table 1).
Inhibition of fIXa binding to fVIII light chain by inhibitor IgG.
Although the fIXa binding site consists of amino acid residues within the A29 and A3 domains,10 the light chain binds to fIXa with moderate affinity (kd =14.8 nmol/L).21 Due to the fact that inhibitors RI, MU, and MS had measurable levels of anti-A2 antibodies (Table 1) that might complicate the interpretation of the results, we chose to examine the effect of inhibitor IgG on fIXa binding to the light chain only. For detection of protein-protein interactions we used a biosensor technique based on the surface plasmon resonance phenomenon which measures protein binding in real time. The change in the refractive index due to association of a fluid-phase ligand with an immobilized ligand is measured by generation of a signal of 200 Arc seconds per 1 ng of protein bound per mm2 of the biosensor cuvette. MoAb NMC-VIII/5 (C2 epitope 2170-23278) was immobilized to the cuvette, and the light chain (20 μg/mL) was maximally bound to 35 fmol/mm2.
To determine if all immobilized light-chain molecules were accessible for fIXa binding, we measured their maximal binding capacity. To prevent the possible fIXa-mediated cleavage of immobilized light chain at Arg1721,22 we used an active-site modified fIXa (fIXa-DEGR), which lacks proteolytic activity19 but retains fVIII binding properties.23
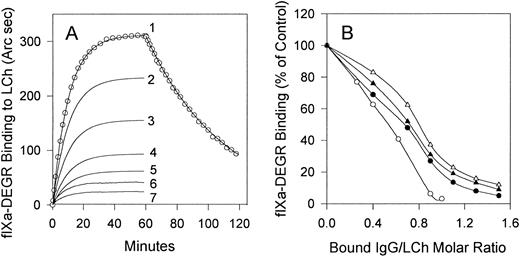
Curve 1 in Fig 4 shows the resonance response of the time course of fIXa-DEGR association with light chain and dissociation of the complex upon replacement of fIXa-DEGR with buffer (shown only for curve 1). No binding of fIXa-DEGR to the cuvette was observed in the absence of light chain (data not shown). The association rate constant (kon = 1.2 × 104 M−1s−1) and the dissociation rate constant (koff = 3.4 × 10−4 s−1) for fIXa-DEGR/light-chain interaction were derived from the best fit of these kinetic data to a model describing single-phase association and dissociation processes using the Fast Fit 1.0b computer program (FISONS). Because the concentration of fIXa-DEGR (1,000 nmol/L) is >25 times the corresponding equilibrium dissociation constant (kd = 28 nmol/L, calculated as koff/kon) for its interaction with light chain, the amount of fIXa-DEGR bound at equilibrium (36.6 fmol/mm2) represents the maximum binding capacity of immobilized light chain. This result shows that the immobilized light chain (35 fmol/mm2) was able to bind fIXa-DEGR with 1:1 stoichiometry. In addition, the similar values of the kd for fIXa-DEGR/light chain determined in our assay (28 nmol/L) and that determined for fIXa/fVIIIa interaction in solution (42 nmol/L)24 suggest that the light-chain binding properties were not significantly altered by immobilization. The absence of phospholipid is probably a major contributor to the relatively high kd values for fVIII -fIX interaction obtained in both our (28 nmol/L) and previous experiments (42 nmol/L).24 This interpretation is consistent with reported kds of 2.3 and 46 nmol/L for fVIIIa/fIXa association in the presence and absence of phospholipid, respectively.24
Effect of stoichiometric titration of the light chain with inhibitors MU, RI, MS, and MoAb CLB-CAg A IgGs on fIXa binding. (A) Association of fIXa-DEGR (1,000 nmol/L) with light chain (35 fmol/mm2) in the presence of 0, 14, 24, 31, 38, 46, and 52 fmol/mm2 bound RI IgG is shown in curves 1 through 7, respectively. The resonance response curves recorded upon addition of fIXa-DEGR were corrected by subtraction of the resonance signal produced by bound IgG, and therefore the curves show the resonance signals solely produced by binding of fIXa to immobilized light chain (curve 1) or to IgG/light chain complexes (curves 2 through 7). (B) Binding of fIXa in the presence of MoAb CLB-CAg A IgG (○) or inhibitor IgG from MU (•), RI (▴), or MS (▵) prebound to immobilized light chain at the indicated molar ratio was determined from the equilibrium binding of fIXa-DEGR in the presence of each IgG as in (A).
Effect of stoichiometric titration of the light chain with inhibitors MU, RI, MS, and MoAb CLB-CAg A IgGs on fIXa binding. (A) Association of fIXa-DEGR (1,000 nmol/L) with light chain (35 fmol/mm2) in the presence of 0, 14, 24, 31, 38, 46, and 52 fmol/mm2 bound RI IgG is shown in curves 1 through 7, respectively. The resonance response curves recorded upon addition of fIXa-DEGR were corrected by subtraction of the resonance signal produced by bound IgG, and therefore the curves show the resonance signals solely produced by binding of fIXa to immobilized light chain (curve 1) or to IgG/light chain complexes (curves 2 through 7). (B) Binding of fIXa in the presence of MoAb CLB-CAg A IgG (○) or inhibitor IgG from MU (•), RI (▴), or MS (▵) prebound to immobilized light chain at the indicated molar ratio was determined from the equilibrium binding of fIXa-DEGR in the presence of each IgG as in (A).
Inhibitor or MoAb IgGs were bound to the light chain at varying IgG/light-chain molar ratios calculated from the resonance signals produced by bound IgG (not shown), which was followed by replacement of the IgG solution with buffer containing 1,000 nmol/L fIXa-DEGR. The binding of fIXa-DEGR to light chain in the presence or absence of bound RI IgG approached equilibrium after 50 minutes, and the fIXa-DEGR binding progressively decreased upon saturation of light chain with RI IgG (Fig 4A). Similar results were obtained for IgG from MU, MS, and MoAb CLB-CAg A (not shown). Because of the multiple binding components, these experiments more closely represent a stoichiometric titration rather than equilibrium binding. In a control experiment, dissociation of bound IgG was less than 3% during 50 minutes, which showed that the IgG/LCh ratio did not change significantly during fIXa-DEGR binding to LCh.
FIXa-DEGR binding to light chain was inhibited more than 95% by MoAb CLB-CAg A at a stoichiometric ratio (Fig 4B). As the MoAb binds to A3 residues 1804-1820, an fIX binding site, the binding of MoAb and fIXa to light chain is mutually exclusive. MU, RI, and MS IgGs maximally inhibited fIXa-DEGR binding by 95%, 91%, and 88%, respectively, at IgG/light chain ratios of 1.5 (Fig 4B). The nonstoichiometric ratios for the human inhibitors may be due to the additional presence of lower concentrations of antibodies directed against regions of A3-C1 other than the fIXa binding site.
Effect of synthetic A3 domain peptide 1804-1819 on inhibitor IgG binding to light chain.
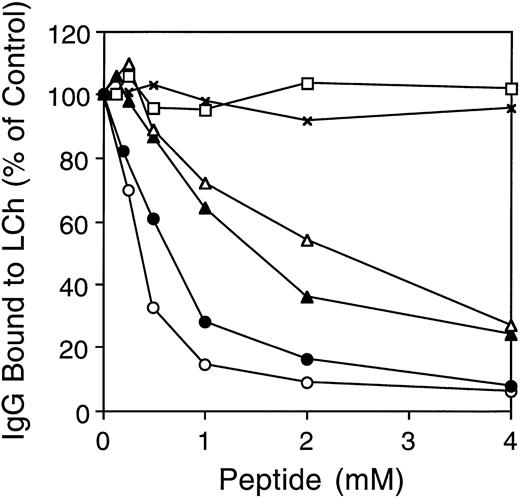
As the inhibitory effect of MoAb CLB-CAg A is caused by direct competition with fIXa for fVIII binding,10 we tested the hypothesis that the human anti–A3-C1 antibodies have the same mechanism of inhibition. We used synthetic A3 peptide 1804-1819 to compete for human IgG binding to light chain. MoAb CLB-CAg A (10 nmol/L) or inhibitor IgGs MU (50 nmol/L), RI (1,000 nmol/L), and MS (1,000 nmol/L) were incubated with increasing concentrations of peptide for 1 hour at 37°C before addition to the biosensor cuvette containing light chain immobilized on MoAb NMC-VIII/5. The above antibody concentrations were chosen as each inhibited the factor Xase assay (see below) by approximately 80%. The concentration of anti-fVIII antibodies was different in each IgG; therefore, the total IgG required in our experiments also varied widely. Stoichiometric titration, measured as above, was achieved within 30 minutes at all peptide concentrations. The binding of human inhibitor MU, RI, and MS, or MoAb CLB-CAg A IgG to light chain was inhibited ≥73% in a dose-dependent mannner by the peptide (Fig5). The maximal inhibition of binding was 94%, 92%, 76%, and 73% for CLB-CAg A, MU, RI, and MS IgG, respectively. The less complete inhibition of MS and RI binding by the peptide may be due to insufficient peptide concentrations or to the presence of a minor population of antibodies with an epitope(s) outside the 1804-1819 sequence. The millimolar concentrations of peptide required for inhibition in each case suggests that the peptide-antibody affinities are low.
Effect of fVIII synthetic peptides on antibody binding to immobilized light chain. Increasing concentrations of A3 peptide 1804-1819 were preincubated with IgG from MU (•), RI (▴), MS (▵), or MoAb CLB-CAg A (○) for 1 hour at 37°C, and the mixture (200 μL) was added to the biosensor cuvette containing light chain immobilized as in Fig 4. In the control experiments, binding of MU in the presence of peptide 417-428 (x), amino acids QRIGRKYKKVRF, or the randomized version of 1804-1819 (□) was determined as above. Antibody binding in the presence of peptide is expressed as the percentage of antibody binding when no peptide was added.
Effect of fVIII synthetic peptides on antibody binding to immobilized light chain. Increasing concentrations of A3 peptide 1804-1819 were preincubated with IgG from MU (•), RI (▴), MS (▵), or MoAb CLB-CAg A (○) for 1 hour at 37°C, and the mixture (200 μL) was added to the biosensor cuvette containing light chain immobilized as in Fig 4. In the control experiments, binding of MU in the presence of peptide 417-428 (x), amino acids QRIGRKYKKVRF, or the randomized version of 1804-1819 (□) was determined as above. Antibody binding in the presence of peptide is expressed as the percentage of antibody binding when no peptide was added.
A randomized, soluble peptide (PVKETYKFNKKTVFNV) of the fVIII sequence 1804-1819 was used as a control for MU and CLB-CAgA (not shown) binding to LCh. There was no inhibition up to 4 nmol/L peptide. Because peptide 1804-1819 contains four positively charged Lys residues and one negatively charged Glu residue, its effect on antibody binding may be caused by the overall positive charge. This possibility was excluded by lack of inhibition of MU (Fig 5; RI and MS, negative but not shown) in the above assay by an unrelated control peptide of A2 domain residues 417-428, containing six positively charged and two negatively charged amino acids.
MU IgG inhibits fVIII activity in a chromogenic factor Xase assay.
To determine if anti–A3-C1 antibodies are able to inhibit fVIII function in a purified system, we tested their effect in a chromogenic factor Xase assay using purified components. This assay (Materials and Methods) measures the ability of activated fVIII (fVIIIa) in the presence of phosphatidylserine/phosphatidylcholine vesicles (PSPC) to act as a cofactor for fIXa in the activation of factor X.16Because fVIIIa activity cannot be directly measured in this assay, we determined the initial rate of factor X activation using a chromogenic peptide substrate, S-2765. Under our conditions, this rate was linearly proportional to the fVIIIa activity.
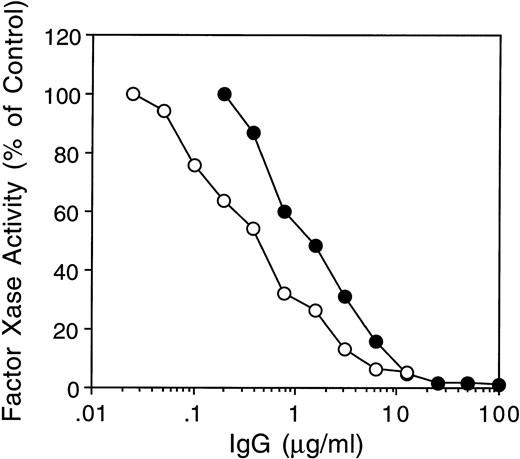
Only inhibitor MU IgG was tested because it had the lowest ratio of anti-A2 to anti–light-chain antibodies (2.5 × 10−5) and no other anti–heavy-chain antibodies by IP assays. Due to further depletion of anti-C2 antibodies (Materials and Methods), the ratio of anti-C2 to anti–light chain antibodies was decreased to 6.7 × 10−5 (Table 1). The MU IgG used in this experiment therefore contained more than 99.9% anti–A3-C1 antibodies. MU and CLB-CAg A IgGs both inhibited the factor Xase assay by more than 90% (Fig 6).
Inhibition of fVIII activity in the factor Xase assay by MoAb CLB CAg A and inhibitor MU. FVIII (2 nmol/L) was preincubated with increasing concentrations of inhibitor MU (•) or MoAb CLB-CAg A (○) IgG for 30 minutes at 37°C followed by determination of fVIII activity in the chromogenic factor Xase assay as described in Materials and Methods.
Inhibition of fVIII activity in the factor Xase assay by MoAb CLB CAg A and inhibitor MU. FVIII (2 nmol/L) was preincubated with increasing concentrations of inhibitor MU (•) or MoAb CLB-CAg A (○) IgG for 30 minutes at 37°C followed by determination of fVIII activity in the chromogenic factor Xase assay as described in Materials and Methods.
DISCUSSION
The greater neutralization of many inhibitor plasmas by the fVIII light chain than by C26 suggested that these plasmas may contain inhibitor antibodies with epitopes in A3-C1. To examine this possibility, we first demonstrated by an IP assay that 17 of 18 human inhibitor IgGs bound to a recombinant 35S-A3-C1 polypeptide. The IgG from three such inhibitors with >10-fold higher anti–light chain than anti–heavy chain antibody titers was further depleted of anti-C2 domain antibodies to prevent their interference in subsequent assays. One IgG (MU) with barely detectable anti–heavy chain antibodies completely inhibited the intrinsic factor Xase assay; therefore, some anti–A3-C1 antibodies are likely to have inhibitor activity.
MoAb CLB-CAg, A which binds to the A3 domain peptide 1804-1818, a fIX binding site, also inhibits fVIII activity.10 No other binding site for a physiological ligand of fVIII has been localized to A3-C1. Therefore, we tested the possibility that some human anti–A3-C1 antibodies inhibit fVIII-fIX binding, which is required for assembly of the factor Xase complex. In an assay of fIXa binding to light chain, the three inhibitor IgGs bound to light chain inhibited subsequent fIXa binding by ≥88%, compared with 97% by CLB-CAg A. The inhibition of the human and MoAb CLB-CAg A IgG binding to light chain by synthetic peptide 1804-1819 suggested that the epitopes of these antibodies overlap the fIX binding site. These data are consistent with a model of direct inhibition of fIX binding to fVIII by these antibodies.
We found that peptide 1804-1819 inhibited binding of inhibitor MU IgG and MoAb CLB-CAg A to light chain by 88% to 90%. This result implies that MU anti–A3-C1 IgG is mainly directed against the fIX binding site, which is consistent with the observed 1.1:1.0 molar ratio of bound MU IgG to light chain. In contrast, binding of the inhibitor IgGs RI and MS to light chain was inhibited only 76% and 73%, respectively, by the peptide. This may be due to insufficient concentrations of peptide for complete inhibition of binding or to the presence of a minor population of anti–A3-C1 antibodies that do not bind to the fIXa site.
Although we tested only three anti–A3-C1 inhibitors, all of which competed for fIX binding to fVIII, this may be a common inhibitory mechanism as suggested by our earlier finding that 20 of 34 inhibitor plasmas were more completely neutralized by light chain than by its C2 domain.6 This remains to be confirmed by characterization of additional inhibitors.
ACKNOWLEDGMENT
We thank Dr Jan van Mourik (Netherlands Red Cross, Amsterdam) for providing the MoAb CLB-CAg A, which was crucial for our work.
D.Z. and E.L.S. made equal contributions to the experimental work.
Address reprint requests to Dorothea Scandella, PhD, American Red Cross, 15601 Crabbs Branch Way, Rockville, MD 20855.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.