Abstract
Acute leukemia with t(4;11)(q21,q23) translocation results from the in-frame fusion of the MLL to the AF4/FEL gene. In previous studies, we and others demonstrated that AF4 transcripts are present in a variety of hematopoietic and nonhematopoietic human cells. To further study the wild-type and leukemia fusion AF4, we used glutathione S-transferase (GST)-fusion proteins as immunogens to produce rabbit polyclonal antibodies that were specific for normal and chimeric AF4 proteins. Using Western blotting analysis, we demonstrated that the AF4 gene encodes proteins with apparent molecular weight of 125 and 145 kD. A 45-kD protein coprecipitated with AF4 protein in immunoprecipitation. Also, the anticipated MLL-AF4–encoded 240-kD protein was detected in all cell lines with t(4;11) translocations; fusion proteins were present in lesser quantity than the wild-type AF4. The proteins recognized by the antibodies are of the predicted sizes of the AF4 and MLL-AF4–encoded proteins based on previous DNA sequencing analysis. The MLL-AF4 fusion protein had a similar subcellular distribution as AF4. Both t(4;11) and non-t(4;11) leukemic cells showed a similar pattern of punctate nuclear staining in all cell lines tested using confocal immunofluorescence microscopy. AF4 antibodies should be useful for further elucidation of the function of AF4 in normal cellular physiology, as well as the function of MLL-AF4 in leukemogenesis. The antibodies should also be helpful for the diagnosis of the MLL-AF4 fusion proteins in t(4;11) leukemias.
AF4, ALSO KNOWN AS FEL, is a gene which was first described as a fusion partner with MLL in the t(4;11) acute leukemia.1-3 Based on analysis by Northern blotting, AF-4 mRNAs were found to be widely expressed in hematopoietic cells and normal human tissues.4,5 cDNA sequence analysis of the AF-4 gene showed that it encodes a serine/proline-rich protein with a predicted size of 130 to 140 kD containing guanosine triphosphate (GTP)-binding and putative nuclear-localization sequences (NLS).2 Studies have demonstrated that a related gene, LAF4, isolated from Burkitt’s lymphoma, shares high degree of sequence homology with AF4. LAF4 was shown to possess DNA binding ability and transcriptional activation potential.6 Both AF4 and LAF4 were shown to be evolutionary conserved in vertebrates suggesting an important functional role of the genes. Recently, a third gene (FMR2) has been recognized as a member of AF4/LAF4 gene family. FMR2 maps to X chromosome at position Xq28. Mutations of FMR2 are associated with mild hereditary mental retardation.7,8 Members of the homologous AF4/LAF4/FMR2 gene family are expected to have transcriptional activation functions. We previously found that another DNA sequence located on chromosome 5q31, probably is a member of this gene family.9
Clinical studies have demonstrated that MLL (also known as HRX and ALL-1) rearrangements in infant ALL are associated with very poor prognosis.4 At the molecular level, important sequence characteristics and putative functional motifs of MLL and MLL-AF4 fusion genes have been defined. Molecular analysis of the MLL gene indicates that it contains two central zinc-fingers, a C-terminal region with homology to Drosophila trithorax gene, three N-terminal A/T hook regions, a domain with homology to methyltransferase, and a proline-rich region.3,10-12 The t(4;11) translocation separates the second and third DNA binding domain of MLL from the zinc finger region.10 The AT-hook and proline-rich regions of MLL are fused in-frame to the region in AF-4, which contains the NLS and GTP-binding activity, resulting in a chimeric mRNA of 12.5 kb that encodes a predicted fusion protein of 240 kD.2,4 Rabbit polyclonal antibodies specific to N-terminal fragments of MLL-encoded proteins expressed in Escherichia colihave been recently reported.13 The antibodies recognized the MLL and MLL/AF-4–encoded proteins in a leukemic cell line and cells transfected with portions of MLL. A monoclonal antibody specific for N-terminal epitopes of MLL protein has also been reported. This antibody detected MLL-AF4 fusion protein in Western blotting.14 Recently, Nilson et al15 reported rabbit antibodies against human AF4 protein; an unexpected 116-kD protein was detected in cell lysates by western blottings with no detection of the MLL-AF4 fusion proteins. The major purpose of our study was to identify and characterize the proteins encoded by AF4 and MLL-AF4 genes by developing antibodies specific for AF4 proteins using molecular and immunological approaches.
MATERIALS AND METHODS
Cell lines and construction of expression vectors.
The RS4;11 cell line was established in our laboratory16and is available from ATCC (American Type Culture Collection, Rockville, MD). Two t(4;11) cell lines, B117 and AN4;114 have been described. Another t(4;11) line, Sem-k2, was a gift from Dr F.E. Cotter (Department of Haematology and Oncology, Institute of Child Health, London, UK). Other cell lines have been described and used in our laboratory for a number of years including Nalm-6, KM3, BLIN-1 (B-cell lines), Raji (B-cell lymphoma), Molt-4, CEM (T-cell lines), M418 (human neuroblastoma), MG-63 (human osteosarcoma), and K562 (myeloid cell line).4 The cells were grown and maintained in RPMI 1640 tissue culture medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. A 3.3-kb cDNA designated PL12 that contained most of AF4 gene open reading frame was isolated from a human placenta cDNA library.9 Two regions of the PL12 clone, one from the 5′ end (base pair 612-1110, designated C9) and another from the 3′ end (base pair 1942-2242, designated C15) of the RS4;11 breakpoint and other breakpoints (Fig 1) were amplified by polymerase chain reaction (PCR) using standard protocols. The C9 and C15 clones encode polypeptides of 18 kD and 10 kD, respectively. The selection of these regions from AF4 gene for construction of expression vectors was based on the “plotstructure analysis” using the GCG computer program (Genetics Computer Group, Madison, WI). Both regions showed high surface probability, low hydrophobicity, and high antigenic index, suggesting a strong potential for stimulating immune responses. PCR primers were selected using the oligos 4s computer software (National Bioscience, Plymouth, MN) and engineered to include EcoR1 and BamH1 restriction sites for direct in-frame insertion into the GST expression vector (Pharmacia, Piscataway, NJ). The PCR products were gel purified and ligated into TA cloning vectors (Invitrogen, San Diego, CA). The inserts were sequenced from both ends into the vector sequences to assure proper orientation. The GST vectors with C9 and C15 inserts were expected to encode a 44- and a 36-kD protein, respectively (GST, 26 kD; inserts, 18 kD and 10 kD).
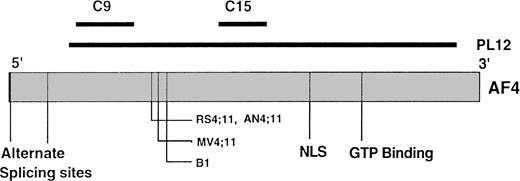
Schematic representation of predicted AF4 gene structure. The relative location and size of C9, C15, and PL12 as compared with AF4 gene are shown as dark lines; alternate splicing sites, breakpoints of t(4;11) translocations, nuclear localization sequences (NSL), and GTP binding motifs of AF4 are indicated. The figure was constructed based on the data from Morrissey et al,2 Frestedt et al,9 Tkachuk et al,11 and Hilden et al.22
Schematic representation of predicted AF4 gene structure. The relative location and size of C9, C15, and PL12 as compared with AF4 gene are shown as dark lines; alternate splicing sites, breakpoints of t(4;11) translocations, nuclear localization sequences (NSL), and GTP binding motifs of AF4 are indicated. The figure was constructed based on the data from Morrissey et al,2 Frestedt et al,9 Tkachuk et al,11 and Hilden et al.22
Expression and purification of GST-fusion proteins.
GST expression vectors with or without the inserts were inserted into DH5 alpha bacteria (GIBCO/BRL, Grand Island, NY) using standard bacterial transformation procedures. The transformed colonies were selected and grown in ampicillin-containing (100 μg/mL) LB medium at 37°C overnight with vigorous shaking. GST-fusion proteins were induced with 0.1 mmol/L IPTG (Isopropyl-B-D-thiogalactopyranoside, Sigma Chemical Co, St Louis, MO) for 3 to 4 hours. The bacteria were harvested by centrifugation, washed and resuspended in PBST-100, pH 7.4 (phosphate-buffered saline, 1% Triton-X100, 1 mmol/L EDTA) and sonicated (Branson Ultrasonic Corp, Brandury, CT) to release the proteins. The presence of the fusion proteins in the cell lysates were determined by analyzing the samples in 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). GST and GST-fusion proteins were affinity purified by passing the bacterial lysates through a glutathione sepharose 4B (Pharmacia) column and washed thoroughly with PBST-100. The bound proteins were eluted with 100 mmol/L NaCl solution containing 25 mmol/L reduced glutathione (Sigma). The proteins were concentrated to 2 to 3 mg/mL using a Centricon-10 protein concentrator (Amicon, Bedford, MA) and buffer exchanged to 100 mmol/L HEPES buffer using a PD-10 desalting column (Pharmacia). The protein concentration was determined by a protein assay reagent (Pierce, Rockford, IL). After affinity purification, GST-fusion proteins were further purified by preparative electrophoresis. The protein samples were separated in 10% SDS-PAGE gels and surface-stained with Commassie blue staining solution for 2 to 3 minutes. The bands of interest were cut from the gels and eluted into SDS-PAGE electrode buffer using an electro-elution device. SDS contained in the protein solution was removed by acetone precipitation method of Hager and Burgess.18
Production and purification of antibodies.
Two hundred microliters (1 mg/mL) of the 44-kD or 36-kD GST-fusion protein obtained from preparative electrophoresis were emusified with an equal volume of complete Freund’s adjuvant and injected into two rabbits at multiple sites. Booster injections with the decreasing amounts of antigen emusified with incomplete Freund’s adjuvant were performed at a 2-week interval over a period of 84 days. The immune responses of the animals were monitored by indirect enzyme-linked immunosorbent assay (ELISA)19 and Western blottings. The rabbits were killed and the sera collected when the ELISA titers of the immune sera against the immunizing antigen reached 1:5,000 or greater. Our initial attempts to immunize the rabbits with glutathione sepharose 4B purified GST-fusion proteins in denatured or native forms failed to generate useful antibodies. This initial problem led us to further purify the proteins by preparative electrophoresis to obtain a higher proportion of full-length fusion protein for immunizations. Rabbits immunized with these highly purified and denatured antigens stimulated strong antibody responses as determined by immunoblotting and ELISA using native and denatured proteins as coating antigens. Specific antibodies in the sera were purified by affinity chromatography. Briefly, 2 mg of purified GST, GST-C9, or GST-C15 fusion proteins were conjugated to 1 mL Affigel-10 (Bio-Rad, Hercules, CA) following manufacturer’s recommendations. Protein-conjugated Affigel-10 was packed into a minicolumn and equilibrated with PBS. The antiserum diluted fourfold with PBST (PBS containing 0.1% Tween-20) was passed through the column containing Affigel-10 conjugated GST to remove antibodies against GST. The eluent was then passed several times through the column containing conjugated GST-C9 or GST-C15 fusion protein, washed extensively with PBST, and eluted with 50 mmol/L glycine buffer (pH 2.8). The eluted antibody was neutralized by 1.89 mol/L Tris buffer (pH 8.9) in the collection tubes. Purified antibodies were designated anti-C–9 and anti-C15 and stored at 4°C in a solution containing 0.5% BSA, 0.4 mol/L L-arginine, and 0.02% sodium azide. IgG fractions of preimmune sera were purified by a protein A column (Pierce).
Western blotting.
Cultured cell lines in log phase were harvested by low speed centrifugation (1,500g), washed twice with PBS, and subjected to sonication at maximum intensity for 10 seconds. The cell lysates were solubilized in SDS-PAGE sample buffer, heated in boiling water for 2 minutes, and separated on SDS-PAGE gels under reducing-conditions. After electrophoresis, the proteins in the gels were electrophoretically transferred onto hydrated nitrocellulose membranes for 2 hours. The membranes were blocked with 5% nonfat milk in PBS for 2 hours at 37°C or at 4°C overnight. The membranes were probed with diluted primary antibodies in blocking buffer at room temperature for 1 hour. For detection of AF4 and MLL-AF4–encoded proteins, affinity purified anti-C15 and anti-C9 antibodies (50 to 100 μg/mL) were diluted 1:100 to 1:200 and used in Western blotting. Mouse antihuman β tubulin monoclonal antibody (Sigma) used as an internal control in some experiments was diluted according to supplier’s recommendations. After incubating with primary antibodies, the blots were washed either under high or low stringent conditions. Unless specified, all Western blottings were performed under high stringent washing conditions. Under high stringent conditions, the blots were washed three times (15 to 20 minutes/each) with PBS containing 0.05% Tween-20. Low stringent wash consisted of three washes (5 to 8 minutes/each) with PBS containing 0.01% Tween-20. After washing, the blots were incubated with goat antirabbit or goat antimouse IgG conjugated to horseradish peroxidase (HRP, Amersham, Alington, IL) for 45 to 60 minutes at room temperature. The blots were washed as before and incubated with enhanced chemiluminescence (ECL) reagents (Amersham) for 1 minute. The blots were blotted dry with filter papers, enclosed in transparent plastic sheets, and exposed to autoradiographic film. Other antibodies used in this study were rabbit anti-LAF4 (A gift from Dr Louis M. Staudt, Metabolism Branch, National Cancer Institute, National Institutes of Health, Bethesda, MD) and monoclonal anti-MLL antibody (a gift from Dr Lisa H. Butler, John Radcliffe Hospital, Oxford, UK).
Blocking experiment.
The specificity of purified anti-C15 and anti-C9 were accessed by blocking with their corresponding immunizing antigens in Western blotting format. Total cell lysate of Km3 was separated in 8% SDS-PAGE (20 to 40 μg/lane) and electrophoretically transferred to nitrocellulose membrane, cut into strips, and incubated with different preparations of primary antibodies. Affinity-purified stock solutions (50 to 100 μg/mL) of anti-C15 and anti-C9 were diluted 1:100 with PBST containing 5% nonfat milk and 80 μg of corresponding immunizing antigens in a final volume of 10 mL. Preimmune IgG and the same amount of antibodies blocked with GST (80 μg/sample) were used as controls. After 30 minutes incubation at room temperature, the nitrocellulose strips were incubated with primary antibodies and the rest of the Western blotting procedures were performed as above.
Immunoprecipitation and pulse chase analysis.
A total of 1 × 107 cells were harvested by low speed centrifugation and washed twice with cold Hank’s balanced salt solution. The cells were resuspended in 5 mL RPMI medium containing 5% dialized FBS (Gibco) and 0.2 mCi 35S (ICN, Costa Mesa, CA) and incubated at 37°C with gentle shaking for 3 hours. The labeled cells were washed twice with cold Hank’s balanced salt solution and lysed with 600 μL of lysis buffer (PBS containing 1% Triton X-100, 0.5% SDS, 0.5% deoxycholate, 1% BSA, leupeptin [10 μg/mL] and 0.2 mmol/L phenylmethyl sulfonyl fluoride [PMSF], Sigma). The cell lysate was incubated in ice for 10 minutes and centrifuged in a microcentrifuge at top speed for 10 minutes to remove insoluble materials. Three hundred microliter cell lysate was precleared by incubating with 1 μg rabbit IgG and 30 μL protein G plus/protein A-agarose (CALBIOCHEM, Cambridge, MA) for 1 hour at room temperature. The supernatant was incubated with 0.5 to 1 μg anti-C15 and 30 μL protein A/G agarose at 4°C for 3 hours. After incubation, the agarose beads were washed three times with lysis buffer. The immunoprecipitates were solubilized in 40 μL SDS-PAGE sample buffer, separated by 10% SDS-PAGE, and visualized by autoradiography. Pulse chase labeling was performed based on the methods of Bonifacino.20 Briefly, 5 × 107 cells were incubated with 5 mL methionine-free RPMI medium with 5% dialized FBS (pulse medium) for 30 minutes at 37°C to deplete intracellular methionine. The cells were pelleted and incubated in 5 mL fresh warmed pulse medium containing 1 mCi/mL 35S for 0.5 to 1 hour. An aliquot of 1 × 107 cells for time 0 were collected, and the remaining cells were washed once with Hank’s balanced salt solution, resuspended in 20 mL complete RPMI medium supplemented with excessive L-methionine (15 mg/mL) and incubated at 37°C for the chase time 1, 2, 3, and 6 hours. At each time point, equal amount of cells were washed, lysed, precleared, and immunoprecipitated with anti-C15 as described above.
Confocal immunofluorescence microscopy.
Cultured cells were harvested by low speed centrifugation and resuspended in complete RPMI 1640 medium. The cells (1 to 2 × 105/mL) were fixed onto microscopic slides by cytospin. The slides were air dried and fixed in absolute methanol solution at −20°C for 10 minutes. The fixed cells were blocked in blocking solution (2% BSA, 10 mg/mL goat IgG in PBS) for 30 minutes followed by incubation with affinity-purified anti-C15 (1:40) at room temperature for 1 hour. The slides were washed extensively with PBS and incubated with goat antirabbit IgG Fab2 conjugated to fluorescein isothiocyanate (FITC) (Sigma) for 1 hour and washed extensively as described. Stained slides were mounted in glycerol mounting buffer (PBS, 10%; p-phenylenediamine, 10 mg/mL; and Glycerol, 90%)21 and examined by a Bio-Rad MR C600 confocal microscope. The images were processed using Adobe photoshop software (Adobe Systems Inc, San Jose, CA).
RESULTS
Specificity of anti-C15 and anti-C9 antibodies.
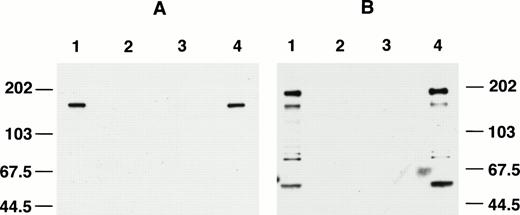
The specificity of affinity-purified anti-AF4 antibodies was first evaluated by Western blotting. As shown in a high stringency experiment (see Materials and Methods), affinity-purified anti-C15 recognized a single protein band of about 145 kD (Fig2A, lane 1) in Western blotting against total Km3 cell lysate. The signal was absent using preimmune serum (lane 2) and was completely blocked by incubating the antibody with immunizing antigen before Western blotting (lane 3). Lane 4 shows lack of blocking with GST. Under low stringent washing conditions, anti-C15 also binds to a 80-kD protein in Western blotting; however, the cross-reactivity could be removed by extensive washing. In contrast to anti-C15, affinity-purified anti-C9 detected four major proteins in Western blotting (Fig 2B, lane 1) with an estimated molecular size of 184, 145, 75, and 50 kD. All four signals were completely blocked by the immunizing antigen (Fig 2B, lane 3). Preimmune IgG did not show any reactivities against the cell lysate (lane 2) and GST failed to block immunoreactivity of the antibody (lane 4).
Blocking of anti-C15 (A) and anti-C9 (B) by immnunizing antigens. Km3 cell lysate was separated in 8% SDS-PAGE and electrophoretically transferred onto a nitrocellulose membrane. The blots were cut into strips and Western blotting was performed using various preparations of primary antibodies. (A) Lane 1, anti-C15 versus cell lysate; lane 2, preimmune IgG; lane 3, anti-C15 blocked by immunizing antigen; lane 4, anti-C15 blocked with GST. (B) Lane 1, anti-C9; lane 2, preimmune IgG; lane 3, anti-C9 blocked by immunizing antigen; lane 4, anti-C9 blocked by GST.
Blocking of anti-C15 (A) and anti-C9 (B) by immnunizing antigens. Km3 cell lysate was separated in 8% SDS-PAGE and electrophoretically transferred onto a nitrocellulose membrane. The blots were cut into strips and Western blotting was performed using various preparations of primary antibodies. (A) Lane 1, anti-C15 versus cell lysate; lane 2, preimmune IgG; lane 3, anti-C15 blocked by immunizing antigen; lane 4, anti-C15 blocked with GST. (B) Lane 1, anti-C9; lane 2, preimmune IgG; lane 3, anti-C9 blocked by immunizing antigen; lane 4, anti-C9 blocked by GST.
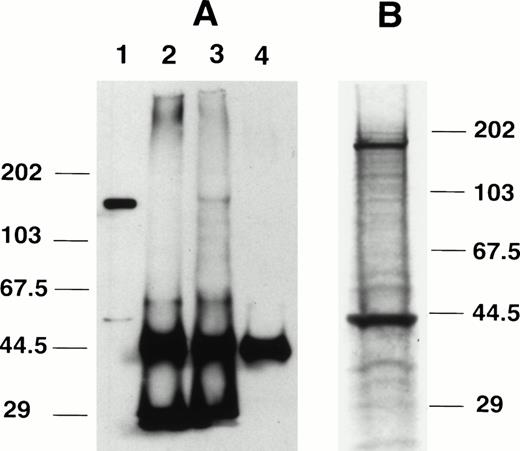
Immunoprecipitation was performed using anti-C15 antibody. After immunoprecipitation of Km3 lysate with anti-C15, a protein of about 145 kD was detected in the immunoprecipitates by Western blotting using the same antibody (Fig 3A, lane 3). The protein band was not seen using preimmune serum (lane 2) or without lysate (lane 4). The protein signal seen in Fig 3A, lane 3, was identical to that of nonprecipitated lysate (lane 1). We also conducted experiments to evaluate proteins that were physically associated with AF4. Km3 cells were radiolabeled with 35S and the cell lysate was immunoprecipitated by anti-C15. In results shown in Fig 3B, the expected protein of about 145 kD and a second 45-kD protein were detected (Fig 3B). The 45-kD protein was not detected in Western blottings suggesting that the 45-kD protein was coprecipitated with the larger protein.
Immunoprecipitation using anti-C15. (A) Immunoprecipitation followed by Western blotting. Lane 1, Km3 cell lysate without immunoprecipitation; lane 2, unlabeled Km3 lysate immunoprecipitated with preimmune IgG; lane 3, Km3 lysate immunoprecipitated with anti-C15; lane 4, anti-C15 with no lysate. The protein samples were separated in 8% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-C15. (B) Immunoprecipitation of radiolabeled proteins. 35S-labeled Km3 lysate (300 μL) was precleared with preimmune IgG and incubated with 1 μg anti-C15 for 3 hours at 4°C. The immune complex was pelleted by protein A/G beads, washed, separated by 10% SDS-PAGE, and autoradiographed.
Immunoprecipitation using anti-C15. (A) Immunoprecipitation followed by Western blotting. Lane 1, Km3 cell lysate without immunoprecipitation; lane 2, unlabeled Km3 lysate immunoprecipitated with preimmune IgG; lane 3, Km3 lysate immunoprecipitated with anti-C15; lane 4, anti-C15 with no lysate. The protein samples were separated in 8% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-C15. (B) Immunoprecipitation of radiolabeled proteins. 35S-labeled Km3 lysate (300 μL) was precleared with preimmune IgG and incubated with 1 μg anti-C15 for 3 hours at 4°C. The immune complex was pelleted by protein A/G beads, washed, separated by 10% SDS-PAGE, and autoradiographed.
More than one AF4 protein is detected in Western blotting.
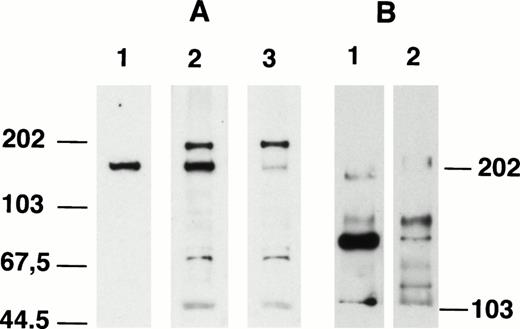
Because the immunizing antigens of anti-C9 and anti-C15 were from the 5′ and 3′ end of AF4 gene, respectively, both antibodies should recognize the full-length AF4 protein in total cell lysate. As shown in Fig 4A, a side-by-side comparison of anti-C15 and anti-C9 showed that anti-C15 recognized a single protein band in the cell lysates and anti-C9 bound to several proteins. One of the proteins detected by anti-C9 (lane A3) showed the same migration as that detected by anti-C15 (lane A1). When anti-C9 and anti-C15 were mixed (lane A2) and used for Western blotting, the AF4 protein signal was stronger than the signals detected by either antibody alone, while other protein bands detected by anti-C9 remained comparable. These results strongly suggest that both antibodies recognized the same AF4 protein. When the Western blottings were performed using lower percentage gels (Fig 4B), the AF4 protein migrated as doublets (145 kD and 125 kD), which were detected by both anti-C15 (lane B1) and anti-C9 (lane B2). Repeated experiments showed that anti-C15 generally reacted stronger with the 125-kD band, which is most likely due to the results of alternate splicing of AF4 mRNA. In previous studies, a 10.5-kb and a 12-kb mRNA transcript of AF4 gene was observed.2,4 An alternate explanation for the doublets is the possibility of precursor-product relationship of the doublets. To evaluate this possibility, pulse chase analysis was conducted. Despite repeated attempts, we were able to show the doublets in autoradiographes using low percentage gels only after long periods of incubation (data not shown). A time course study of cell lysates labeled with 35S and precipitated by anti-C15 indicated that the doublets were observed in autoradiographes only in cell lysates that have been labeled for a longer period of time (>3 hours). The extented labeling time required to show the doublets prevented us from obtaining interpretable results of pulse chase analysis, which requires a short labeling window of 30 minutes to 1 hour.20 Low immunoprecipitation efficiency of anti-C15 may also be in part responsible for the observations.
Comparison of Western blotting patterns of anti-C15 and anti-C9. (A) Km3 lysate (20 μg/lane) was separated in 8% SDS-PAGE, transferred to nitrocellulose paper, and probed with anti-C15 (lane 1), anti-C15 plus anti-C9 (lane 2), and anti-C9 (lane 3). (B) K562 lysate (20 μg/lane) was separated in 4% SDS-PAGE, transferred to nitrocellulose paper, and probed with anti-C15 (lane 1) and anti-C9 (lane 2).
Comparison of Western blotting patterns of anti-C15 and anti-C9. (A) Km3 lysate (20 μg/lane) was separated in 8% SDS-PAGE, transferred to nitrocellulose paper, and probed with anti-C15 (lane 1), anti-C15 plus anti-C9 (lane 2), and anti-C9 (lane 3). (B) K562 lysate (20 μg/lane) was separated in 4% SDS-PAGE, transferred to nitrocellulose paper, and probed with anti-C15 (lane 1) and anti-C9 (lane 2).
Anti-C15 and anti-C9 recognize MLL-AF4 and AF4-MLL reciprocal fusion proteins, respectively.
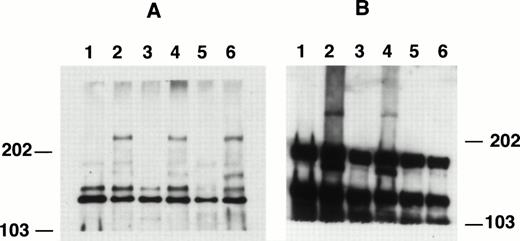
When the cell lysates were run on low percentage SDS-PAGE gels (4% to 5%), a protein band with a calculated molecular weight of 240 kD was detected by anti-C15 in cell lines containing MLL-AF4 fusion gene (Fig 5). This protein was consistently detected in all cell lines with known t(4;11) translocations (Fig 5A, lanes 2, 4, and 6), but not in those without the translocations (lanes 1, 3, and 5). These data indicate that the 240-kD protein is the MLL-AF4–encoded fusion protein and is consistent with the predicted protein sizes.2 A side-by-side comparison of anti-C15 and the anti-MLL/HRX antibody (HRX107)14 in Western blotting showed that a 240-kD protein band were detected by both antibodies (data not shown). In these low percentage gels, anti-C15 also generally detected the AF4 doublet in cells used (Fig 5A). Again, as shown in Fig4B, the smaller protein species showed stronger signals than the larger species.
Western blotting patterns of anti-C15 and anti-C9 in 4% SDS-PAGE. Cell lysates were separated by SDS-PAGE, electrophoretically transferred to nitrocellulose membranes, and probed with affinity-purified anti-C15 (A) or anti-C9 (B). Lane 1, Nalm-6; lane 2, RS4;11; lane 3, Km3; lane 4, Sem-k2; lane 5, K562; lane 6, B1.
Western blotting patterns of anti-C15 and anti-C9 in 4% SDS-PAGE. Cell lysates were separated by SDS-PAGE, electrophoretically transferred to nitrocellulose membranes, and probed with affinity-purified anti-C15 (A) or anti-C9 (B). Lane 1, Nalm-6; lane 2, RS4;11; lane 3, Km3; lane 4, Sem-k2; lane 5, K562; lane 6, B1.
We also evaluated anti-C9 antibody for study of the fusion proteins. Figure 5B shows the immunoblotting patterns of anti-C9 against total cell lysates in 4% gel. In addition to 180 kD and 145 kD proteins, the antibody also recognized a high molecular weight protein (>240 kD) in cells with t(4;11) translocations, RS4;11 and SEM-k2 (lanes 2 and 4, respectively). The B1 cells, which were previously shown to have low to absent levels of 14 kb der4 RNA transcript,4 were found to have a very weak band as shown in Fig 5B, lane 6. Although the exact size of this high molecular weight protein cannot be determined due to the lack of proper molecular weight markers, by comparing its migration rate with that of the 240 kD MLL-AF4 fusion protein, the size of the protein was estimated in the range of 300 to 350 kD. Because the high molecular weight protein signal is much weaker than that of AF4 protein, the blot in Fig 5B was intentionally overexposed to show the high molecular weight band. Because anti-C9 was raised using N-terminal polypeptide of AF4 as immunogen, it might expect to recognize AF4 and reciprocal AF4-MLL proteins, but not the MLL-AF4 fusion proteins. In view of the observation that the high molecular weight protein detected by anti-C9 was seen only with t(4;11) lines and that the size of the protein falls in the calculated size range of AF4-MLL fusion protein {(430 (MLL) + 140 (AF4) − 240 (MLL-AF4) = 335 kD (AF4-MLL)}, we suspect that anti-C9 recognizes the AF4-MLL reciprocal fusion protein in all t(4;11) lines tested.
AF4 and MLL-AF4 fusion proteins are localized in the nucleus, but not in the cytoplasm.
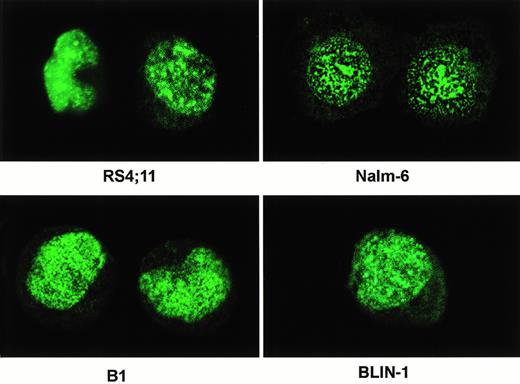
Purified anti-C15 was used to evaluate localization of the AF4 and MLL-AF4–encoded fusion proteins in immunofluorescence studies. All cell lines stained with the antibody showed a strong punctate fluorescence staining in nucleus, but not in cytoplasm (Fig 6). The immunostaining patterns were generally similar in all cells tested and there was no significant difference in the patterns and distribution of the labeling for cell lines with or without t(4;11) translocations. Further study will be required to establish whether the AF4 and MLL-AF4 proteins colocalize in the same subcellular compartment. It is also important to determine whether wild-type AF4 and the fusion proteins function in distinct cellular compartments.
Indirect immunofluorescence microscopy. Cultured cells in log phase were harvested and adjusted to 1 to 2 × 105/mL. The cells were attached to microscope slides by cytospin. The cells were probed with anti-C15 followed by goat antirabbit IgG conjugated to FITC. The image of the stained cells was obtained by a Bio-Rad MR C600 confocal microscope and processed using Adobe photoshop software. The cell lines used are indicated.
Indirect immunofluorescence microscopy. Cultured cells in log phase were harvested and adjusted to 1 to 2 × 105/mL. The cells were attached to microscope slides by cytospin. The cells were probed with anti-C15 followed by goat antirabbit IgG conjugated to FITC. The image of the stained cells was obtained by a Bio-Rad MR C600 confocal microscope and processed using Adobe photoshop software. The cell lines used are indicated.
DISCUSSION
In this study, we have produced and characterized specific antibodies to AF4-encoded proteins. We have also provided direct evidence that AF4-encoded proteins are present in a variety of human leukemic and nonleukemic cells, as demonstrated by immunoblottings and immunofluorescence microscopy. In Western blottings, anti-C15 and anti-C9 antibodies consistently recognized a protein of about 145 KD in all cell lines tested. Under high stringency conditions of washing, the anti-C15 antibody recognized a single 145-kD AF4 protein in Western blottings, which migrated as doublets when lower percentage gels (4%) were used, the antibody also detected a 240-kD protein in all cell lines with t(4;11) translocations. The molecular weight of the 240- and 145-kD proteins are consistent with predicted sizes of MLL-AF4 and AF4-encoded proteins based on previous DNA sequencing and Northern blotting analysis.2,4 22 Previous studies demonstrated the presence of 12 kb and 10.5 kb AF4 mRNA transcripts in cell lines tested. The 12-kb and 10.5-kb RNA transcripts identified in the previous studies are most likely to be the results of alternate splicing of AF4 mRNA, which subsequently encode the 145-kD and 125-kD proteins. The 12.5-kb mRNA transcript previously detected in all cell lines with t(4;11) translocations corresponds to the 240-kD protein detected in this study. Coprecipitation of the 45-kD protein with AF4 protein suggests the possibility that it may exert its biological function through interaction with a second protein.
The anti-C9 that was raised against N-terminal end of AF4 protein detected the protein of about 145 kD. However, it also bound to a 184-kD, a 75-kD, and a 50-kD protein in Western blottings. It is not clear whether these proteins are AF4-related. Thus far, mRNA species corresponding to these proteins have not been identified. It was of interest that anti-C9 reacted to a high molecular weight protein (>240 kD) in RS4;11 and Sem-k2 cells with t(4;11) translocations, while the protein was absent from non-t(4;11) cells. These observations suggest that anti-C9 is able to detect AF4-MLL reciprocal fusion protein if it is present.
Molecular analysis of the MLL and AF4 genes by our group and others3,10,11,12 22 indicates that the N-terminal portion of MLL fuses to C-terminal portion of AF4. The C15 clone used for expression and immunization was chosen to be downstream of all known breakpoints. Thus, anti-C15 recognizes the chimeric MLL-AF4 protein in all cell lines tested (B1, RS4;11, and Sem-k2). The MLL-AF4 chimeric proteins in RS4;11, AN4;11, and B1 cells are calculated to contain 2319, 2276, and 2234 amino acid residues, respectively. Fusion proteins seen in the Western blottings migrated as a single band in 4% SDS-PAGE gel (Fig 5) with apparent similar molecular weight, which is consistent with the predicted sizes and showing that the size difference of the proteins was too small to be resolved under the experimental conditions used.
Recently, Joh et al13 have generated rabbit polyclonal antibodies specific for N-terminal epitopes of MLL. These antibodies recognized a 240-kD protein in Western blotting. Butler et al14 produced a monoclonal antibody against a 15 amino acid peptide at N-terminal of MLL gene and the antibody (HRX 107) also recognized the 240-kD MLL-AF4 fusion protein in the Sem-k2 cell. It is of some interest that the quantity of MLL-AF4 fusion protein detected by our Western blotting is significantly less than that of the AF4 protein. The significance of these differences is not clear at present.
A gene previously cloned by Ma and Staudt6 from a cDNA library of a Burkitt’s lymphoma is closely related to the AF4 gene. Both genes share a high degree of homology (75% homology at C-terminal and 62% at N-terminal), contained a proline/serine-rich region and NLSs at virtually the same position. The LAF4 gene encodes a major protein of 135 kD. However, the expression of LAF4-encoded protein was shown to be restricted to lymphoid cells. As predicted from DNA sequence analysis, our Western blotting results indicate that LAF4 encoded a smaller protein than that detected by AF4 antibodies (data not shown).
Results of our experiments indicate that anti-C15 detects the AF4 protein in subcellular compartments in immunofluoresence microscopy. The subcellular localization of AF4 with punctate distribution in the nucleus is similar to that previously described for transcription-associated proteins. Other proteins with similar distribution are LAF4, MLL,13 and several other MLL partners including ENL/LT6,13,23 AF9/LT69,13and ELL.24 ELL is of interest because of the demonstration that the ELL gene encodes a RNA polymerase II enlongation factor.25 Consistent with a role for AF4 and LAF4 in transcriptional regulation is the previous observation by Ma and Staudt that AF4 has domains that activate transcription when fused to GAL4 DNA-binding domain. Of note is the observation that the transactivation domain of AF4 is retained within the MLL-AF4 fusion protein.6 Further studies will be necessary to determine the significance of the punctate compartmentalized staining seen with anti-AF4 antibody and other antibodies to transcription-related molecules. Previous studies with another transcription factor have demonstrated that phosphorylation of hepatic nuclear factor 4 is required for nuclear compartmentalization.26 It has been shown that the compartmentalized nuclear staining of the protein ofDrosophila polycomb (related to Drosophila trithorax) developed homogeneous nuclear staining after the protein was mutated.27
Finally, the observation that the anti-C15 antibody was able to detect the MLL-AF4 fusion proteins in leukemia lines studied demonstrates the potential use of this AF4-specifc antibody as a diagnostic reagent using Western blotting.
Supported in part by an Outstanding Investigator Grant Award No. R35 CA 49721 from the National Cancer Institute (to J.H.K.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to John H. Kersey, MD, University of Minnesota Cancer Center, Box 86 MAYO, 420 Delaware St, SE, Minneapolis, MN 55455.