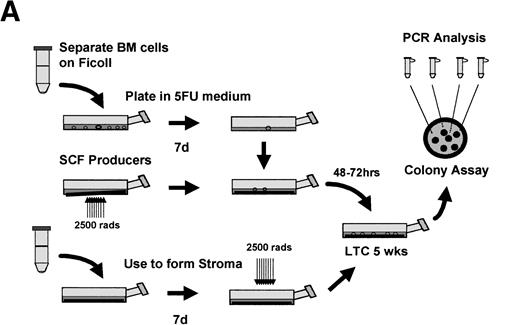
Abstract
Pluripotent hematopoietic stem cells (PHSC) are rare cells capable of multilineage differentiation, long-term reconstituting activity and extensive self-renewal. Such cells are the logical targets for many forms of corrective gene therapy, but are poor targets for retroviral mediated gene transfer owing to their quiescence, as retroviral transduction requires that the target cells be cycling. To try and surmount this problem we have constructed a retroviral producer line that expresses the membrane-bound form of human stem cell factor (SCF) on its cell surface. These cells are capable, therefore, of delivering a growth signal concomitant with recombinant retroviral vector particles. In this report we describe the use of this cell line to transduce a highly quiescent population of cells isolated from adult human bone marrow using the 5-fluorouracil (FU) resistance technique of Berardi et al. Quiescent cells selected using this technique were transduced by cocultivation with retroviral producers expressing surface bound SCF or with the parent cell line that does not. Following coculture, the cells were plated in long-term bone marrow culture for a further 5 weeks, before plating the nonadherent cells in semisolid media. Colonies forming in the semisolid media over the next 14 days were analyzed by polymerase chain reaction for the presence of the retroviral vector genome. Over six experiments, the transduction frequency of the quiescent 5-FU resistant cells using the SCF-expressing producer line averaged about 20%, whereas those transduced using the parent producer line showed evidence of reduced levels or no transduction.
A MAJOR OBSTACLE TO the development of gene therapy protocols for inherited hematopoietic disorders has been the relative difficulty of introducing new genetic material into human pluripotent hematopoietic stem cells (PHSC). Clinical gene marking and large animal studies have suggested that only about 0.1% to 1% of PHSC are routinely targeted.1,2 A major contributory factor to this low efficiency is the quiescence of PHSC3 and the inability of retroviral vectors to stably integrate into the genome of nondividing cells.4,5 Although, some alternative vectors (eg, AAV or HIV-based systems) are capable of infecting such cells, they all have serious attendant problems, making the use of retroviral vectors still an attractive proposition, especially as they remain the most highly developed of the vectors applicable to PHSC gene transfer and have been proven safe enough for some limited clinical usage.6-8 Moreover, as retroviral vectors have been used successfully in murine systems it seems likely that there is no fundamental problem to their application in human gene therapy protocols. To try and circumvent these difficulties, we have developed a number of retroviral producer cells lines that also express human recombinant stem cell factor (SCF)9,10 in its membrane-bound form.11 12 In this way we hoped to promote cell cycling in PHSC simultaneous with their exposure to retroviral vector and so facilitate their transduction.
Given that there is no unequivocal evidence indicating to what cytokines PHSC respond, the choice of SCF was dictated by a number of factors. Firstly, cells expressing high levels of SCF receptor are highly enriched for PHSC.13 Secondly, cells selected by their functional properties as human PHSC were shown to have high levels of surface SCF receptor.3 In the mouse,Steel mutants make no functional SCF with devastating consequences for their hematopoietic system.10,14 In addition, mice bearing the “dickie” allele of Steel make a biologically active soluble form of SCF but fail to make the membrane-bound form,15 yet they have a phenotype very similar to individuals that are unable to make SCF at all, suggesting that the membrane-associated form of the growth factor is more critical for the maintenance and differentiation of PHSC.12
This paper reports the successful transduction of quiescent bone marrow cells selected using the 5-fluorouracil (FU) technique of Berardi et al,3 by using a retroviral producer line expressing human recombinant SCF on its cell surface.
MATERIALS AND METHODS
Retroviral vectors and packaging lines.
Human SCF cDNA was obtained in a Bluescript SK-plasmid.16This was excised and inserted into an expression vector pREP8 (Invitrogen, San Diego, CA), which contained the histidinol resistance gene as a selectable marker. The resulting plasmid pREP8ΔSCF was then used to transfect the amphotropic retroviral producer cell line, 1MI, which is derived from the AM1217 packaging line and produces a retroviral vector (pBABE) carrying the p47phox gene as described previously,18 but lacking a neo selectable marker. The producer cells were transfected using the calcium phosphate precipitation method. Several clones were isolated in Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal calf serum (Sigma, St Louis, MO), containing histidinol (2.5 mmol/L) and were tested for SCF expression by immunofluorescence. The best of these (clone 2) was chosen for the following experiments and designated 1MI-ΔSCF. These were grown in DMEM and 10% fetal calf serum.
Northern and dot blotting.
RNA was extracted from murine retroviral producer fibroblasts by acid phenol extraction, ethanol precipitated, and resuspended in sterile distilled-deionized water that had been treated with diethyl pyrocarbonate. RNA samples (10 μg) were separated by electrophoresis on 1% agarose-formaldehyde gels, transferred to nylon membrane by capillary transfer, hybridized, washed, and exposed to x-ray film all as described previously.18 For dot blots, fresh culture medium was added to confluent cultures of retroviral producer cells. Following a 24-hour incubation, the supernatant was removed and filtered through a 0.45-μm filter. The retroviral supernatant was aliquoted and stored at −70°C. Supernatant was spotted under vacuum onto the surface of a nylon membrane using a dot blot manifold (GIBCO-BRL, Gaithersburg, MD). The filter was cross-linked with ultraviolet light using a Stratalinker (Stratagene Corp, La Jolla, CA) and hybridized as described above for Northern blots.
Proliferation assays.
Bioassay of SCF was performed by 3H thymidine incorporation essentially as described by Callard et al.19 Log-phase TF-1 cells (1 × 106) were washed twice with phosphate-buffered saline (PBS) to remove all traces of cytokine. Washed cells (5 × 105) were maintained in media without cytokines for 24 hours. A total of 1 × 104 (100 μL) and the quiescent cells were then dispensed in triplicate into a 24-well plate. The plates were incubated for 48 hours at 37°C in 5% CO2 in air, after which 1 μCi3H-TdR (specific activity, 5 Ci/mmol; Amersham International, UK) was added to each well. Following a further incubation period of 12 to 18 hours, the labeled cells were harvested onto glass-fiber discs using Dynatech Automash Cell Harvester (LabTech Intl, UK). The incorporated 3H thymidine was counted on a liquid scintillation counter (Wallac 1209 Rack Beta, Wallac, UK) and expressed as disintegrations per minute (dpm) after correction for quenching and efficiency of counting.
For experiments involving cocultivation, 24 hours before addition of TF-1 cells, 105 producer fibroblasts were plated into wells of a 24-well plate and allowed to adhere to the plastic. Following an overnight incubation the cells were irradiated as described elsewhere. In culture experiments using a Transwell (Costar, Cambridge, MA) fibroblasts were added to the plate well as above and the TF-1 cells were added to the insert, which is connected to the culture via a 3-μm semipermeable membrane.
Human bone marrow.
Normal human bone marrow cells were obtained from healthy adult volunteers. Use of these samples was approved by The Research Ethical Committee at Great Ormond Street Hospital and The Institute of Child Health. Light density mononuclear cells were obtained by centrifugation for 20 minutess at 2,000 rpm on Ficoll-paque (Pharmacia, Uppsala, Sweden).
Stromal cells.
Stroma was allowed to form over a period of 2 weeks from bone marrow cells separated on Ficoll, as described above and plated at 1 × 107 cells per T25 flask in modified McCoy’s 5A medium supplemented as below. Stromal monolayers were irradiated with 2,500 rads at 14 days.
CD34+ selection.
CD34+ cells were isolated from bone marrow cells separated on Ficoll by positive selection on a MiniMACs immunomagnetic column (Miltenyi Biotec, Surrey, UK), as described by the manufacturers.
5-FU treatment.
CD34+ cells or unfractionated bone marrow cells were selected in 5 FU, as described by Berardi et al.3 Briefly, cells were incubated for 7 days at 37°C, 5% CO2 in Iscove’s modified Dulbecco’s medium (IMDM; GIBCO) with 10% fetal calf serum (Sigma) supplemented with SCF (100 ng/mL; R&D Systems, Oxford, UK) and interleukin-3 (IL-3; 100 ng/mL) with or without 5-FU (0.6 mg/mL; David Bull Laboratories, Warwick, UK).
Characterization of cell cycle and SCF receptor status.
Cycle status for 5-FU treated cells and untreated cells was determined by 3H-thymidine incorporation. One microcurie of3H-thymidine was added to 200 μL of treated or untreated cells (2 to 4 × 107) and incubated for 24 hours. Cells were then spun down, washed in PBS, and pelleted onto glass slides using a Cytospin (Shandon Scientific, Runcorn, UK). SCF receptor status was determined by staining with antihuman SCF receptor antibody and fluorescein isothiocyanate (FITC)-conjugated anti-mouse Ig (Dako, Bucks, UK ). Photographs were taken on a fluorescence microscope (Olympus). Slides were dipped in photographic emulsion (50% solid emulsion, 2% glycerol in H2O) and developed after 1 week. Slides were counterstained with Wright’s stain or Toluidine Blue. Bright field images of cells were photographed. Counts on labeled cells were performed on 30 random views under oil immersion. Approximately 400 cells were counted.
Transduction with retroviral vector.
Selected or unselected cells were cocultured with the irradiated 1MI-ΔSCF, or the 1MI parent cell line, for 72 hours in supplemented McCoy’s 5A medium. Cells were then obtained, washed in PBS, and resuspended in 10 mL supplemented McCoy’s 5A medium.
Long-term marrow culture.
LTC-IC assays were performed by culturing transduced cells on irradiated stroma for 5 weeks in McCoy’s 5A medium supplemented with 12.5% fetal calf serum (Sigma), 12.5% horse serum (Sigma), 1% sodium bicarbonate (GIBCO), 1% minimum essential medium (MEM), sodium pyruvate (100 mmol/L; GIBCO), 0.4% MEM nonessential amino acids (10 mmol/L; GIBCO), 2 mmol/L L-glutamine (GIBCO), 1% MEM amino acids with L glutamine (GIBCO), 1% MEM vitamins solution (GIBCO), 100 U/mL penicillin (Sigma), 100 μg/mL streptomycin (Sigma), 0.25 μg/mL amphotericin B (Sigma), and 0.03% hydrocortisone (1 mg/mL in DMSO; Sigma). LTMC were incubated at 37°C, 5% CO2, and fed weekly by removal of 50% of the media. Nonadherent cells were spun down and returned to the culture in fresh media.
Methylcellulose assays.
Cells from LTMC were spun down and resuspended in 500 μL Iscove’s medium together with 2.5 mL STEMGEM medium (obtained from Dr J. Hatzfeld, Paris, France) containing 5 U IL-3, 30 U IL-6, 10 U granulocyte-macrophage colony-stimulating factor (GM-CSF), 6 to 12 U erythropoietin (EPO), 18 ng SCF, 30 μL iron-saturated transferrin, 2 mmol/L glutamine, 5 × 10−5 mol/L 2-mercapto-ethanol, 3 mg bovine serum albumin, 30% fetal calf serum, and 1% methylcellulose. Cells were incubated at 37°C, 5% CO2, and saturated humidity for 2 to 4 weeks.
Analysis of retroviral infection.
Efficiency of infection was determined by polymerase chain reaction (PCR) analysis of colonies or single cells picked from methylcellulose assays into lysis buffer (50 mmol/L Tris pH 8.0, 1 mmol/L EDTA, 0.5% Tween 20, 200 μg/mL proteinase K) incubated 1 hour at 55°C, then 10 minutes at 95°C. Successful transfer of the p47phox gene was detected by amplification of a region of the vector that included sequences of both retroviral and p47phox cDNA. A nested PCR strategy was also employed using a forward primer (ATGGCCAACCTTTAACGTCGGATG) derived from the gag region of the retroviral vector and a reverse primer taken from internal p47phox sequence (GTTTTATGGAACTCGTAGATCTCG). The nested upstream primer (GAGCACTGGAGGCCACCCAGT) was taken from the 5′ untranslated region of the p47phox cDNA. The expected size of the product of the first amplification was 454 bp and the final product size was 180 bp. PCR reactions were performed in 50-μL total volume, containing 1 μmol/L primers, 0.1 mmol/L dNTPs (Promega, Madison, WI), 1 × KCL buffer 1.5 mmol/L MgCl2, and 2 U Taq polymerase (Bioline, London, UK). PCR products were run on 2% agarose gels containing 1 μg/mL ethidium bromide in 1× TAE buffer (40 mmol/L Tris pH7.8, 20 mmol/L sodium acetate, 2 mmol/L EDTA) and visualized on a 300-nm ultraviolet transilluminator.
RESULTS
Construction of a retroviral producer cell line (1MI-ΔSCF) expressing human SCF on its cell surface.
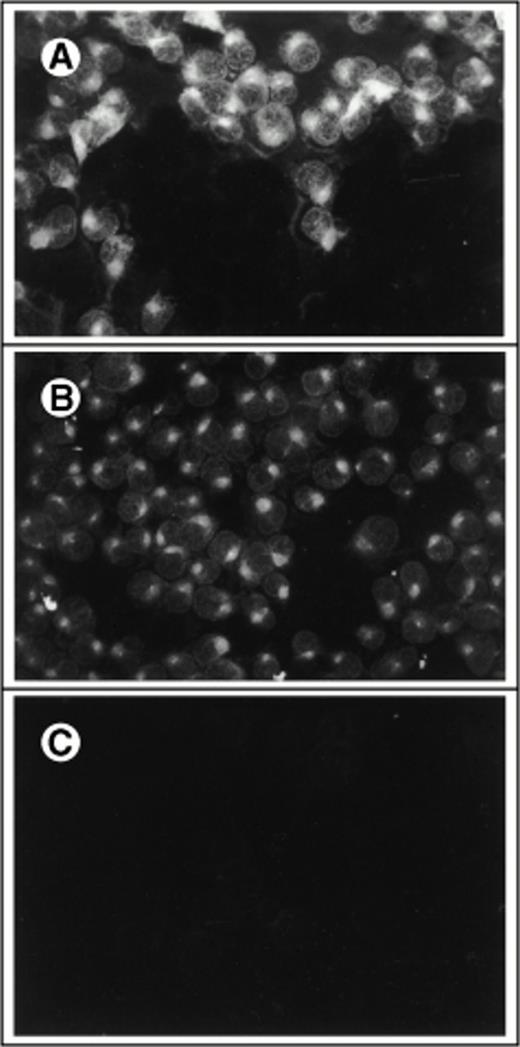
To develop a retroviral producer cell line expressing surface SCF, we first constructed a mammalian expression plasmid encoding the membrane-bound form of human SCF. The membrane-bound isoform of human SCF cDNA is an 814-bp fragment encoding a splice variant lacking exon 6. Exon 6 contains a proteolytic cleavage site that enables the molecule to be cleaved from the cell surface to produce the soluble growth factor. This cDNA was cloned into the expression vector pREP8 (Invitrogen) using HindIII and BamH1 (Fig 1A). pREP8 encodes a marker conferring resistance to the drug histidinol. This plasmid (pREP8-ΔSCF) was then introduced into our retroviral producer cell line 1MI, by calcium phosphate transfection. The 1MI cell line is a retroviral producer line derived from AM1217 by transfection with a retroviral vector encoding p47phox (Fig 1B), the molecule most commonly affected in the autosomal recessive form of chronic granulomatous disease. The transfected cells were then selected in histidinol and individual clones isolated by ring cloning. The resulting clones were tested for their expression of SCF by immunofluorescence using an anti-SCF antibody on permeabilized (Fig 2A through C) and unpermeabilized (Fig2D and E) cells to confirm the surface expression of SCF. Intense fluorescence was visible in transfected cells, shown both before (Fig2A) and after (Fig 2B) selection and cloning. Untransfected cells are just visible in the unselected culture and staining of the parent line is shown in Fig 2C. Note that in the permeabilized cells the staining indicates a major accumulation of SCF in a nuclear-adjacent region, probably the Golgi, whereas in unpermeabilized cells the strongest staining regions are the junctions between cells.
Construction of retroviral producer cells expressing membrane-bound SCF. (A) Schematic of plasmid pREP8-▵SCF. (B) Evolution of cell line 1MI-▵SCF from AM12 packaging line.
Construction of retroviral producer cells expressing membrane-bound SCF. (A) Schematic of plasmid pREP8-▵SCF. (B) Evolution of cell line 1MI-▵SCF from AM12 packaging line.
Immunofluorescent detection of SCF on retroviral producer cells. Fibroblasts were stained with FITC-conjugated antibody to human SCF with (A through C) or without (D and E) permeabilization and viewed under the fluorescence microscope (×400). (A) 1MI cells following transfection but before selection in histidinol; (B) 1MI-▵SCF cells following selection and cloning; (C) untransfected 1MI cells. (B) and (C) were photographed under exactly the same exposure conditions; (D) 1MI-▵SCF cells following selection and cloning; (E) untransfected 1MI cells. (E) Is overexposed to make the cells more visible.
Immunofluorescent detection of SCF on retroviral producer cells. Fibroblasts were stained with FITC-conjugated antibody to human SCF with (A through C) or without (D and E) permeabilization and viewed under the fluorescence microscope (×400). (A) 1MI cells following transfection but before selection in histidinol; (B) 1MI-▵SCF cells following selection and cloning; (C) untransfected 1MI cells. (B) and (C) were photographed under exactly the same exposure conditions; (D) 1MI-▵SCF cells following selection and cloning; (E) untransfected 1MI cells. (E) Is overexposed to make the cells more visible.
Expression of SCF was also analyzed by Northern blotting on RNA extracted from four independently arising clones (Fig 3A). The level of expression of SCF mRNA detected in the Northern blot was also in good agreement with the relative immunofluorescent intensity of the different clones. The retroviral titer of these four clones was also checked to ensure that the 1MI-ΔSCF cells were still able to generate retrovirus. Retroviral supernatants were used to prepare a dot blot, which was hybridized with a radiolabeled p47phox probe. All four clones showed only slight variations in titer from the parent 1MI line and in no case was an increase in titer observed. On the basis of these analyses the clone with the best balance of expression and titer, clone 2, was used for all the subsequent experiments; this cell line was termed 1MI-ΔSCF.
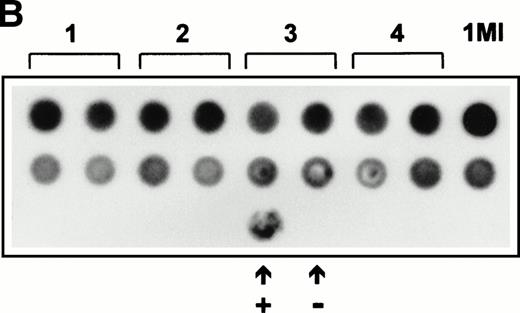
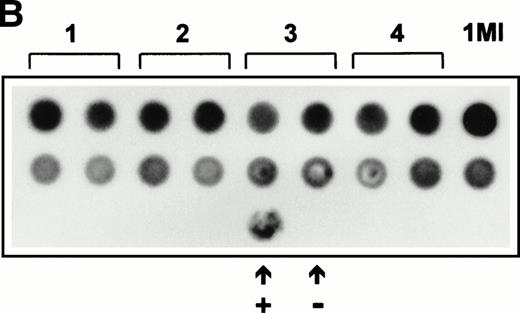
(A) Expression of SCF mRNA by retroviral producer cells. 1MI cells transfected with the plasmid pREP8▵SCF were selected in histidinol and individual clones isolated. RNA was extracted from four independently isolated clones and analyzed by Northern blot. The blot was hybridized to an SCF cDNA probe. Lane C, pREP8▵SCF plasmid DNA digested with BamH1 and HindIII to release the SCF cDNA fragment; lanes 1 through 4, four independent clones of transfected 1MI cells. (B) Retroviral titer of fibroblast clones isolated following transfection with pREP8▵SCF. Culture supernatants were collected from four independent clones of 1MI cells following transfection with plasmid pREP8▵SCF. Twenty-five microliters (top row) or 2.5 μL (bottom row) of supernatant was spotted onto the surface of a nylon filter using a dot blot manifold and hybridized with a probe for p47phox. Lanes 1 through 4, supernatants from four independently isolated clones; (+), retroviral genomic DNA plasmid containing p47phox cDNA ; (−) , wild-type retroviral genomic DNA plasmid. (C) Proliferation assay on TF-cells. Biological activity of the membrane-bound growth factor on the surface of the 1MI-▵SCF cell line was assayed by 3H thymidine incorporation into quiescent TF-1 cells. sSCF, TF-1 cells stimulated with 50 ng/mL recombinant human SCF; 1MI-▵SCF, TF-1 cells cocultured for 24 hours with the 1MI-▵SCF cell line; 1MI-▵SCF + sSCF, TF-1 cells cocultured for 24 hours with the 1MI-▵SCF cell line and 50 ng/mL recombinant human SCF; 1MI-▵SCF + Transwell, TF-1 cells cocultured for 24 hours with the 1MI-▵SCF cell line, where the TF-1 cells were suspended in a Transwell. Incorporation was corrected for background values obtained with unstimulated TF-cells and 1MI-▵SCF cells, as appropriate.
(A) Expression of SCF mRNA by retroviral producer cells. 1MI cells transfected with the plasmid pREP8▵SCF were selected in histidinol and individual clones isolated. RNA was extracted from four independently isolated clones and analyzed by Northern blot. The blot was hybridized to an SCF cDNA probe. Lane C, pREP8▵SCF plasmid DNA digested with BamH1 and HindIII to release the SCF cDNA fragment; lanes 1 through 4, four independent clones of transfected 1MI cells. (B) Retroviral titer of fibroblast clones isolated following transfection with pREP8▵SCF. Culture supernatants were collected from four independent clones of 1MI cells following transfection with plasmid pREP8▵SCF. Twenty-five microliters (top row) or 2.5 μL (bottom row) of supernatant was spotted onto the surface of a nylon filter using a dot blot manifold and hybridized with a probe for p47phox. Lanes 1 through 4, supernatants from four independently isolated clones; (+), retroviral genomic DNA plasmid containing p47phox cDNA ; (−) , wild-type retroviral genomic DNA plasmid. (C) Proliferation assay on TF-cells. Biological activity of the membrane-bound growth factor on the surface of the 1MI-▵SCF cell line was assayed by 3H thymidine incorporation into quiescent TF-1 cells. sSCF, TF-1 cells stimulated with 50 ng/mL recombinant human SCF; 1MI-▵SCF, TF-1 cells cocultured for 24 hours with the 1MI-▵SCF cell line; 1MI-▵SCF + sSCF, TF-1 cells cocultured for 24 hours with the 1MI-▵SCF cell line and 50 ng/mL recombinant human SCF; 1MI-▵SCF + Transwell, TF-1 cells cocultured for 24 hours with the 1MI-▵SCF cell line, where the TF-1 cells were suspended in a Transwell. Incorporation was corrected for background values obtained with unstimulated TF-cells and 1MI-▵SCF cells, as appropriate.
The biological activity of the surface SCF expressed by 1MI-ΔSCF was confirmed by proliferation assay on the TF-1 cell line20(Fig 3C). Compared with soluble human recombinant SCF, similar levels of stimulation were obtained by coculture of TF-1 with 1MI-ΔSCF cells. Addition of soluble SCF to TF-1/1MI-ΔSCF cocultures did not produce any increase in proliferation, suggesting that TF-1 cells were maximally stimulated by 1MI-ΔSCF. Furthermore, coculture of TF-1 with 1MI-ΔSCF cells using a Transwell to separate the target cells and prevent cell-cell contact significantly reduced the proliferative effect of the 1MI-ΔSCF cells.
Isolation of 5-FU–resistant marrow cells.
To obtain a target cell population of highly enriched stem cells with which to test our modified producers we adopted the method of Berardi et al.3 This method is attractive because it selects specifically for quiescent cells in the marrow and makes few assumptions about the surface phenotype of PHSC, which remains somewhat contentious.
Aspirated bone marrow cells were separated on Ficoll and the mononuclear cell fraction isolated. These cells were then incubated for 7 days in medium supplemented with soluble human SCF, IL-3, and 5-FU. A sample of cells from a such a culture at 4 days of incubation is shown in Fig 4A (top right panel), together with a control culture in which the 5-FU was omitted (Fig 4A, top left panel), by way of comparison. A strikingly different picture was observed in the presence of 5-FU with the majority of the surviving cells being terminally differentiated cells such as erythrocytes and polymorphs. Large cells with expanded nuclei and prominent cytoplasm frequent in the untreated cultures were conspicuous by their absence. To show that the cells surviving the 5-FU treatment were quiescent, 24 hours before harvesting, tritiated thymidine was added to the cultures to label any dividing cells. After 7 days of culture in the selective medium, the cells were collected onto microscope slides by Cytospin and dipped in autoradigraphic emulsion. Prominent clusters of silver grains could be seen in untreated control cultures (Fig 4A, middle left panel), whereas cells from the 5-FU–treated culture were unlabeled (Fig 4A, middle right panel). The cells were also analyzed for high-level expression of the SCF receptor, a marker for the 5-FU–resistant cells.3 As shown in Fig 4A, the 5-FU–resistant cells (bottom right panel) all showed bright fluorescence with a FITC-conjugated anti-SCF receptor antibody, whereas the untreated controls (bottom left panel) showed a wide spectrum of intensities, confirming that 5-FU selects a subset of cells with a higher level of expression of SCF receptor.
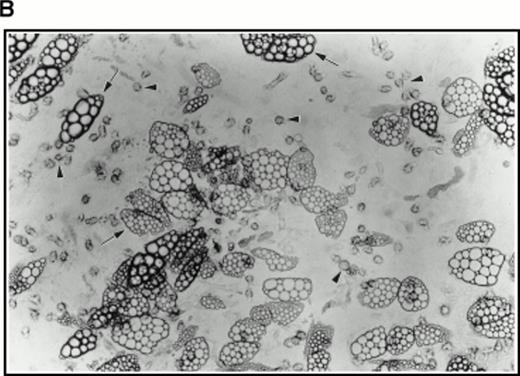
(A) Selection of quiescent bone marrow cells using 5-FU. Bone marrow mononuclear cells were incubated for 7 days in IMDM either supplemented with IL-3, SCF, and 5-FU to kill dividing cells (right-hand panels) or not (left-hand panels). At 4 days of incubation an aliquot was spun onto microscope slides and stained with Wright’s stain (top panels). Twenty-four hours before harvesting, 3H thymidine was added to the cultures. Following 7 days incubation cells were obtained and spun onto microscope slides. The slides were dipped in photographic emulsion and exposed for 7days before being developed, counterstained with Wright’s stain, and visualized under the light microscope (middle panels). (Bottom panels) Cells as above, but stained with antibody to c-kit and viewed under fluorescence. (B) Long-term culture. 5-FU–selected cells were plated on monolayers of bone marrow derived fibroblast as described in Materials and Methods and used to initiate long-term bone marrow cultures. The arrows show areas of extensive lipid deposition characteristic of these types of culture. Arrowheads identify “cobblestone areas,” clusters of developing hematopoietic cells.
(A) Selection of quiescent bone marrow cells using 5-FU. Bone marrow mononuclear cells were incubated for 7 days in IMDM either supplemented with IL-3, SCF, and 5-FU to kill dividing cells (right-hand panels) or not (left-hand panels). At 4 days of incubation an aliquot was spun onto microscope slides and stained with Wright’s stain (top panels). Twenty-four hours before harvesting, 3H thymidine was added to the cultures. Following 7 days incubation cells were obtained and spun onto microscope slides. The slides were dipped in photographic emulsion and exposed for 7days before being developed, counterstained with Wright’s stain, and visualized under the light microscope (middle panels). (Bottom panels) Cells as above, but stained with antibody to c-kit and viewed under fluorescence. (B) Long-term culture. 5-FU–selected cells were plated on monolayers of bone marrow derived fibroblast as described in Materials and Methods and used to initiate long-term bone marrow cultures. The arrows show areas of extensive lipid deposition characteristic of these types of culture. Arrowheads identify “cobblestone areas,” clusters of developing hematopoietic cells.
Very few cells survived this procedure. Indeed, as assayed by Trypan blue staining of a small sample, all visible cells were found to be nonviable. However, when used to initiate long-term cultures on preformed, irradiated stroma, this population of cells proved very active in establishing hematopoiesis as evidenced by extensive lipid deposition and characteristic “cobblestone” areas (Fig 4B).
Induction of cycling in 5-FU–resistant cells.
To successfully transduce the 5-FU–resistant cells it was necessary to induce them to cycle. To test whether this was possible using soluble cytokines, we used a standard mixture of IL-3, IL-6, and SCF, as well as coculturing the cells with the retroviral producer line 1MI and soluble SCF or the 1MI-ΔSCF line alone. To detect cells that divided during the period of exposure to growth factor, we used3H-thymidine incorporation and examined the cells under the light microscope for the deposition of silver grains (Fig 5). Only the cells cocultivated with the 1MI-ΔSCF producers showed evidence for 3H-thymidine incorporation in significant numbers, the average of two experiments being 28.4%. Much lower levels of cell division were detected using the parent cell line, 1MI, even when cells were cocultivated in the presence of added exogenous soluble SCF where only 6.6% of cells showed incorporation of label. Representative views are shown for cells stimulated by 1MI-ΔSCF (Fig 5, top panels) and 1MI (Fig 5, bottom panels). Stimulation, in liquid culture using some other combinations of cytokines, did not give rise to labeled cells (not shown).
3H Thymidine labeling of 5-FU cells. Bone marrow cells selected in 5-FU, IL-3, and SCF were incubated for 48 hours on monolayers of retroviral producer cells and labeled for 24 hours in the presence of 3H thymidine. (Top panels) Cells incubated with producer line 1MI-▵SCF. (Bottom panels) Cells incubated with producer line 1MI and exogenous soluble SCF (100 ng/mL). (Left-hand panels) Photographed at 400× original magnification; (right-hand panels) 1,000× (oil immersion). Arrowheads indicate examples of labeled cells.
3H Thymidine labeling of 5-FU cells. Bone marrow cells selected in 5-FU, IL-3, and SCF were incubated for 48 hours on monolayers of retroviral producer cells and labeled for 24 hours in the presence of 3H thymidine. (Top panels) Cells incubated with producer line 1MI-▵SCF. (Bottom panels) Cells incubated with producer line 1MI and exogenous soluble SCF (100 ng/mL). (Left-hand panels) Photographed at 400× original magnification; (right-hand panels) 1,000× (oil immersion). Arrowheads indicate examples of labeled cells.
Retroviral transduction of quiescent 5-FU–resistant bone marrow cells.
Having isolated a population highly enriched for quiescent stem cells, we then wished to test whether they could be transduced with retroviruses using our 1MI-ΔSCF producer line. A schematic of the experiment is shown in Fig 6A. 5-FU–resistant bone marrow cells, selected as described above, were incubated for 72 hours with retroviral producers either 1MI-ΔSCF or the parent 1MI cell line. Following incubation with the retroviral producers, the cells were plated onto an irradiated, heterologous human stromal monolayer. The cells were allowed to establish a long-term marrow culture and the culture continued for 5 weeks. After this time, cells in the supernatant were obtained and plated for colony assay in semisolid media. Colonies were allowed to develop for 2 to 3 weeks then plucked from the semisolid media, using a glass capillary tube, into individual microcentrifuge tubes containing cell dissociation buffer. This material was then subjected to analysis by PCR using a nested primer strategy. The initial amplification used an upstream primer specific for the gag region of the retroviral vector and a downstream primer specific for the p47phox cDNA insert. A small aliquot of this first reaction was then used for a second round of PCR amplification using the same downstream primer and a second upstream primer also derived from the p47phox cDNA. Four control amplifications were routinely performed, three containing different amounts of the retroviral genome in the form of plasmid DNA and one using genomic DNA from the 1MI producer line that contains an integrated copy of the retroviral genome. A typical example of the colony analysis is shown in Fig 6B and a more complete description of the transduction results in Table 1. It is apparent from these data that 5-FU–resistant cells could be successfully transduced and that a noticeable enhancement was obtained by using the 1MI-ΔSCF producers that express surface SCF. In two out of four experiments the background level of transduction achieved using the 1MI parent line was below our level of detection (<1/30). It seems likely that in the remaining two experiments, where a significant background transduction frequency was observed, the 5-FU failed to kill all the dividing cells in the marrow aspirate. If this background is subtracted from the 1MI-ΔSCF result transduction frequencies of 27% and 30%, respectively, are obtained. This correlates well with the results obtained in the other four experiments, which remained quite consistent, ranging from 13% to 25% of colonies scoring positive.
Retroviral transduction of 5-FU–selected bone marrow cells. (A) Schematic of experimental plan for analysis of retroviral transduction of 5-FU–selected bone marrow cells. (B) PCR analysis of 5-week LTC-IC–derived colonies plated in semisolid media. Individual colonies arising from 5-FU–selected cells plated in 5-week long-term cultures were picked into separate tubes and the genomic DNA subjected to PCR with primers specific for the retroviral genome. The PCR products were separated by electrophoresis on 2% agarose gels, stained with ethidium bromide, and visualized under ultraviolet light. Lanes 1 through 11, colonies picked from semisolid media, following transduction on 1MI-▵SCF cell line; lane 0, DNA (1 μg) from AM12 cell line; lane C, DNA (1μg) from 1MI parent cell line; lane M,HindIII digest of phage λ DNA and HaeIII digest of phage ▹X174 DNA used as size markers. The arrow denotes the expected size of the PCR product specific to the p47phox-containing retroviral genome.
Retroviral transduction of 5-FU–selected bone marrow cells. (A) Schematic of experimental plan for analysis of retroviral transduction of 5-FU–selected bone marrow cells. (B) PCR analysis of 5-week LTC-IC–derived colonies plated in semisolid media. Individual colonies arising from 5-FU–selected cells plated in 5-week long-term cultures were picked into separate tubes and the genomic DNA subjected to PCR with primers specific for the retroviral genome. The PCR products were separated by electrophoresis on 2% agarose gels, stained with ethidium bromide, and visualized under ultraviolet light. Lanes 1 through 11, colonies picked from semisolid media, following transduction on 1MI-▵SCF cell line; lane 0, DNA (1 μg) from AM12 cell line; lane C, DNA (1μg) from 1MI parent cell line; lane M,HindIII digest of phage λ DNA and HaeIII digest of phage ▹X174 DNA used as size markers. The arrow denotes the expected size of the PCR product specific to the p47phox-containing retroviral genome.
DISCUSSION
Stem cells, defined by their functional ability to provide long-term hematopoietic reconstitution, are difficult to isolate. Many in vitro assay systems and flow cytometry approaches have been developed to substitute for this cumbersome and time consuming methodology, but do not appear to identify true PHSC.21 For example, highly efficient transduction of LTC-IC or CD34+ populations by retroviruses has been observed in a number of cases,2,22-24but this has translated into very poor levels of retrovirally marked cells in transplanted individuals.2,8,23,25 The 5-FU technique of Berardi et al,3 which takes a functional approach to the isolation of stem cells by exploiting their quiescence, has proven straightforward to perform, does not require the use of sophisticated equipment, and is highly efficient. On the basis of the experiments described here though we should include a caveat that on some occasions up to one third of the cells purified with a single dose of 5-FU may still be cycling. This could probably be improved by adding fresh 5-FU during the selection period, or by preselecting for CD34+ cells, so that the density of cells in the selective media is significantly reduced. Preliminary results of ours have indicated that the latter strategy is indeed more efficient at removing cycling cells (N.W. and C.C., unpublished observation, January 1998)
An unfortunate consequence of the quiescence of the 5-FU–selected cells (and presumably all PHSC) is that it makes them difficult targets for retroviral transduction. We have sought to overcome this by stimulating these cells to divide simultaneously with their exposure to retrovirus. The stimulating growth factor we have used is SCF, which creates something of a paradox, as SCF is used in the 5-FU selection process, yet it is also clear that the cells that survive selection also express SCF receptors3 (and Fig 4).
SCF has classically been described as a survival factor rather than as a mitogen per se,26 though SCF has been shown to induce cycling directly in human CD34+ cells,27 and particular attention has been drawn to its ability to synergize with other growth factors to amplify their response.28-31 Some lines of evidence indicate that SCF does, indeed, play a role in the cycling of PHSC. For example, Bodine et al have shown that repopulating cells in the mouse are highly enriched in a “kit bright” fraction13 and that the absolute number of PHSC can be influenced by SCF.32 Crucially, Miller et al have shown that the SCF receptor is implicated in stem-cell cycling in a competitive repopulating mouse model using W/Wvmice.33
The murine Steel-dickie mutant provides evidence to suggest that it is the membrane-associated form of SCF rather than the soluble form that is vital for stem-cell function. These mice carry an interstitial deletion that render them incapable of synthesising a membrane-bound growth factor, whilst retaining a biologically active soluble form. Despite this they still display an anemia that is virtually identical to those animals that bear complete deletions of the SCF gene.11,34 The membrane-bound growth factor has also been shown to support early hematopoiesis more effectively than the soluble growth factor both with cell lines35 and with cultured primary murine marrow.12 Indeed, an increased level of retroviral gene transfer to rhesus monkey PHSC has been reported using membrane-bound SCF in a system similar to our own.36 One potentially crucial difference, however, is that these authors chose to modify a murine stromal cell line (derived from a steel mouse), thus losing any benefit of delivering retrovirus from the same cells as those stimulating growth. Additionally, stromal cells might be expected to exert potent inhibitory effects on stem-cell cycling, as in the normal stromal environment stem cells are predominantly quiescent.
Other investigators have been able to highlight the greater potency and longer lasting effects of the membrane-bound growth factor on target cells.37 This may originate from the fact that SCF receptors are rapidly internalized and only replaced by de novo protein synthesis,38 a process that may be much less efficient when the growth factor is itself anchored.
Whether SCF is capable on its own of inducing G0 stem cells to cycle remains unclear. A significant factor in the context of the experiments described here is the IL-6 known to be produced by retroviral packaging cell lines.39 The effects of murine IL-6 production could possibly account for the variable background level of transduction seen in the absence of SCF, though this is hardly consistent with the poor levels of gene transfer to PHSC seen by others, where recombinant IL-6 has often been included as a stimulatory factor. In addition, we have not performed transduction experiments under serum-free conditions, which still leaves open the possibility that the membrane-bound SCF is interacting with cytokines, known or as yet unknown, present in serum. We are continuing to refine the transduction protocol to reliably achieve higher levels of gene transfer to 5-FU–resistant cells but hope that the levels observed thus far still represent a step further on the road to gene therapy for disorders necessitating gene transfer to PHSC.
ACKNOWLEDGMENT
Our thanks to Prof Sunitha Wickramasinghe and Dr Vivien Hodges for their appraisal of this manuscript. Thanks also to Immunex Corp (Seattle, WA) for making available the SCF cDNA, Prof David Linch (UCL) for the kind gift of IL-3, to Prof Mary Collins (UCL) for supplying the AM12 packaging line, and to Profs Jacques and Antoinette Hatzfeld for the STEMGEM and advice on long-term culture.
Supported by the Medical Research Council, the Birth Defects Foundation, and the Chronic Granulomatous Disorder Research Trust.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Colin Casimir, PhD, Department of Haematology, Imperial College School of Medicine, St Mary’s Campus, Norfolk Place, London W2 1PG, UK; e-mail: c.casimir@ic.ac.uk.