Abstract
Contact system activation, in vitro, is triggered by activation of factor XII (FXII) on binding to an activator, such as negatively charged surfaces. A putative surface-binding site of FXII has been located within the amino acid residues 1-28 by identifying the epitope recognized by a monoclonal antibody (MoAb), B7C9, which inhibits kaolin-induced clotting activity. To further elucidate the role of the amino terminal binding site in the regulation of FXII activation, we have characterized a FXII recombinant protein (rFXII-▵19) deleted of the amino acid residues 3-19, which are encoded by the second exon of FXII gene. A plasmid encoding for rFXII-▵19 was constructed and expressed in HepG2 cells by using vaccinia virus. Purified rFXII-▵19 migrated as a single band of Mr 77,000 on sodium dodecyl sulfate (SDS)-polyacrylamide gel, did not bind to MoAb B7C9 immobilized on Protein A-Sepharose, thus confirming that it lacked the epitope for this MoAb, and had no amidolytic activity towards the chromogenic substrate S-2302 in the absence of activator. rFXII-▵19 specific clotting activity was lower (44%) than that of native FXII. The activation rate of rFXII-▵19 by kallikrein in the absence of dextran sulfate was about four times higher than that of full-length FXII and was increased in the presence of dextran sulfate. However, rFXII-▵19 underwent autoactivation in the presence of dextran sulfate. Labeled rFXII-▵19 bound to kaolin, which binding was equally well inhibited by either, rFXII-▵19 or full-length FXII (IC50 = 7.2 ± 2.2 nmol/L for both proteins). Accordingly, a synthetic peptide corresponding to FXII amino acid residues 3-19 did not inhibit the binding of labeled full-length FXII to kaolin. rFXII-▵19 generated a similar amount of FXIIa- and kallikrein-C1–inhibitor complexes in FXII-deficient plasma in the presence of kaolin, as did full-length FXII; but generated less factor XIa-C1–inhibitor complexes (50%) than full-length FXII. This impaired factor XI activation by rFXII-▵19a was also observed in a purified system and was independent of the presence of high molecular weight kininogen. Furthermore, the synthetic peptide 3-19, preincubated with factor XI, inhibited up to 30% activation of factor XI both in the purified system as well as in plasma. These results together indicate that amino acid residues 3-19 of FXII are involved in the activation of factor XI and do not contribute to the binding of FXII to negatively charged surfaces.
HUMAN FACTOR XII (FXII or Hageman factor), a serine-protease produced by the liver, circulates in plasma as a single-chain inactive zymogen (molecular weight [Mr] 80,000). On activation, the zymogen is converted to a two-chain active protease, activated FXII (FXIIa), which can activate several plasma cascade systems including the contact system, the intrinsic pathway of coagulation, fibrinolysis, and the complement system.1-3 Like other serine proteases belonging to plasma cascade systems, FXII consists of several structural domains, which starting from the amino terminus are: a leader peptide, a fibronectin domain type II, an epidermal growth factor–like domain, a fibronectin domain type I, a second epidermal growth factor–like domain, a kringle domain, a proline-rich region and the catalytic domain.4-6
Activation of FXII, in vitro, requires interaction with negatively charged surfaces to induce a conformational change rendering single-chain FXII more susceptible to proteolytic cleavage at amino acid residues 353-354.7-9 Cleavage at this site yields the two-chain enzyme FXIIa (Mr 80,000), which consists of a heavy chain (Mr 52,000) and a light chain (Mr 30,000) containing the active site.10,11 In vitro, proteolytic cleavage of single-chain FXII may occur via autoactivation or, more efficiently, by kallikrein.12-14 The pathophysiologic activating surface(s) is still unknown. FXII has been demonstrated to bind to endothelial cells and to neuthrophils,15-17 but it remains to be established if these cells may serve as activators of FXII in vivo. However, artificial negatively charged surfaces such as kaolin, ellagic acid, sulfatide micelles, and high molecular weight dextran sulfate, have been extensively used to study activation of FXII and of the contact system in vitro.14,18-21 The putative surface-binding site of FXII has been identified by mapping the epitope for monoclonal antibody (MoAb) B7C9, which inhibits kaolin-induced clotting activity of FXII.22 Initial studies with this MoAb pointed to the amino acid residues at positions 134-153 as being the binding site, which assignment was based on the recognition of the kallikrein-cleaved FXII fragments by immobilized B7C9.22Later studies with a FXII cDNA expression library in λgt11, located the epitope for MoAb B7C9, and hence a surface-binding region, between residues 1 and 28.23 However, there are no experimental data, for example as obtained with recombinant FXII molecules lacking the surface-binding site or with synthetic peptide mimicking its sequence, supporting the function of the putative binding site, except for a study showing that recombinant FXII proteins with gross deletions of various specific regulatory domains display reduced, but not absent, binding to negatively charged surfaces.24
To elucidate the role of the amino terminal binding site in the regulation of FXII activation, we have prepared and characterized a FXII recombinant protein, rFXII-Δ19, deleted of the amino acid residues 3-19, ie, the residues encoded by the second exon of FXII gene. The recombinant FXII protein though lacking (one of) the proposed binding site for negatively charged surfaces, bound normally to these surfaces, but showed an impaired clotting activity due to an impaired capability to interact with FXII substrate factor XI.
MATERIALS AND METHODS
General.
Restriction endonucleases, T4 DNA ligase, and the Klenow fragment of DNA polymerase I were purchased from New England Biolabs GmbH (Schwalbach/Taunus, Germany). Calf intestine alkaline phosphatase and T4 polynucleotide kinase were from Boehringer Mannheim (Mannheim, Germany). All of the enzymes were used according to manufacturer’s instructions. 125I was from Amersham Intl (Buckinghamshire, UK). Culture medium and fetal calf serum were obtained from GIBCO Laboratories (Breda, The Netherlands). Cyanogen bromide (CNBr)-activated Sepharose 4B, Protein G-Sepharose, Protein A-Sepharose, and dextran sulfate (Mw 500,000) were from Pharmacia Fine Chemicals (Uppsala, Sweden), trypsin and soybean trypsin inhibitor (SBTI) from Sigma Chemical Co (St Louis, MO). Factor XII- and factor XI-deficient plasma were obtained from George King Biomedical Inc (Overland Park, KS), hexadimethrine bromide (Polybrene) was obtained from Janssen Chimica (Beerse, Belgium). The chromogenic substrates H-D-Pro-Phe-Arg-p-nitroanilide (S-2302) and pyro-Glu-Pro-Arg-p-nitroanilide (S-2366) were purchased from Chromogenix AB (Molndal, Sweden). Factor XI was from Kordia Laboratory Supplies, Leiden, The Netherlands.
Construction of plasmid carrying rFXII-Δ19 sequence.
To obtain a plasmid carrying the sequences encoding for rFXII-Δ19 (a FXII protein deleted of amino acid residues 3-19, corresponding to the second exon of FXII gene) the plasmid pBFXII25 was digested with HincII and Nco I restriction endonucleases, the cDNA fragment comprising nucleotides 48-552 (numbering according to Citarella et al25) was isolated and digested with the restriction endonuclease MaeIII to remove a fragment containing nucleotides 48-126 corresponding to the second exon of FXII gene. The cDNA fragment containing nucleotides 127-552 was ligated into pBFXII,HincII-Nco I digested, together with a synthetic deoxyribonucleotide to restore the correct reading frame and the proper cleavage site after the leader peptide. The synthetic deoxyribonucleotide was obtained by annealing the two chemically synthesized complementary deoxyribonucleotides, 5′-AACACTTTCGATTCCAGTTCTCACT-3′ and 3′-TTGTGAAAGCTAAGGTCAAGAGTGACAGTG-5′. As confirmed by sequence analysis according to Sanger,26 the FXII cDNA inserted in pBFXII-Δ19 was 1,768 bp in length and coded for the complete leader peptide (amino acid residues −19 to +2 ) followed by amino acid residues 20-596. Recombinant vaccinia viruses carrying the cDNA sequence coding for rFXII-Δ19 (vFXII.Δ19) were obtained as described.25
Production of recombinant FXII proteins.
The human hepatoma cell line HepG2, grown to subconfluence in Dulbecco’s modified Eagle’s medium containing 10% (vol/vol) fetal calf serum, was infected with the recombinant viruses carrying the sequences coding for rFXII (full-length FXII) or rFXII-Δ19 at a multiplicity ratio of 10 plaque-forming units/cell. Six hours thereafter, medium was replaced with the same medium but without fetal calf serum, and the infected cells were incubated for 48 hours, before harvesting medium.
Electrophoretic analysis of immunoprecipitated recombinant proteins.
Vaccinia virus-infected HepG2 cells were labeled with 50 μCi [L-35S] methionine/mL medium for 6 hours at 37°C. After radiolabeling, media were harvested and immunoprecipitated with different MoAbs bound to protein A-Sepharose. The proteins immunoprecipitated by immobilized MoAbs were electrophoresed on 12% (wt/vol) sodium dodecyl sulfate (SDS)-polyacrylamide gel under reducing conditions and visualized by autoradiography using x-ray films.
MoAbs.
Several MoAbs against different regions of FXII were used to purify, quantify, and characterize the recombinant FXII proteins. MoAbs KOK5 and F1, which recognize epitopes localized in the fibronectin type II and in the kringle domain, respectively, have been described (manuscript submitted).27 MoAb B7C922 was kindly provided by Dr Robin A. Pixley (Temple University, Philadelphia, PA). MoAbs OT2 and F3 are directed against epitopes localized on the light chain of FXII.24,28 MoAbs were precipitated from hybridoma-conditioned medium by 50% (wt/vol) ammonium sulfate, extensively dialyzed against phosphate-buffered saline (PBS), and affinity-purified on Protein G-Sepharose according to the manufacturer’s instructions. MoAbs were biotinylated as described.29
Purification of proteins.
Recombinant rFXII-Δ19 protein was purified from HepG2 cell-conditioned medium by affinity chromatography using MoAb OT2 coupled to CNBr-activated Sepharose 4B according to a procedure described earlier.24 The purified rFXII-Δ19 preparation was analyzed by electrophoresis on 10% to 15% SDS-polyacrylamide gels.
FXII and prekallikrein were immunopurified from 150 mL of human citrated plasma.27,30 Prekallikrein was converted into kallikrein as described.27 High molecular weight kininogen (HK), prepared according to Kerbiriou and Griffin,31 was a gift from Dr B.N. Bouma (Academic Hospital, Utrecht, The Netherlands). All of the protein preparations used were more than 95% homogeneous as determined by SDS-polyacrylamide gel electrophoresis.
The amount of FXII and rFXII-Δ19 present in purified preparations was assessed both by measuring the absorbance at 280 nm using the absorbance coefficient A 1%cm 14.2 and by enzyme-linked immunosorbent assay (ELISA) as described.24rFXII-Δ19 preparations contained less than 0.1% of native FXII as was assessed using a sensitive sandwich ELISA with MoAb B7C9 as the capturing antibody and biotinylated MoAb F3 as the detecting antibody.
Purified FXII and rFXII-Δ19 were labeled with 125I by the chloramine-T method. Free label was removed by dialysis against PBS (10 mmol/L sodium phosphate, 140 mmol/L NaCl, pH 7.4), containing 0.1% (wt/vol) Tween-20. The specific activity of the labeled preparations was 4.2 × 107 cpm per μg of FXII and 8.6 × 107 cpm per μg of rFXII-Δ19, respectively. Labeled proteins were electrophoresed on SDS-polyacrylamide gel and visualized by autoradiography to assess the quality of preparation.
Peptides.
Synthetic peptides were synthesized at the synthetic peptides facility of the Amsterdam-Leiden Institute for Immunology by Dr J.W. Drijfhout (Department of Immunohaematology and Blood Bank, University Hospital Leiden, Leiden, The Netherlands). Synthetic peptides were made on Abimed 422 multiple peptide synthesizer (Abimed, Langenfeld, Germany) as described.32 The peptides were isolated and purified by repeated ether precipitations, dissolved in 10% acetic acid, and lyophilized. The amino acid content was checked by reverse phase high-performance liquid chromatography (HPLC). Finally, the peptides were resuspended in distilled water and stored in aliquots at −70°C. The concentration of synthetic peptides was determined by BCA protein assay reagent (Pierce, Rockford, IL).
Clotting assay for FXII proteins.
FXII-specific coagulant activity was determined by a modification of the activated partial thromboplastin time as described.24To study the effect of MoAb B7C9 on the clotting activity, the purified proteins (400 nmol/L) were preincubated 15 minutes with serial dilutions of MoAb B7C9 in PBS after which three dilutions of the mixture were assayed for the remaining clotting activity.
FXII binding to kaolin.
Binding of labeled FXII proteins to kaolin was determined as described33 with minor modifications. Briefly, 50 μL of fivefold dilutions of nonlabeled proteins in PBS 0.1% (wt/vol) Tween-20 (PT) were added to 50 μL of 125I-FXII or125I-rFXII-Δ19 (0.025 pmol/mL) in 0.5 mL polypropylene tubes and then incubated with 50 μL of kaolin (0.032 mg/mL in PT) for 10 minutes at room temperature. The tubes were centrifuged for 2 minutes at 10,000g, and the pellets were counted for radioactivity. Binding was expressed as the percentage of the total counts added. Under these experimental conditions, no significant binding of labeled proteins to the tubes (less than 0.5%) was observed. The efficiency of the inhibition of the binding was analyzed by computing the regression line of the binding curve and establishing the amount of the competitor able to inhibit the binding of the tracer molecule to 50% (IC50).33
FXII activation by kallikrein.
The kinetics of activation of FXII proteins by kallikrein were determined as described.14 Briefly, 25 μL of FXII (125 nmol/L) were added to 25 μL of dextran sulfate (DS500) (50 μg/mL) and 25 μL of buffer to yield final concentrations of 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.1% (wt/vol) Tween-20, pH 7.8, in Dynatech (Plochingen, Germany) microplates (96 wells). After a 5-minute preincubation at 37°C, 25 μL of prewarmed kallikrein (12.5 nmol/L) were added to start the reaction. After various time intervals, the amount of FXIIa formed was determined from the rate of hydrolysis of the chromogenic substrate S-2302: 50 μL of assay mixture (S-2302 1 mmol/L, SBTI 0.1 mg/mL in Tris 50 mmol/L, NaCl 150 mmol/L, pH 7.8) were added to the wells and the increase in absorbance at 405 nm was recorded at time intervals of 2 minutes by a Bio-Kinetics Reader (Biotek Instruments Inc) set at 37°C. Under these conditions, the rates of increase in absorbance were constant for at least 10 minutes and were used to calculate the amount of FXIIa on the basis of a calibration curve obtained with known amounts of fully activated FXII. In control experiments, it was established that the amount of SBTI added was sufficient to prevent conversion of the chromogenic substrate by kallikrein at a concentration of 100 nmol/L.
FXII autoactivation.
Autoactivation of FXII proteins was studied in a 96-well microtiter plate: 25 μL of FXII proteins (400 nmol/L), 25 μL of twofold serial dilutions of DS500 and 50 μL of buffer yielding final concentrations of 50 mmol/L Tris-HCl, 50 mmol/L NaCl, 0.1% (wt/vol) Tween-20, pH 7.8, were incubated for 60 minutes at 37°C. Thereafter, 50 μL of assay mixture (see above) were added to the wells; the increase in absorbance at 405 nm was recorded at time intervals of 2 minutes and the amount of FXIIa calculated as described above.
Activation of contact system in plasma.
The capability of rFXII-Δ19 protein to activate contact system was assessed by measuring the generation of FXIIa-C1–inhibitor, kallikrein-C1–inhibitor, and factor XIa-C1–inhibitor complexes in FXII-deficient plasma reconstituted with either native FXII or the recombinant protein. To this, 20 μL of serial dilutions of FXII proteins in PT were added to 40 μL of FXII-deficient plasma and then incubated with 20 μL of kaolin (5 mg/mL in PT) for 20 minutes at 37°C. The reaction was stopped by addition of 120 μL of stop solution (PBS, 0.1 mg/mL SBTI, 0.05% [wt/vol] Polybrene), followed by centrifugation for 2 minutes at 10,000g to discard the kaolin pellet. The amount of FXIIa-C1–inhibitor and kallikrein-C1–inhibitor complexes generated in EDTA-plasma was determined by ELISA developed as a modification of previously described radioimmunoassays (Minnema M, et al, manuscript submitted). Sample dilutions were tested in duplicate and results were calculated by comparison with an inhouse standard that consisted of normal pooled human plasma fully activated by DS500 (50 μg/mL). Generation of factor XIa-C1–inhibitor complexes was determined by an ELISA using kaolin-activated reference plasma.34
To study the effect of the synthetic peptide 3-19 on the activation of factor XI in plasma, 10 μL of serial dilutions of peptides in PT (16 mmol/L to 0.2 mmol/L) were incubated with 10 μL of purified factor XI (160 nmol/L) overnight at 4°C. Thereafter, 40 μL of factor XI-deficient plasma were added and contact activation was started by adding 20 μL of kaolin (5 mg/mL). After a 20-minute incubation at 37°C, the reaction was stopped by adding 120 μL of stop solution followed by centrifugation to discard the kaolin pellet and the amount of protease-C1–inhibitor complexes generated was measured.
Factor XI activation by FXII proteins.
The rate of factor XI activation by FXIIa was determined as described35 with minor modifications. Briefly, FXII and rFXII-Δ19 (50 nmol/L) were first activated by incubation with DS500 (1 μg/mL) in 50 mmol/L Tris-HCl, 50 mmol/L NaCl, 0.1% (wt/vol) Tween-20 for 90 minutes at 37°C, after which time the activation of the enzymes was tested (to be the same) by measuring the amidolytic activity toward the chromogenic substrate S-2302. A total of 25 μL of the mixture 1 to 20 diluted (containing 2.5 nmol/L FXIIa [rFXII-Δ19a] and 0.05 μg/mL of DS500) were added to 25 μL of purified human factor XI (40 to 80 nmol/L) preincubated 5 minutes at room temperature with 25 μL of HK (16 to 33 nmol/L, respectively) in Dynatech (Plochingen, Germany) microplates (96 wells). The buffer final concentrations were 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.1% (wt/vol) Tween-20, pH 7.8. After a 15-minute incubation at 37°C, the reaction was stopped by adding 25 μL of MoAb OT2 (100 nmol/L) (which blocks the amidolytic activity of FXIIa30) and the rate of factor XI activation was measured by adding 50 μL of the chromogenic substrate S-2366 (1 mmol/L) and recording the increase in absorbance at 405 nm at time intervals of 5 minutes. Factor XIa generation was quantitated from a standard curve prepared using purified factor XIa. In control experiments, it was established that the amount of MoAb OT2 added was sufficient to prevent conversion of the chromogenic substrate by FXIIa at a concentration of 10 nmol/L.
RESULTS
Production and characterization of recombinant FXII proteins.
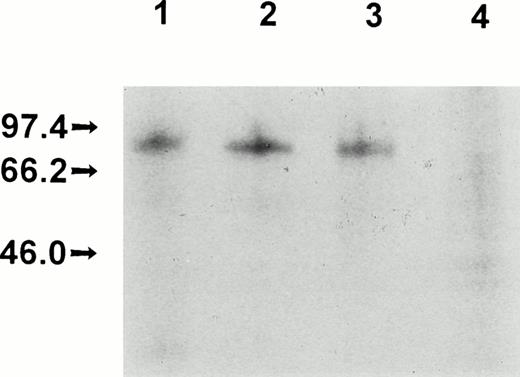
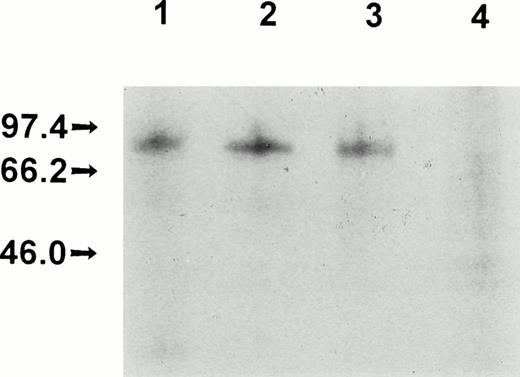
Production, purification, and a partial functional characterization of the recombinant full-length FXII (rFXII) have been detailed in previous publications by this laboratory.24,25 The recombinant deletion mutant rFXII-Δ19 was produced by infecting HepG2 cells with recombinant vaccinia viruses carrying the FXII cDNA deleted of the sequences of the second exon encoding for amino acid residues 3-19. rFXII and rFXII-Δ19 were labeled in vivo and were immunoprecipitated from the culture media with two protein A Sepharose-bound MoAbs directed against the heavy chain region of FXII. As shown in Fig 1, both proteins were immunoprecipitated with MoAb KOK5 and rFXII-Δ19 showed the expected molecular size (Mr, approximately 77,000). However, rFXII-Δ19 was not recognized by MoAb B7C9, which is in agreement with a previous report showing the amino acid residues 1-28 to contain the epitope for MoAb B7C9.23 The recombinant protein rFXII-Δ19 was purified from the culture medium of infected HepG2 cells by immunoaffinity-chromatography. Purified rFXII-Δ19 was homogeneous and consisted of a single peptide chain as assessed by SDS-polyacrylamide gel electrophoresis (not shown). Purified rFXII-Δ19 did not show a catalytic activity towards the chromogenic substrate S-2302 before activation. On activation by limited proteolysis with trypsin,24 rFXII-Δ19 showed a specific amidolytic activity (ΔA − min−1nmol−1= 15.1 ± 0.9) similar to that of native FXII (Table 1). A similar activity was found when rFXII-Δ19 was optimally activated by kallikrein in the presence of DS500 (data not shown).
Electrophoretic analysis of recombinant FXII proteins. Vaccinia virus-infected HepG2 cells were metabolically labeled and the culture media were immunoprecipitated and electrophoresed as indicated in Materials and Methods. Culture media from HepG2 cells infected with vFXII (lane 1) or with vFXII.▵19 (lane 2) immunoprecipitated with MoAb KOK5; culture media from HepG2 cells infected with vFXII (lane 3) or with vFXII.▵19 (lane 4) immunoprecipitated with MoAb B7C9.
Electrophoretic analysis of recombinant FXII proteins. Vaccinia virus-infected HepG2 cells were metabolically labeled and the culture media were immunoprecipitated and electrophoresed as indicated in Materials and Methods. Culture media from HepG2 cells infected with vFXII (lane 1) or with vFXII.▵19 (lane 2) immunoprecipitated with MoAb KOK5; culture media from HepG2 cells infected with vFXII (lane 3) or with vFXII.▵19 (lane 4) immunoprecipitated with MoAb B7C9.
Clotting activity of rFXII-Δ19.
Because MoAb B7C9 was shown to inhibit kaolin-induced FXII clotting activity, we asked whether rFXII-Δ19, lacking the amino acid residues recognized by MoAb B7C9, had an impaired clotting activity. Clotting activity was determined using a modified kaolin-activated partial thromboplastin time. As shown in Table 1, full-length recombinant FXII, rFXII, had a specific clotting activity comparable to that of plasma FXII, whereas rFXII-Δ19 specific clotting activity was 44% of that of native FXII. Furthermore, on preincubation with MoAb B7C9 (at a protein:MoAb ratio 1:10), the clotting activity of plasma FXII and rFXII was inhibited to 55%, whereas the clotting activity of rFXII-Δ19, although decreased itself, was not affected (data not shown) consistent with the observation (see above) that this protein lacks the epitope for MoAb B7C9.
Binding of rFXII-Δ19 to kaolin.
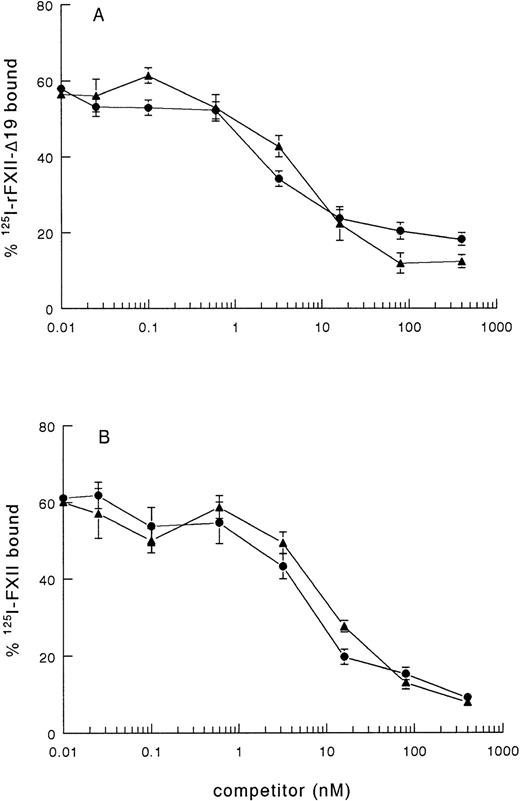
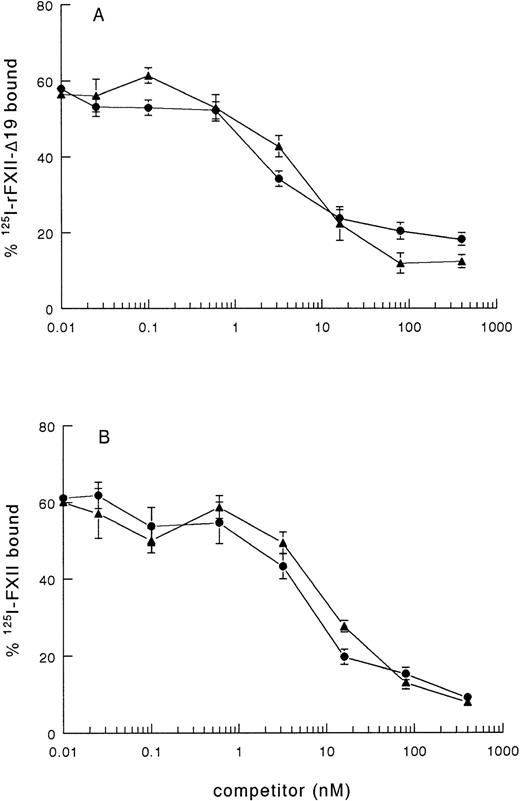
As rFXII-Δ19, lacking amino acid residues 3-19, possibly lacks a proposed binding site for negatively charged surfaces, the impaired clotting activity of rFXII-Δ19 might be due to impaired binding to kaolin. Therefore, we investigated the binding of radiolabeled rFXII-Δ19 to kaolin. 125I-rFXII–Δ19 did bind to kaolin, which binding was inhibited in a similar dose-dependent manner by either unlabeled FXII or rFXII-Δ19 (Fig 2A), but not by bovine serum albumin used as control (not shown). As well, rFXII-Δ19 inhibited the binding of 125I-FXII to kaolin (Fig 2B). The IC50 was the same for FXII and rFXII-Δ19 (IC50 = 7.2 ± 2.2 nmol/L), which indicated that both proteins bound to kaolin with the same efficiency. These results suggested that amino acid residues 3-19 were not involved in the binding of FXII to kaolin. Further, a synthetic peptide corresponding to residues 3-19 did not inhibit the binding of labeled FXII to kaolin even at a 106-fold molar excess (not shown).
Binding of 125I-FXII and125I-rFXII-▵19 to kaolin and inhibition by unlabeled proteins. A volume of 50 μL of fivefold dilutions of nonlabeled proteins in PBS 0.1 % (wt/vol) Tween-20 (PT) was added to 50 μL of125I-rFXII-▵19 (A) or 125I-FXII (B) (0.025 pmol/mL) and then incubated with 50 μL of kaolin (0.032 mg/mL in PT) for 10 minutes at room temperature. The tubes were centrifuged for 2 minutes at 10,000g and the pellets were counted for radioactivity. Binding was expressed as the percentage of the total counts added and are the means ± standard error (SE) of two independent experiments in duplicate. Symbols: FXII (•), rFXII-▵19 (▴).
Binding of 125I-FXII and125I-rFXII-▵19 to kaolin and inhibition by unlabeled proteins. A volume of 50 μL of fivefold dilutions of nonlabeled proteins in PBS 0.1 % (wt/vol) Tween-20 (PT) was added to 50 μL of125I-rFXII-▵19 (A) or 125I-FXII (B) (0.025 pmol/mL) and then incubated with 50 μL of kaolin (0.032 mg/mL in PT) for 10 minutes at room temperature. The tubes were centrifuged for 2 minutes at 10,000g and the pellets were counted for radioactivity. Binding was expressed as the percentage of the total counts added and are the means ± standard error (SE) of two independent experiments in duplicate. Symbols: FXII (•), rFXII-▵19 (▴).
Activation of rFXII-Δ19 by kallikrein.
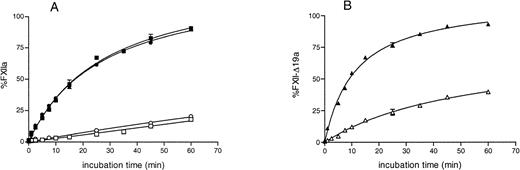
To elucidate further the cause of the diminished specific clotting activity of rFXII-Δ19, we studied the activation of the purified recombinant protein by kallikrein, together with that of rFXII and plasma FXII, in the presence or in the absence of a negatively charged surface, such as DS500.
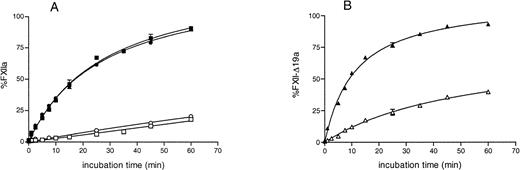
The activation rate of recombinant full-length FXII by kallikrein alone was low and, similar to that of plasma FXII, was enhanced in the presence of DS00 (Fig 3A). The activation rate of rFXII-Δ19 by kallikrein was about four times higher than that of rFXII in the absence of DS500 and was enhanced in the presence of DS500, reaching levels similar to that of rFXII (Fig 3B). These results indicated that rFXII-Δ19 had a configuration more susceptible to cleavage by kallikrein than full-length FXII and was efficiently activated by this enzyme in the presence of negatively charged surfaces.
Activation of purified recombinant FXII proteins by kallikrein. FXII recombinant proteins (125 nmol/L) were activated with kallikrein (12.5 nmol/L) as described in Materials and Methods in the presence (filled symbols) or in the absence (open symbols) of DS500. At several time intervals, the reaction was stopped by adding 50 μL of assay mixture containing SBTI and S-2302, and the increase in absorbance at 405 nm was immediately recorded. Results are expressed as the percentage of the maximum amount of FXIIa present in the wells after full activation and represent the mean ± SE of three experiments. The absence of standard error bars indicates that the variation was too little to portray visually. Symbols: plasma FXII (•), rFXII (▪), rFXII-▵19 (▴).
Activation of purified recombinant FXII proteins by kallikrein. FXII recombinant proteins (125 nmol/L) were activated with kallikrein (12.5 nmol/L) as described in Materials and Methods in the presence (filled symbols) or in the absence (open symbols) of DS500. At several time intervals, the reaction was stopped by adding 50 μL of assay mixture containing SBTI and S-2302, and the increase in absorbance at 405 nm was immediately recorded. Results are expressed as the percentage of the maximum amount of FXIIa present in the wells after full activation and represent the mean ± SE of three experiments. The absence of standard error bars indicates that the variation was too little to portray visually. Symbols: plasma FXII (•), rFXII (▪), rFXII-▵19 (▴).
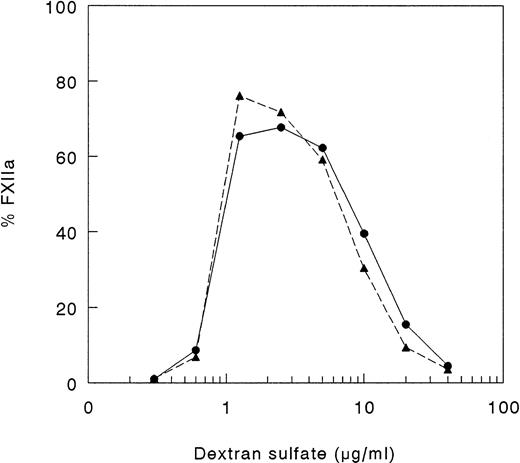
Autoactivation of rFXII-Δ19.
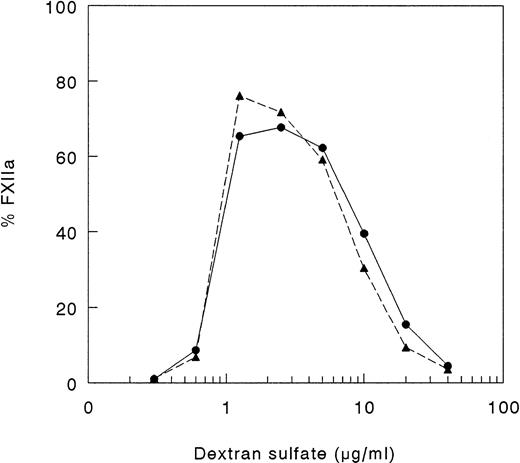
Because, in vitro, proteolytic cleavage of single-chain FXII may be mediated not only by kallikrein, but also by autoactivation, we studied the capability of the recombinant FXII protein to undergo autoactivation in the presence of DS500. As shown in Fig 4, FXII and rFXII-Δ19 did undergo autoactivation in the presence of DS500. The rate of autoactivation and optimum DS500 concentration able to induce autoactivation (1 to 2 μg/mL) were similar for FXII and rFXII-Δ19 and were consistent with the proposed template model.14 36
Autoactivation of FXII proteins. A total of 25 μL of FXII proteins (400 nmol/L), 25 μL of twofold serial dilutions of DS 500 and 50 μL of buffer yielding final concentrations of 50 mmol/L Tris-HCl, 50 mmol/L NaCl, 0.1% (wt/vol) Tween-20, pH 7.8, were incubated for 60 minutes at 37°C. Thereafter, 50 μL of assay mixture containing S2302 were added and the increase in absorbance at 405 nm was immediately recorded. Results are expressed as percentage of total FXII activity and represent the mean of two experiments. Symbols: FXII (•), rFXII-▵19 (▴).
Autoactivation of FXII proteins. A total of 25 μL of FXII proteins (400 nmol/L), 25 μL of twofold serial dilutions of DS 500 and 50 μL of buffer yielding final concentrations of 50 mmol/L Tris-HCl, 50 mmol/L NaCl, 0.1% (wt/vol) Tween-20, pH 7.8, were incubated for 60 minutes at 37°C. Thereafter, 50 μL of assay mixture containing S2302 were added and the increase in absorbance at 405 nm was immediately recorded. Results are expressed as percentage of total FXII activity and represent the mean of two experiments. Symbols: FXII (•), rFXII-▵19 (▴).
Contact system activation in plasma by rFXII-Δ19.
The results of the above experiments indicated that rFXII-Δ19 was able to bind to negatively charged surfaces, to undergo autoactivation, and to be activated by kallikrein. All together, these results indicated that the amino acid residues 3-19 do not harbor the binding site for negatively charged surfaces and the impaired clotting activity of rFXII-Δ19 was not due to reduced binding to kaolin. Therefore, we hypothesized that the impaired clotting activity of this protein might be due to an impaired interaction with FXII substrates, ie, factor XI or prekallikrein.
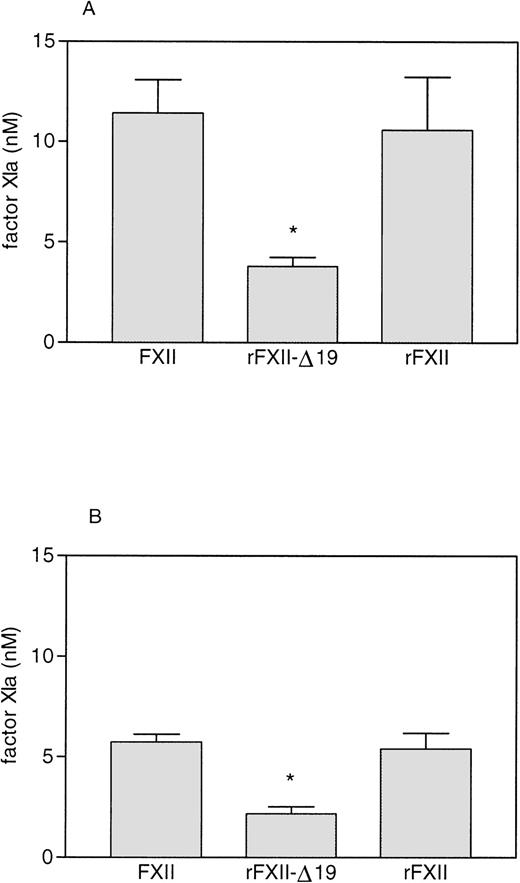
To answer this question, we investigated the activation of prekallikrein and factor XI in FXII-deficient plasma, reconstituted with rFXII-Δ19. The activation of the contact system proteases was evaluated by measuring the generation of FXIIa-, factor XIa-, and kallikrein-C1–inhibitor complexes. We found that equal amounts of FXII and rFXII-Δ19, added to FXII-deficient plasma, in the presence of kaolin induced the generation of a similar amount of FXIIa-C1–inhibitor complexes, indicating that the two proteins were equally well activated in plasma. However, whereas the amount of kallikrein-C1–inhibitor complexes generated also was the same, the generation of factor XIa-C1–inhibitor complexes was 50% lower in FXII-deficient plasma reconstituted with rFXII-Δ19 than in that reconstituted with FXII, thus indicating that rFXII-Δ19 efficiently activated prekallikrein, but was not able to efficiently activate factor XI (Fig 5).
Activation of contact system in plasma by FXII proteins. A volume of 20 μL of FXII proteins (40 nmol/L) in PT was added to 40 μL of FXII-deficient plasma and then incubated with 20 μL of kaolin (5 mg/mL in PT) for 20 minutes at 37°C. The reaction was stopped by addition of 120 μL of stop solution (PBS, 0.1 mg/mL SBTI, 0.05% [wt/vol] Polybrene), followed by centrifugation for 2 minutes at 10,000g to discard the kaolin pellet. The amount of FXIIa-C1–inhibitor (▧), kallikrein-C1-inhibitor (░), and factor XIa-C1–inhibitor (▪) complexes generated in EDTA-plasma was determined as described in Materials and Methods. Results are expressed as nmol/L and represent the means ± SD of three independent experiments each performed in duplicate (n = 6). * P < .05 as compared with the amount of factor XIa-C1–inhibitor complexes generated by FXII.
Activation of contact system in plasma by FXII proteins. A volume of 20 μL of FXII proteins (40 nmol/L) in PT was added to 40 μL of FXII-deficient plasma and then incubated with 20 μL of kaolin (5 mg/mL in PT) for 20 minutes at 37°C. The reaction was stopped by addition of 120 μL of stop solution (PBS, 0.1 mg/mL SBTI, 0.05% [wt/vol] Polybrene), followed by centrifugation for 2 minutes at 10,000g to discard the kaolin pellet. The amount of FXIIa-C1–inhibitor (▧), kallikrein-C1-inhibitor (░), and factor XIa-C1–inhibitor (▪) complexes generated in EDTA-plasma was determined as described in Materials and Methods. Results are expressed as nmol/L and represent the means ± SD of three independent experiments each performed in duplicate (n = 6). * P < .05 as compared with the amount of factor XIa-C1–inhibitor complexes generated by FXII.
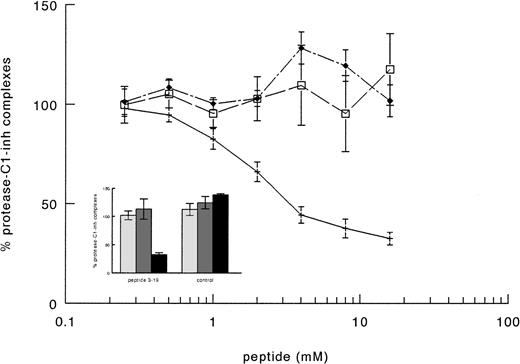
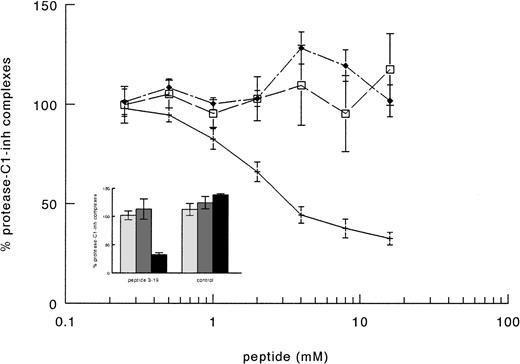
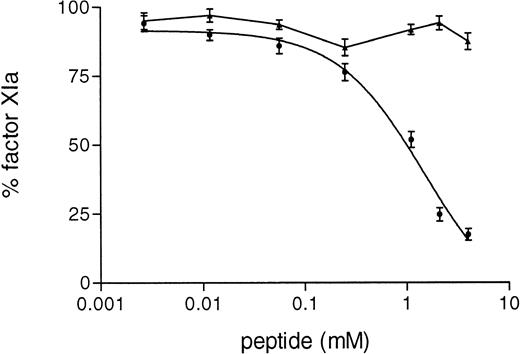
The supposed role of residues 3-19 in the activation of factor XI by FXIIa was further substantiated by experiments in which purified factor XI was preincubated with a synthetic peptide corresponding to this sequence of FXII. After preincubation with the synthetic peptide, FXI was added to factor XI-deficient plasma and activation of contact system was induced by adding kaolin. The synthetic peptide 3-19 (net charge −0.82 at pH 7) inhibited the generation of FXIa-C1–inhibitor complexes in FXI-deficient plasma in a dose-dependent manner, the IC50 being 3.1 ± 0.7 mmol/L (Fig 6). On the contrary, the synthetic peptide 3-19 did not have any effect on the generation of FXIIa-C1–inhibitor and kallikrein-C1–inhibitor, further suggesting that residues 3-19 are involved in the activation of factor XI by FXIIa. The synthetic peptide BVEAAAASAISVA (net charge −1.01 at pH 7), used as an irrelevant control, showed no effects on FXIa-C1–inhibitor complexes, as well as on FXIIa- and kallikrein-C1–inhibitor complexes (Fig 6).
Inhibition of factor XI activation in plasma by synthetic peptides. A total of 10 μL of serial dilutions (16 mmol/L to 0.2 mmol/L) of peptides in PBS 0.1% (wt/vol) Tween-20 (PT) were incubated with 10 μL of purified factor XI (160 nmol/L) overnight at 4°C. Thereafter, 40 μL of factor XI-deficient plasma were added and contact activation was started by adding 20 μL of kaolin (5 mg/mL). After a 20-minute incubation at 37°C, the reaction was stopped by adding 120 μL of stop solution followed by centrifugation to discard kaolin pellet. The amount of FXIIa-C1-inhibitor (□), kallikrein-C1–inhibitor (⧫), and factor XIa-C1-inhibitor (+––+) complexes generated in EDTA-plasma was determined as described in Materials and Methods. Results are expressed as percentage of the amount of complexes generated in the absence of peptides and represent the means ± SE of two different experiments in duplicate. In the inset, the results obtained with an irrelevant peptide, tested as control at 8 mmol/L, are shown. Symbols: FXIIa-C1–inhibitor (▧), kallikrein-C1-inhibitor (░), and factor XIa-C1–inhibitor (▪) complexes.
Inhibition of factor XI activation in plasma by synthetic peptides. A total of 10 μL of serial dilutions (16 mmol/L to 0.2 mmol/L) of peptides in PBS 0.1% (wt/vol) Tween-20 (PT) were incubated with 10 μL of purified factor XI (160 nmol/L) overnight at 4°C. Thereafter, 40 μL of factor XI-deficient plasma were added and contact activation was started by adding 20 μL of kaolin (5 mg/mL). After a 20-minute incubation at 37°C, the reaction was stopped by adding 120 μL of stop solution followed by centrifugation to discard kaolin pellet. The amount of FXIIa-C1-inhibitor (□), kallikrein-C1–inhibitor (⧫), and factor XIa-C1-inhibitor (+––+) complexes generated in EDTA-plasma was determined as described in Materials and Methods. Results are expressed as percentage of the amount of complexes generated in the absence of peptides and represent the means ± SE of two different experiments in duplicate. In the inset, the results obtained with an irrelevant peptide, tested as control at 8 mmol/L, are shown. Symbols: FXIIa-C1–inhibitor (▧), kallikrein-C1-inhibitor (░), and factor XIa-C1–inhibitor (▪) complexes.
Activation of factor XI by FXII proteins.
Factor XI circulates in plasma noncovalently complexed with HK and forms a ternary complex with FXIIa in the presence of negatively charged surfaces.37-40 The results of the experiment performed in plasma (see Fig 5) indicated that the recombinant protein rFXII-Δ19 had an impaired clotting activity due to the fact that it had an impaired capability to activate factor XI, but did not give any indication about the role of FXII residues 3-19 in the formation of the ternary complex. Therefore, we studied the influence of HK on the activation of purified factor XI by FXIIa and rFXII-Δ19.
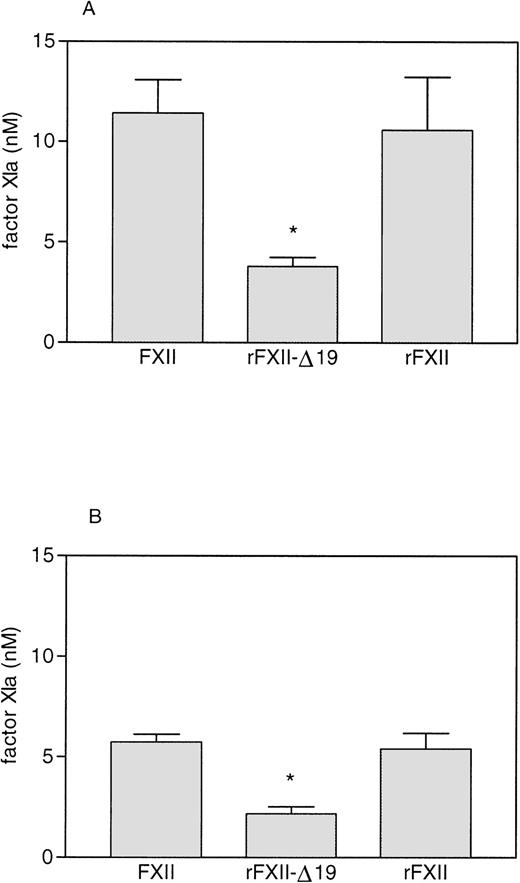
As shown in Fig 7, preincubation with HK increased the rate of activation of factor XI by both full-length FXII (either recombinant FXII or plasma FXII) and rFXII-Δ19. The FXIa generated by rFXII-Δ19a was significantly lower than that generated by an equal amount of FXIIa. However, the relative amount of factor XI activated by rFXII-Δ19 compared with full-length FXII was very similar either when HK was present or absent (33.2% and 37.7%, respectively), thus indicating that the impaired capability of rFXII-Δ19 to activate factor XI was independent on the presence of HK.
Factor XI activation by FXII proteins. FXII and rFXII-▵19 (50 nmol/L) were first activated by incubation with DS500 (1 μg/mL) as described in Materials and Methods. Thereafter, 25 μL of the mixture 1 to 20 diluted were added to 25 μL of purified human factor XI (40 to 80 nmol/L) preincubated 5 minutes at room temperature with 25 μL of HK (16 to 33 nmol/L, respectively) (A) or with 25 μL of buffer (B). After a 15-minute incubation at 37°C, the reaction was stopped by adding 25 μL of MoAb OT2 (100 nmol/L) and the rate of factor XI activation was measured by adding 50 μL of the chromogenic substrate S-2366 (1 mmol/L) and recording the increase in absorbance at 405 nm. Results, expressed as the nmol/L of factor XIa generated, represent the means ± SD of four different experiments. * P< .005 as compared with the amount of factor XI activated by FXII and rFXII.
Factor XI activation by FXII proteins. FXII and rFXII-▵19 (50 nmol/L) were first activated by incubation with DS500 (1 μg/mL) as described in Materials and Methods. Thereafter, 25 μL of the mixture 1 to 20 diluted were added to 25 μL of purified human factor XI (40 to 80 nmol/L) preincubated 5 minutes at room temperature with 25 μL of HK (16 to 33 nmol/L, respectively) (A) or with 25 μL of buffer (B). After a 15-minute incubation at 37°C, the reaction was stopped by adding 25 μL of MoAb OT2 (100 nmol/L) and the rate of factor XI activation was measured by adding 50 μL of the chromogenic substrate S-2366 (1 mmol/L) and recording the increase in absorbance at 405 nm. Results, expressed as the nmol/L of factor XIa generated, represent the means ± SD of four different experiments. * P< .005 as compared with the amount of factor XI activated by FXII and rFXII.
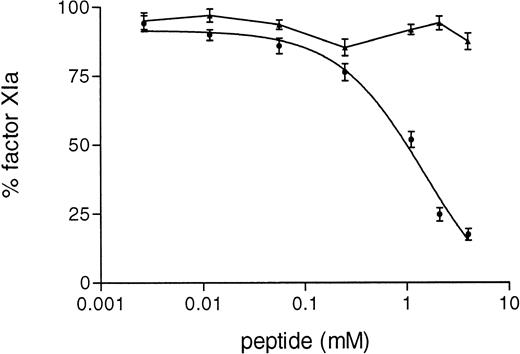
Furthermore, in the purified system we observed that the synthetic peptide 3-19 inhibited in a dose-dependent manner the activation of factor XI by FXIIa (IC50 = 1.66 ± 0.8 mmol/L), whereas the activation of factor XI by rFXII-Δ19 was not inhibited at all (Fig 8).
Inhibition of FXI activation by synthetic peptides. A total of 15 μL of serial dilutions (4.5 mmol/L to 0.2 mmol/L) of peptide in PT were incubated with 15 μL of purified factor XI (80 nmol/L) for 6 hours at 4°C. Thereafter, 15 μL of HK (33 nmol/L) were added and after 5 minutes at room temperature, factor XI activation was started by adding 25 μL of FXII (2.5 nmol/L) activated as in Materials and Methods. After a 15-minute incubation at 37°C, the reaction was stopped by adding MoAb OT2 and the rate of factor XI activation was determined by adding 50 μL of the chromogenic substrate S-2366 (1 mmol/L) and recording the increase in absorbance at 405 nm. Results are expressed as the percentage of factor XIa formed by either protein, FXII (•) or rFXII-▵19 (▴) in the absence of peptide and represent the means ± SD of three different experiments.
Inhibition of FXI activation by synthetic peptides. A total of 15 μL of serial dilutions (4.5 mmol/L to 0.2 mmol/L) of peptide in PT were incubated with 15 μL of purified factor XI (80 nmol/L) for 6 hours at 4°C. Thereafter, 15 μL of HK (33 nmol/L) were added and after 5 minutes at room temperature, factor XI activation was started by adding 25 μL of FXII (2.5 nmol/L) activated as in Materials and Methods. After a 15-minute incubation at 37°C, the reaction was stopped by adding MoAb OT2 and the rate of factor XI activation was determined by adding 50 μL of the chromogenic substrate S-2366 (1 mmol/L) and recording the increase in absorbance at 405 nm. Results are expressed as the percentage of factor XIa formed by either protein, FXII (•) or rFXII-▵19 (▴) in the absence of peptide and represent the means ± SD of three different experiments.
DISCUSSION
Since FXII protein is coded by 14 exons corresponding, in part, to the structural domains at the amino acid level, it is possible to construct recombinant deletion mutants of FXII that comprise the serine protease domain and one or more of the other structural domains of the intact molecule.24 25 In this report, we describe the production and characterization of a recombinant FXII protein, rFXII-Δ19, deleted of the amino acid residues 3-19, ie, the residues that are encoded by the second exon of FXII gene and that contain a putative binding site for negatively charged surfaces. Our results, however, indicate that this sequence is not involved in the binding of FXII to negatively charged surfaces, but comprises a site for the interaction of FXII with its substrate factor XI.
rFXII-Δ19, produced by using vaccinia virus as expression system, was first characterized by two MoAbs directed against the heavy chain region of FXII, MoAbs KOK5 and B7C9. The recombinant protein, consisting of one polypeptide chain with the expected electrophoretic mobility, was immunoprecipitated by MoAb KOK5 from the culture media of HepG2 cells infected with the recombinant virus, which indicated that the recombinant protein was correctly folded and secreted. On the contrary, MoAb B7C9 failed to immunoprecipitate rFXI-Δ19.
A putative surface-binding site of FXII has been localized within amino acid residues 134-153 and/or 1-28 by mapping the epitope for MoAb B7C9, which inhibits kaolin-induced FXII clotting activity to 60%.22,23 Because the clotting activity of kaolin-bound FXII was not inhibited by MoAb B7C9, it was suggested that MoAb B7C9 recognized the binding site of FXII for negatively charged surfaces, as this binding site may not be accessible on kaolin-bound FXII.22 Our results indicated that the recombinant FXII protein deleted of amino acid residues 3-19 did not bind MoAb B7C9: rFXII-Δ19, indeed, was not immunoprecipitated by immobilized MoAb B7C9, its clotting activity was not affected by preincubation with this MoAb, and it was not detected in an ELISA with MoAb B7C9 as a capturing antibody and MoAb F3 (against the light chain region) as a detecting antibody. Hence, the epitope for MoAb B7C9 resides within the amino acid residues 3-19 of FXII molecule. Yet, rFXII-Δ19, lacking this region, normally bound to kaolin and to dextran sulfate, as it was evident from the following observations: 125I-rFXII-Δ19 bound to kaolin equally well as 125I-FXII; unlabeled rFXII-Δ19 inhibited the binding of 125I-FXII to kaolin with the same efficiency of unlabeled FXII; the activation rate of rFXII-Δ19 by kallikrein was increased in the presence of dextran sulfate; dextran sulfate supported the autoactivation of rFXII-Δ19 at the same concentration and at the same rate as that of plasma FXII. These results do not support a role of amino acid residues 3-19 in the binding of FXII to negatively charged surfaces.
Consistent with the observation that MoAb B7C9 inhibits the clotting activity of FXII, rFXII-Δ19, lacking the epitope for MoAb B7C9, had a lower clotting activity (44%) than full-length FXII but, as discussed above, this impaired clotting activity cannot be explained by lack of binding to kaolin.
It is well accepted that the proteolytic enzymes of the cascade systems in plasma have a catalytic region as well as binding site(s) for their substrates necessary to efficiently catalyze the conversion of the substrate to the product. Our results indicating that rFXII-Δ19 did bind to negatively charged surfaces (as well as FXII) and was efficiently activated by kallikrein, led us to predict that the impaired clotting activity of this recombinant protein was due to either (1) an impaired enzymatic activity or (2) an impaired interaction with one of FXII substrates: prekallikrein or factor XI. The former possibility could be ruled out by the fact that the specific amidolytic activity of activated rFXII-Δ19 was comparable to that of activated plasma FXII and that rFXII-Δ19 did undergo autoactivation. On the contrary, the finding that in the presence of kaolin, equal amounts of kallikrein-C1–inhibitor complexes were generated in FXII-deficient plasma by either FXII or rFXII-Δ19, whereas only 50% of FXIa-C1–inhibitor complexes were formed in the presence of rFXII-Δ19 compared with FXII, indicated that the impaired clotting activity of rFXII-Δ19 was due to an impaired interaction with factor XI. This conclusion was supported by the observation that in a purified system rFXII-Δ19 also activated factor XI twice less efficiently than full-length FXII independent on the presence of HK (Fig 7) and that a synthetic peptide corresponding to amino acid residues 3-19 inhibited in a dose-dependent fashion the activation of factor XI in plasma (Fig6), as well as in the purified system (Fig 8).
Baglia et al35 41-44 have demonstrated the presence of various binding sites for enzymes, substrate, and cofactor in the different domains of the factor XI molecule such as a HK-binding site in the A1 domain, a factor XIa substrate-binding site for factor IX in the A2 domain, an enzyme-binding site for thrombin in the A1 domain and a binding site for FXIIa in the A4 domain. Studying the effects of the factor XI A4 domain peptide Gly326-Lys357 on the activation of factor XI by FXIIa and on the amidolytic activity of FXIIa through the chromogenic substrate S-2302, the investigators found that the A4-derived peptide was a noncompetitive inhibitor of macromolecular substrate (factor XI) conversion, whereas it is a competitive inhibitor of small peptide (S-2302) hydrolysis. They postulated that the factor XI A4-derived peptide binds to a small peptide substrate-binding site near the catalytic site of FXIIa that does not appear to function as a macromolecular substrate-binding site. They proposed that the putative small peptide substrate-binding site near to the active site of FXIIa (recognized by the chromogenic substrate S-2302 and by the factor XI A4-derived peptide) represents a secondary substrate-binding site. This site may serve to positioning the factor XI cleavage site (Arg369-Ile370) near the catalytic site of FXIIa once factor XI is already anchored to FXIIa via a macromolecular substrate-binding site, which may well exist elsewhere in the FXIIa molecule. Our results support this model and provide indirect evidence for the existence of two different substrate-binding sites. We speculate that the FXII amino acid residues 3-19 may, indeed, represent the macromolecular substrate-binding site for factor XI as the presence of these residues increases the efficiency of factor XI cleavage by FXIIa. Cleavage of factor XI apparently may occur without this site, which may explain that the deletion of amino acid residues 3-19 did not completely abolish the clotting activity of FXII, but reduced it to 44% of that of plasma FXII. This reduction by 56% is in agreement with the finding that maximum inhibition of FXII clotting activity by MoAb B7C9 is 60%, only. Accordingly, the generation of factor XI-C1–inhibitor complexes was only partially (50%) inhibited in FXII-deficient plasma reconstituted with rFXII-Δ19 recombinant protein.
In conclusion, we have produced and characterized a recombinant FXII protein, which lacks the amino acid residues 3-19, encoded by the second exon of FXII gene. The binding capacity of this recombinant protein to negatively charged surfaces, its capacity to undergo autoactivation, and its susceptibility for activation by kallikrein were similar to those of full-length FXII. However, the recombinant FXII deletion mutant displayed a reduced clotting activity, which correlated with an impaired capability to activate factor XI. Therefore, we suggest that residues 3-19 at the amino terminus of FXII molecule do not contribute to the surface-binding site of FXII, but play a role in the activation of factor XI.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Prof C. Erik Hack, c/o Publication Secretariat, Central Laboratory of The Netherlands Red Cross Blood Transfusion Service, Sanquin Blood Supply Foundation, Department Pathophysiology of Plasma Proteins, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands.

![Fig. 5. Activation of contact system in plasma by FXII proteins. A volume of 20 μL of FXII proteins (40 nmol/L) in PT was added to 40 μL of FXII-deficient plasma and then incubated with 20 μL of kaolin (5 mg/mL in PT) for 20 minutes at 37°C. The reaction was stopped by addition of 120 μL of stop solution (PBS, 0.1 mg/mL SBTI, 0.05% [wt/vol] Polybrene), followed by centrifugation for 2 minutes at 10,000g to discard the kaolin pellet. The amount of FXIIa-C1–inhibitor (▧), kallikrein-C1-inhibitor (░), and factor XIa-C1–inhibitor (▪) complexes generated in EDTA-plasma was determined as described in Materials and Methods. Results are expressed as nmol/L and represent the means ± SD of three independent experiments each performed in duplicate (n = 6). * P < .05 as compared with the amount of factor XIa-C1–inhibitor complexes generated by FXII.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/11/10.1182_blood.v92.11.4198/5/m_blod42311005y.jpeg?Expires=1765991249&Signature=TljF2uML9ZnnOPdvdoAzSvaXZZ7P6ZLJLD4ZBqRL9leMyvCoaeNizXswRvzdAdoyJvKFw1up55wohr8QVdzBziMkVnAVv~m51LtpS3RBm2o8ZvHbxExr5~THrt5l3qJzcQkDILLpEa5eo9HDvIvq-RnGP-6Spg14Ec5RB-tUpJj2ssIdR14GMUn-KLx9-E4oAN4HCTMkMyhTNRjUjRHwbRittO6e2uPPERSAPdfkMn7-GSJNaQSg~-On4N3zy7P~vEFJip~HB03NNjni6SoADpDU3z-VCETyudXPEyRfpTo3pT9RpFp7u7GXwKFMfjG39J3MMwm2oGyR7Z7OWmRaIA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 5. Activation of contact system in plasma by FXII proteins. A volume of 20 μL of FXII proteins (40 nmol/L) in PT was added to 40 μL of FXII-deficient plasma and then incubated with 20 μL of kaolin (5 mg/mL in PT) for 20 minutes at 37°C. The reaction was stopped by addition of 120 μL of stop solution (PBS, 0.1 mg/mL SBTI, 0.05% [wt/vol] Polybrene), followed by centrifugation for 2 minutes at 10,000g to discard the kaolin pellet. The amount of FXIIa-C1–inhibitor (▧), kallikrein-C1-inhibitor (░), and factor XIa-C1–inhibitor (▪) complexes generated in EDTA-plasma was determined as described in Materials and Methods. Results are expressed as nmol/L and represent the means ± SD of three independent experiments each performed in duplicate (n = 6). * P < .05 as compared with the amount of factor XIa-C1–inhibitor complexes generated by FXII.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/11/10.1182_blood.v92.11.4198/5/m_blod42311005y.jpeg?Expires=1765991250&Signature=aF8S13cxyOUpIzCwGLZVuVsWOe-RksAeAFeLxBXKddp7jGcKH4SNCNQKDr~d7xiKXbuFsZQtrQ-e9U5F7uFnG78aGM99HECTtM~phlbzgzmAtXdHcd-ZNpd~GAFPphDqA2upKqLX5kd5qSZQM4BHXyZo-uJOL1xPmq1S1Hnf0V7~-RTGQlIbFFZhowicCGZYgdTP6FEyYepk5EsiwFl0Lic-Y69TSv~ugvGZXPLTmXq79YFQ7rIj3vQNXSgdVbf8j7VsPoeW4II0HLszgs5A3ZHFzAkPuWPSrY8AUbo8cP-ISpFAtKLxohxY0ETjFX-9H7resXaI0MwgqUH2cF2~qA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)