Abstract
Our previous work showed that the nuclear scaffold (NS) protease is required for apoptosis of both thymocytes and chronic lymphocytic leukemic (CLL) lymphocytes. Because partial sequencing of one of the subunits of the NS protease revealed homology to the proteasome, we tested the effects of classical proteasome inhibitors on apoptosis in CLL cells. Here we report that proteasome inhibition caused high levels of DNA fragmentation in all patients analyzed, including those resistant to glucocorticoids or nucleoside analogs, in vitro. Proteasome inhibitor-induced DNA fragmentation was associated with activation of caspase/ICE family cysteine protease(s) and was blocked by the caspase antagonist, zVADfmk. Analysis of the biochemical mechanisms involved showed that proteasome inhibition resulted in mitochondrial dysregulation leading to the release of cytochrome c and a drop in mitochondrial transmembrane potential (▵Ψ). These changes were associated with inhibition of NFκB, a proteasome-regulated transcription factor that has been implicated in the suppression of apoptosis in other systems. Together, our results suggest that drugs that target the proteasome might be capable of bypassing resistance to conventional chemotherapy in CLL.
CHRONIC LYMPHOCYTIC leukemia (CLL) is an illness characterized by an accumulation of monoclonal mature B cells in the peripheral blood. Although CLL is the most common leukemia in the Western world, little is known about the biology of the disease. Treatment schemes rely heavily on glucocorticoids, chlorambucil, and nucleoside analogs, and we and others have shown that all of these agents trigger apoptosis in CLL cells in vitro, suggesting that induction of apoptosis may account for their therapeutic efficacy. Furthermore, recent work has shown that apoptosis in vitro correlates with Rai stage,1,2 and rates of apoptosis detected following fludarabine treatment correlate with clinical response in vivo.3 However, despite the initial effectiveness of these drugs in patients with low-grade disease, resistant cells ultimately emerge, leaving no effective treatment options available. It is possible that drug-resistant CLL cells possess intrinsic defect(s) in their ability to undergo apoptosis.
Protease activation is required for completion of the apoptotic program in all cellular and cell-free systems interrogated to date.4,5 Of central importance are members of the ICE/caspase family of aspartate-specific cysteine proteases, which appear to function at the core of the “effector” machinery for cell death. Caspase activation can either be directly promoted by oligomerization of certain caspase-associated cell surface “death” receptors (Fas, TNF-RI)6 or by intramitochondrial events that lead to the release of the electron transport chain intermediate, cytochrome c.7 Precisely how caspases promote the downstream features of apoptosis is not clear, but studies with specific peptide-based active site inhibitors indicate that they are required for all of the major biochemical events observed in apoptotic cells, including changes in cellular morphology, loss of plasma membrane asymmetry (exposure of phosphatidylserine on the outer leaflet), and DNA fragmentation.8
We and others have obtained evidence that certain noncaspase proteases are also required for DNA fragmentation and apoptosis. Specifically, we have shown that peptide-based active site inhibitors of a Ca2+-dependent nuclear protease, termed the nuclear scaffold (NS) protease, block glucocorticoid- and nucleoside analog–induced DNA fragmentation in CLL lymphocytes.9Although the molecular characteristics of the NS protease are at present unclear, preliminary evidence obtained by another laboratory10 suggests that it is structurally and functionally related to the 26S multicatalytic protease complex (MPC), otherwise known as the proteasome. This possible similarity may explain why NS protease inhibitors block DNA fragmentation, because previous studies have implicated the proteasome in the programmed cell death of intersegmental muscles in the moth, Manduca sexta,11 and more recent work in isolated mouse thymocytes12 and neuronal cells13 has shown that proteasome inhibitors block caspase activation and other downstream events associated with apoptosis in these cells.
The results presented above suggested to us that the effects of NS protease inhibitors in CLL cells might be due to proteasome inhibition. To directly address this possibility, we tested the effects of several specific proteasome inhibitors on caspase activation and DNA fragmentation in isolated CLL lymphocytes, expecting that they would suppress apoptotic cell death. On the contrary, here we report that proteasome inhibition resulted in extraordinarily high levels of DNA fragmentation in all patient isolates analyzed, including those found to be completely resistant to glucocorticoid-induced apoptosis. Analysis of the biochemical mechanisms involved showed that the effects are linked to inhibition of NFκB, a transcription factor implicated in the maintenance of cell survival in other model systems.14-17
MATERIALS AND METHODS
Materials.
The esterified peptide caspase inhibitor, Z-VAD (OMe)fmk, the fluorigenic caspase substrate, DEVD-AMC, and the mouse anti-PARP monoclonal antibody C2-10 were purchased from Enzyme Systems Products, Inc (Dublin, CA). A peptide inhibitor of the NS protease, Z-APFcmk, and the caspase antagonist, Boc-Asp-chloromethylketone (BDcmk) were purchased from Bachem Bioscience (King of Prussia, PA). Monoclonal antibodies for caspase-3, p53, p27, and c-Jun were purchased from Transduction Laboratories (Lexington, KY). A monoclonal antibody to c-Fos and the proteasome inhibitors lactacystin and MG-132 were obtained from Calbiochem (San Diego, CA). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were from Amersham Corp (Arlington Heights, IL).
Patients, cell isolation, and incubation criteria.
All patients fulfilled the National Cancer Institute’s (NCI) criteria for the diagnosis of CLL. Some of the patients had received prior therapy, although none within the last 6 months before experimentation. Immunophenotyping by dual-parameter flow cytometry showed coexpression of CD5 with B-cell antigen and isotypic light chain expression. Clinical staging was based on the system described by Rai.18 Freshly isolated peripheral blood was fractionated by Ficoll-Hypaque (Winthrop Pharmaceuticals, New York, NY) sedimentation at 4°C. Nonadherent mononuclear cells were then immediately suspended in complete RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 10 mmol/L HEPES (pH 7.5), and antibiotics at a cellular concentration of 1 to 2 × 106 cells/mL. Cell viability was assessed by Trypan blue exclusion and exceeded 95% following the isolation procedure.
Granulocyte-colony stimulating factor (G-CSF)–mobilized progenitor cells were obtained from pheresis samples by magnetic cell sorting (MACS). The pheresis samples were resuspended in 50 mL of cold RPMI medium, and two “soft-spins” (200g, 10 minutes) were performed to remove platelets. Cells were labeled with anti-CD34 antibody and isolated with a commercial CD34 isolation kit (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. A MACS buffer consisting of Ca2+/Mg2+-free Hanks’ buffered salt solution (HBSS) containing 0.6% ACD-A (Baxter, Deerfield, IL), 0.5% bovine serum albumin (BSA; Sigma, St Louis, MO), pH 6.5, was used throughout staining and separation to prevent cell clumping while maintaining optimal progenitor viability. Cells were separated on VS-positive selection columns using a VarioMACS according to the manufacturer’s instructions. Cell purity was assessed by flow cytometry using CD34-phycoerythrin (PE) and CD45-fluorescein isothiocyanate (FITC) as described previously.19
DNA fragmentation analysis.
Quantification of apoptosis by propidium iodide (PI) staining and fluorescence-activated cell sorting (FACS) analysis was performed as described previously.20 Following incubation with various agents in vitro, cells were pelleted by centrifugation and resuspended in phosphate-buffered saline (PBS) containing 50 μg/mL PI, 0.1% Triton X-100, and 0.1% sodium citrate. Samples were stored at 4°C for 16 hours and vortexed before FACS analysis (FL-3 channel).
Cytochrome c release measurements.
Release of cytochrome c from mitochondria was measured by immunoblotting essentially as described previously.21 Cells were incubated in the absence or presence of 10 μmol/L APFcmk, 10 μmol/L MG-132, or 10 μmol/L methylprednisolone for 4 hours, obtained by centrifugation, and gently lysed for 30 seconds in an ice-cold buffer containing 250 mmol/L sucrose, 1 mmol/L EDTA, 0.1% digitonin, and 25 mmol/L Tris, pH 6.8. Lysates were centrifuged for 2 minutes at 12,000g, supernatants were mixed with 2× Laemmli’s reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and extracts from equal numbers of cells (10 to 20 × 106) were resolved by 15% SDS-PAGE. Polypeptides were transferred to nitrocellulose membranes (0.2 μm; Schleicher & Scheull, Keene, NH), and cytochrome c was detected by immunoblotting with a monoclonal antibody (clone 7H8.2C12; purchased from Pharmingen, San Diego, CA).
Caspase activity assay.
Protease activity measurements were conducted as described previously.9 Cells were lysed in 1 mL of a buffer containing 25 mmol/L HEPES (pH 7.4), 5 mmol/L EDTA, 2 mmol/L dithiothreitol, and 10 μmol/L digitonin for 15 minutes on ice. The lysates were clarified by centrifugation (12,000g), and supernatants were incubated with 50 μmol/L Asp-Glu-Val-Asp-aminomethylcoumarin (DEVD-AMC; Enzyme Systems Products, Inc at 37°C in the dark. Relative activities were then measured in a spectrofluorimeter (400 nm excitation, 505 nm emission); blanks included supernatants processed as outlined above without dye and supernatants preincubated with BDcmk (25 μmol/L).
Mitochondrial membrane potential measurements.
The potential-sensitive fluorochrome JC-1 (Molecular Probes, Eugene, OR) was used to measure ΔΨmito. Cells were obtained by centrifugation and incubated with 10 μmol/L JC-1 for 15 minutes at 37°C in the dark. Cells were washed in PBS and analyzed by FACS on the FL-2 channel (FACScan; Becton Dickinson, Mountain View, CA).
Annexin V binding.
Exposure of surface phosphatidylserine was quantified by surface annexin V staining as described previously.22 This assay was used as a DNA fragmentation-independent endpoint to confirm the involvement of apoptosis in the mechanism of cell death. Cells were resuspended in binding buffer containing 1 μg/mL FITC-conjugated annexin V (Nexins Research B.V., Hoeven, The Netherlands) and incubated for 30 minutes at 4°C, and cells were analyzed by flow cytometry (FACScan, Becton Dickinson).
Immunoblotting.
For detection of caspase-3, p53, p27, Fos, and Jun, cells were lysed for 1 hour at 4°C in a buffer containing 150 mmol/L NaCl, 1% Triton X-100, a cocktail of protease inhibitors (Complete Mini tablets; Boehringer-Mannheim, Indianapolis, IN), and 25 mmol/L Tris (pH 7.5). Debris was sedimented by centrifugation for 5 minutes at 12,000g, and the supernatants were solubilized for 5 minutes at 100°C in Laemmli’s SDS-PAGE sample buffer containing 100 mmol/L dithiothreitol.
For analysis of PARP cleavage samples were denatured in a urea/SDS buffer as follows: cells were incubated with various agents for 16 hours, obtained by centrifugation, and resuspended in 25 μL ice-cold PBS. Cells were subsequently disrupted by addition of 100 μL of a buffer containing 6 mol/L urea, 2% SDS, 10% glycerol, 5 mmol/L EDTA, 5% 2-mercaptoethanol, and 100 mmol/L Tris (pH 6.8) followed by pipeting through a 1-mL Pipetman tip, and samples were sonicated for 20 seconds at high power to sheer DNA. Samples were then incubated for 15 minutes at 65°C and centrifuged for 2 minutes at 12,000gbefore they were loaded onto 8% SDS-PAGE gels.
Polypeptides were resolved at 100 V on 8% to 12% gels and electrophoretically transferred to 0.2-μm nitrocellulose membranes (Schleicher & Schuell Inc) for 1 hour at 100 V. Membranes were blocked for 1 hour a TBS-T buffer (25 mmol/L Tris, pH 8.0, 150 mmol/L NaCl, and 0.05% Tween-20) containing 3% (wt/vol) nonfat dried milk. Blots were then probed overnight with primary antibody and developed using a horseradish peroxidase-coupled anti-mouse secondary antibody by enhanced chemiluminescence (Supersignal; Pierce Chemical Co, Rockford, IL) according to the manufacturer’s instructions.
Electrophoretic mobility shift (EMSA) assays.
Isolated nuclei were prepared by lysis with Triton X-100 and centrifugation through a glycerol cushion as described previously.23 Nuclear protein was extracted using a high salt, detergent-free buffer containing 20 mmol/L HEPES (pH 7.9), 400 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L dithiothreitol, and 1 mmol/L phenylmethylsulfonyl fluoride, for at least 20 minutes on ice. Extracts were centrifuged at 4°C for 5 minutes at 12,000g, and protein content in supernatants was measured by the Bradford method. A consensus double-stranded NFκB probe (obtained from Promega Inc, Madison, WI) was end-labeled using T4 polynucleotide kinase and γ-32P-ATP. Five to 20 μg of nuclear extract was then incubated in binding buffer (supplied by the manufacturer) containing 1 μg/mL poly dI:dC (Promega), ensuring that the final salt concentration was between 50 and 100 mmol/L. Reactions were incubated for 20 minutes at room temperature, and 1 μL of end-labeled probe was added. Samples were incubated for 30 minutes before addition of loading buffer (Promega) and electrophoresis on 4% nondenaturing polyacrylamide gels that were prerun in 0.5×Tris-borate-EDTA (TBE) buffer for 30 minutes at 100 V before use. Gels were run at 100 V in 0.5× TBE and dried, and DNA-protein complexes were detected by autoradiography.
Statistical analyses.
Mean values and standard deviations were calculated with Microsoft Excel (Microsoft Inc, Redmond, WA). Significance was evaluated using two-tailed paired Student’s t-tests with SPSS software (SPSS Inc, Chicago, IL).
RESULTS
Effects of proteasome inhibitor on DNA fragmentation and surface exposure of phosphatidylserine.
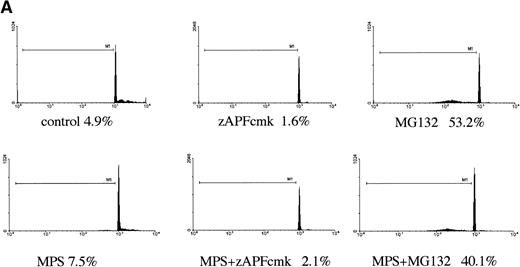
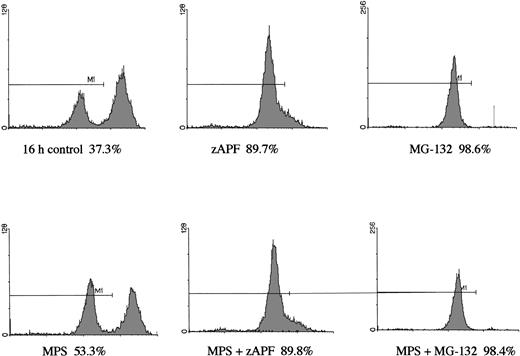
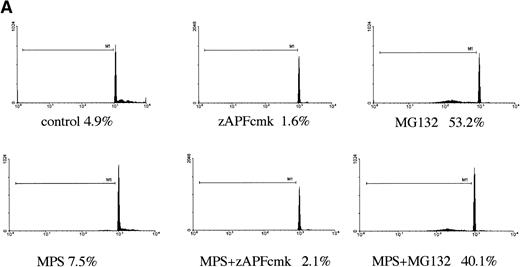
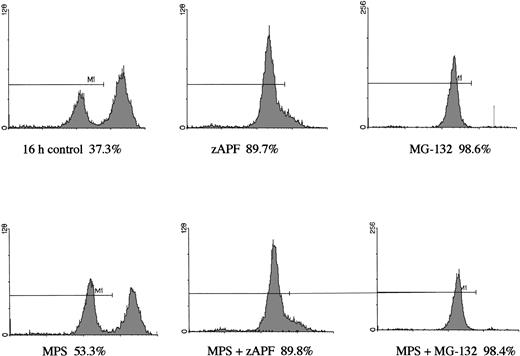
In a previous study we showed that a specific inhibitor of the NS protease completely suppressed apoptosis-associated DNA fragmentation in CLL cells treated with glucocorticoid or the nucleoside analog, fludarabine.9 Because proteasome inhibitors block apoptosis in thymocytes and neuronal cells,12,13 and another group has reported that the NS protease is homologous to the proteasome,10 we tested the effects of proteasome inhibitors on DNA fragmentation, measured by PI staining and FACS analysis, in CLL cells to determine whether the effects of zAPFcmk might be attributed to the proteasome. Levels were compared with those observed in response to treatment with glucocorticoid hormone. Our patient isolates fell into three general catetories: (1) those exhibiting relatively high (mean = 40%, n = 10) levels of apoptosis upon in vitro culture in the absence of hormone (“spontaneous”); (2) those exhibiting low spontaneous apoptosis but strong (mean = 40%, n = 28) increases in DNA fragmentation in response to glucocorticoid treatment (“sensitive”); and (3) those exhibiting low spontaneous apoptosis and low levels of glucocorticoid-induced DNA fragmentation (mean = 10%, n = 21) (“resistant”; Table 1). Strikingly, and contrary to our expectations, treatment with MG-132, a peptide-based proteasome antagonist, promoted high levels of DNA fragmentation in all three categories of cells (Table 1). Proteasome inhibitors were effective in all patient isolates analyzed (n = 59). Similar results were obtained with another, structurally distinct proteasome inhibitor, lactacystin (data not shown). Proteasome inhibitors also induced surface phosphatidylserine exposure, another downstream event in apoptosis that is thought to be independent of endonuclease activation (Fig 1B). Importantly, preincubation with zAPFcmk blocked MG-132–induced DNA fragmentation (Fig 1A), indicating that these inhibitors exert their effects on different biochemical activities.
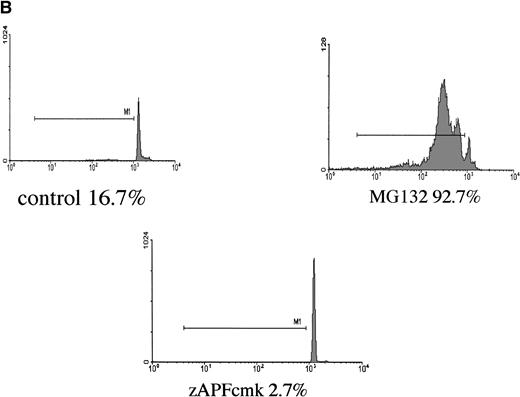
Induction of apoptosis by proteasome inhibition in CLL patient isolates. (A) DNA fragmentation analysis. Cells from a representative glucocorticoid-sensitive patient (see Table 1) were preincubated in the presence of 25 μmol/L zAPFcmk or 200 μmol/L zVADfmk for 1 hour and then treated with 10 μmol/L MG132, and DNA fragmentation was measured at 16 hours by PI staining and FACS analysis. (B) Surface phosphatidylserine exposure. Cells were incubated in absence or presence of 10 μmol/L methylprednisolone or 10 μmol/L MG132 with or without 200 μmol/L zVADfmk, and surface PS exposure was quantitated by staining with annexin-FITC and measured by FACS analysis. Results characteristic of three independent experiments with different patient isolates.
Induction of apoptosis by proteasome inhibition in CLL patient isolates. (A) DNA fragmentation analysis. Cells from a representative glucocorticoid-sensitive patient (see Table 1) were preincubated in the presence of 25 μmol/L zAPFcmk or 200 μmol/L zVADfmk for 1 hour and then treated with 10 μmol/L MG132, and DNA fragmentation was measured at 16 hours by PI staining and FACS analysis. (B) Surface phosphatidylserine exposure. Cells were incubated in absence or presence of 10 μmol/L methylprednisolone or 10 μmol/L MG132 with or without 200 μmol/L zVADfmk, and surface PS exposure was quantitated by staining with annexin-FITC and measured by FACS analysis. Results characteristic of three independent experiments with different patient isolates.
Effects of MG-132 on DNA fragmentation in normal hematopoietic cells.
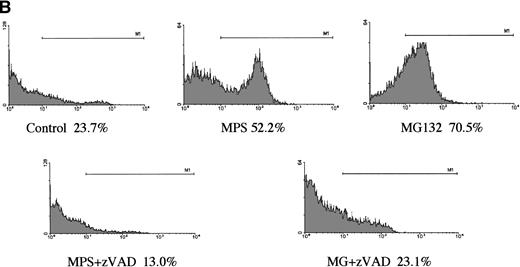
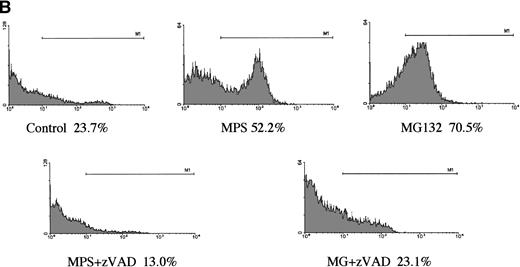
In a preliminary attempt to determine whether MG-132’s proapoptotic effects were selective for CLL cells, we analyzed the effects of the proteasome antagonist on DNA fragmentation in normal peripheral blood lymphocytes and in G-CSF–mobilized CD34+CD45+hematopoietic progenitor cells. Normal lymphocytes were killed by the compound, but the kinetics of the response were markedly delayed and the maximal response shifted from 16 hours to 48 hours (Fig 2A). Surface staining with specific B- and T-cell markers indicated that MG-132 was substantially more toxic to normal T cells than to B cells (data not shown). In contrast to the attenuated responses of lymphocytes, mobilized stem cells were highly sensitive to MG-132 (Fig 2B). Thus, proteasome inhibitors are capable of inducing apoptosis in certain normal as well as transformed hematopoietic cells.
Effect of proteasome inhibitors on normal lymphocytes. (A) Effects on normal lymphocytes. Isolated peripheral blood lymphocytes from normal donors were incubated in the presence of 10 μmol/L MG132 or 10 μmol/L methylprednisolone with or without 25 μmol/L zAPFcmk for 16 hours, and apoptosis was assessed by PI staining and FACS analysis. Results of one experiment are representative of three independent replicates. (B) Effects on hematopoietic progenitor cells. G-CSF–mobilized CD34+CD45+ progenitor cells were incubated in the absence or presence of 25 μmol/L zAPFcmk or 10 μmol/L MG-132 and DNA fragmentation was measured after 16 hours by PI staining and FACS analysis.
Effect of proteasome inhibitors on normal lymphocytes. (A) Effects on normal lymphocytes. Isolated peripheral blood lymphocytes from normal donors were incubated in the presence of 10 μmol/L MG132 or 10 μmol/L methylprednisolone with or without 25 μmol/L zAPFcmk for 16 hours, and apoptosis was assessed by PI staining and FACS analysis. Results of one experiment are representative of three independent replicates. (B) Effects on hematopoietic progenitor cells. G-CSF–mobilized CD34+CD45+ progenitor cells were incubated in the absence or presence of 25 μmol/L zAPFcmk or 10 μmol/L MG-132 and DNA fragmentation was measured after 16 hours by PI staining and FACS analysis.
Effects of proteasome inhibition on caspase activation.
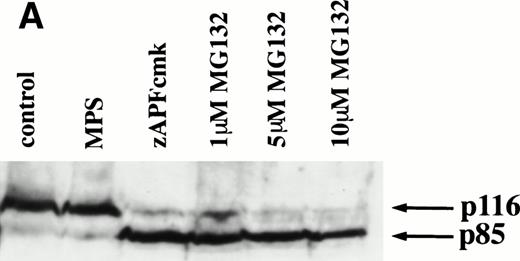
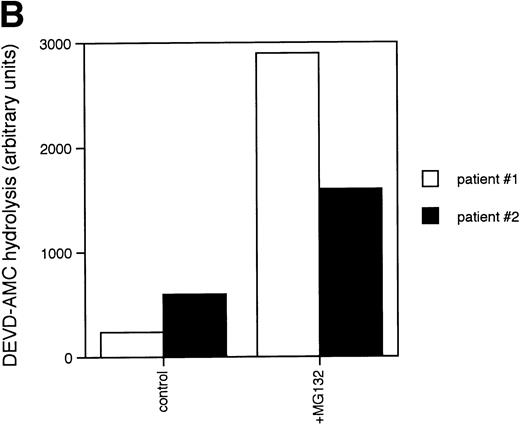
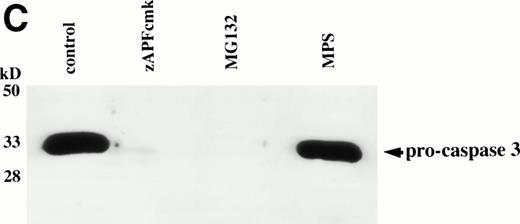
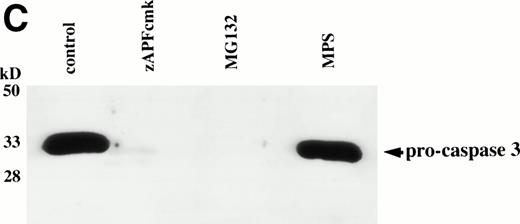
Caspases are a family of cysteine proteases that are thought to act at the core of the apoptotic pathway. Our previous work9 and that of others24 25 has confirmed that caspases are required for drug-induced apoptosis in CLL cells. We therefore investigated whether or not caspases were also required for apoptosis induced by proteasome inhibitors by four independent approaches. First, induction of apoptosis by proteasome inhibitors was also associated with specific cleavage of the caspase substrate, PARP, as detected by immunoblotting (Fig 3A). This occurred in all patient samples analyzed (n = 4), including one that did not respond to glucocorticoid treatment (Fig 3A). Second, proteasome inhibitors promoted hydrolysis of a specific caspase substrate (DEVD-AMC; Fig 3B). Third, proteasome inhibitors induced proteolytic processing of the inactive form of caspase-3 (procaspase-3; Fig 3C), providing more direct evidence for activation of caspase-3 and presumably other caspases in the response. Finally, the caspase inhibitor zVADfmk completely blocked MG-132–induced DNA fragmentation (Fig 1A) and surface exposure of phosphatidylserine (Fig 1B), confirming that caspase activation was required for proteasome inhibitor–induced apoptosis.
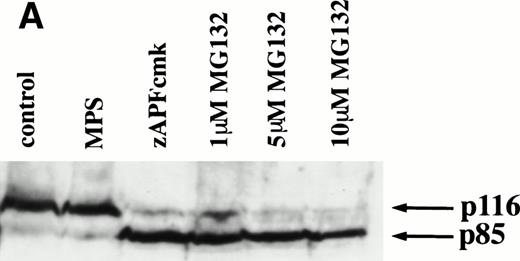
Caspase activation by proteasome inhibitors. (A) Cleavage of the caspase substrate, PARP, by treatment with proteasome inhibitor. Cells were treated with either 10 μmol/L methylprednisolone, 25 μmol/L zAPFcmk, or various doses of MG132. PARP was detected by immunoblotting. Intact (p116) and fragmented (p85) forms of PARP are indicated by arrows. (B) Effect of MG132 on DEVDase activity. Cells were treated in the absence or presence of 10 μmol/L MG132 for 8 hours, and hydrolysis of the caspase substrate DEVD-AMC was measured in a spectrofluorimeter. Results are from two experiments with independent patient isolates. Cells treated with 10 μmol/L BDcmk, a caspase inhibitor, did not show DEVDase activity more than 50 U above baseline levels. DNA fragmentation from these patients was measured in parallel. These patients are not included in Table 1. Patient no. 1: control = 18.0, MG132 = 96.9; patient no. 2: control = 1.4, MG132 = 32.5. (C) Activation of caspase-3 by proteasome inhibitor. Cells were treated with either 25 μmol/L zAPFcmk, 10 μmol/L MG132, or 10 μmol/L methylprednisolone for 16 hours and procaspase-3 was detected by immunoblotting. Results of one experiment representative of three replicates with independent patient isolates.
Caspase activation by proteasome inhibitors. (A) Cleavage of the caspase substrate, PARP, by treatment with proteasome inhibitor. Cells were treated with either 10 μmol/L methylprednisolone, 25 μmol/L zAPFcmk, or various doses of MG132. PARP was detected by immunoblotting. Intact (p116) and fragmented (p85) forms of PARP are indicated by arrows. (B) Effect of MG132 on DEVDase activity. Cells were treated in the absence or presence of 10 μmol/L MG132 for 8 hours, and hydrolysis of the caspase substrate DEVD-AMC was measured in a spectrofluorimeter. Results are from two experiments with independent patient isolates. Cells treated with 10 μmol/L BDcmk, a caspase inhibitor, did not show DEVDase activity more than 50 U above baseline levels. DNA fragmentation from these patients was measured in parallel. These patients are not included in Table 1. Patient no. 1: control = 18.0, MG132 = 96.9; patient no. 2: control = 1.4, MG132 = 32.5. (C) Activation of caspase-3 by proteasome inhibitor. Cells were treated with either 25 μmol/L zAPFcmk, 10 μmol/L MG132, or 10 μmol/L methylprednisolone for 16 hours and procaspase-3 was detected by immunoblotting. Results of one experiment representative of three replicates with independent patient isolates.
Effects of proteasome inhibitor on mitochondrial function.
Disruption of mitochondria leading to the release of the electron transport chain intermediate, cytochrome c, has recently been implicated in caspase activation in other model systems.26Although the mechanisms underlying cytochrome c release are unclear, the event is associated with a drop in transmembrane potential (ΔΨ), which may facilitate the opening of transmembrane pores in the mitochondrial membrane that would allow passage of cytochrome c and other proapoptotic factors from the organelle.27 To determine whether this pathway of caspase activation was induced by proteasome inhibitors in CLL cells, we measured the effects of MG-132 on cytochrome c release in digitonin-permeabilized cells. MG-132 promoted rapid release of cytochrome c from mitochondria in intact CLL cells (Fig 4, lane 3), effects that were also observed in cells treated with zAPFcmk (Fig 4, lane 4) and to a lesser extent with glucocorticoid (Fig 4, lane 5). At this time point, control cells did not exhibit either increased levels of cytosolic cytochrome c (Fig4, compare lanes 1 and 2) or significant caspase activation (Fig 3B). The effects of zAPFcmk on cytochrome c release are consistent with earlier experments that showed that NS protease inhibition results in caspase activation9 (Fig 3). Proteasome inhibition also resulted in a drop in mitochondrial membrane potential, measured with the potential-sensitive dye, JC-1 (Fig 5). These results indicate that proteasome inhibitors promote caspase activation via direct or indirect effects on mitochondria.
Proteasome inhibition leads to release of cytochrome c from mitochondria. Cells were incubated in the absence (control) or presence of 10 μmol/L MG132, 25 μmol/L zAPFcmk, or 10 μmol/L methylprednisolone for 6 hours, and cytosolic cytochrome c was measured in digitonin-permeabilized cells by immunoblotting. Lane 1, 0 hours control; lane 2, 6 hours control; lane 3, MG132; lane 4, zAPFcmk; lane 5, methylprednisolone. Results are typical of three independent experiments with different CLL isolates.
Proteasome inhibition leads to release of cytochrome c from mitochondria. Cells were incubated in the absence (control) or presence of 10 μmol/L MG132, 25 μmol/L zAPFcmk, or 10 μmol/L methylprednisolone for 6 hours, and cytosolic cytochrome c was measured in digitonin-permeabilized cells by immunoblotting. Lane 1, 0 hours control; lane 2, 6 hours control; lane 3, MG132; lane 4, zAPFcmk; lane 5, methylprednisolone. Results are typical of three independent experiments with different CLL isolates.
Effects of proteasome inhibition on mitochondrial membrane potential. Cells were incubated in the absence or presence of 10 μmol/L methylprednisolone with or without 25 μmol/L zAPFcmk or 10 μmol/L MG132. Mitochondrial membrane potential was assessed by the potential sensitive fluorochrome JC-1 and quantitated by FACS analysis.
Effects of proteasome inhibition on mitochondrial membrane potential. Cells were incubated in the absence or presence of 10 μmol/L methylprednisolone with or without 25 μmol/L zAPFcmk or 10 μmol/L MG132. Mitochondrial membrane potential was assessed by the potential sensitive fluorochrome JC-1 and quantitated by FACS analysis.
The proteasome is known to degrade many proteins implicated in the control of cell survival, including p53,28Fos,29 Jun,30 Myc,31p27,32 and IκBα33 (a protein inhibitor of the transcription factor NFκB34). We did not observe any detectable alteration in the levels of p53, Fos, Jun, Myc, or p27 in CLL cells treated with MG-132 compared with cells incubated in medium alone (data not shown), suggesting that these polypeptides may not participate in proteasome inhibitor-induced apoptosis in this system. On the other hand, quantification of NFκB activity by EMSA revealed that both zAPFcmk and MG-132 drastically reduced the levels of active NFκB in isolated nuclei from CLL cells (Fig 6). Thus, blockade of the NFκB survival pathway may be responsible for triggering the disruption of mitochondrial function and caspase activation in these cells.
Inhibition of NFκB activity by NS protease and proteasome inhibitors. Cells were incubated for 6 hours in the absence (control) or presence of 25 μmol/L zAPFcmk or 10 μmol/L MG132, and NFκB activity was measured in isolated nuclear extracts by EMSA using an NFκB consensus element DNA probe. Lane 1, control extracts with excess unlabeled probe (specificity control); lane 2, 6 hours control; lane 3, zAPFcmk; lane 4, MG-132. Results of one experiment typical of over 20 independent replicates.
Inhibition of NFκB activity by NS protease and proteasome inhibitors. Cells were incubated for 6 hours in the absence (control) or presence of 25 μmol/L zAPFcmk or 10 μmol/L MG132, and NFκB activity was measured in isolated nuclear extracts by EMSA using an NFκB consensus element DNA probe. Lane 1, control extracts with excess unlabeled probe (specificity control); lane 2, 6 hours control; lane 3, zAPFcmk; lane 4, MG-132. Results of one experiment typical of over 20 independent replicates.
DISCUSSION
In spite of the development of nucleoside analogs (fludarabine and cladribine) that have led to much better management of disease burden in CLL patients, CLL cells ultimately develop resistance to all currently available therapies, possibly because of apoptosis suppression. Our data show that the proteasome controls a central step in the maintenance of cell survival in CLL cells, such that inhibitors are capable of inducing apoptosis in all of them. The results support and extend independent work recently published by another group, who reported that the proteasome inhibitor lactacystin can promote radiation- and tumor necrosis factor (TNF)-induced apoptosis in CLL cells.35 The mechanism underlying the responses involves mitochondrial alterations leading to the release of cytochrome c and loss of mitochondrial membrane potential. The mitochondrial alterations are associated with caspase protease activation, as measured by specific cleavage of hallmark endogenous (PARP) and exogenous (DEVD-AMC) caspase substrates and proteolytic processing of procaspase-3. It is encouraging that we were not able to identify a single patient isolate exhibiting de novo resistance to proteasome inhibition among a fairly large (n = 59) panel, some of which (n = 21) showed marked resistance to glucocorticoid-induced apoptosis. However, our work does not address the issue of whether or not CLL cells can develop resistance to these agents under other conditions. Elimination of proteasome function in yeast results in lethality,36 but recent work suggests that mammalian cells contain another protease complex that can compensate for loss of proteasome function in cells chronically exposed to proteasome inhibitors.37
Although the precise mechanism(s) precipitating the mitochondrial changes await further investigation, our preliminary efforts indicate that the effects of MG-132 are tightly linked to suppression of NFκB activity and not to stabilization of several other proteasome-regulated factors (p53, Fos, Jun, p27) that have been implicated in the control of apoptosis in other systems. Independent results obtained recently by another group support the idea that suppression of NFκB leads to apoptosis in CLL.35 The principal mechanism regulating NFκB activation involves an inhibitor protein, IκBα, that binds to NFκB and prevents its translocation to the nucleus.38Stimulation of cells with NFκB-activating signals results in phosphorylation of IκBα and its coupling to ubiquitin,33 a small (8 kD) polypeptide that forms polymers that serve to target proteins for destruction by the proteasome.39 In other B-cell model systems, constitutive NFκB activity is essential for cell survival40 and inhibition of NFκB using protease inhibitors or mutant forms of IκBα that cannot be degraded by the proteasome facilitates apoptosis in a number of different cell types.15-17Interestingly, the immunosuppressive effects of glucocorticoid hormones are linked to an inhibition of NFκB activity,41-44suggesting that suppression of NFκB may also be required for glucocorticoid-induced apoptosis. Our ongoing efforts are focused on further characterizing the role of NFκB in the maintenance of survival in CLL.
Even though some studies had shown that proteasome inhibitors trigger apoptosis in tumor cell lines,45,46 the observation that proteasome inhibition results in apoptosis in CLL was surprising to us. Proteasome inhibitors block glucocorticoid-induced apoptosis in immature thymocytes12 and the death of neuronal cells deprived of neurotrophins,13 and ubiquitin-dependent pathways appear involved in developmentally regulated cell death in the hawkmoth Manduca sexta and in radiation-induced apoptosis in tumor cells.47 Furthermore, we had shown that inhibitors of the NS protease, a putative proteasome homolog, completely block DNA fragmentation in CLL isolates.9 However, NS protease inhibitors did promote several other features of apoptosis, including caspase activation, mitochondrial dysregulation, and PS exposure, suggesting that their effects did overlap.9 Interestingly, a peptide inhibitor of the protease complex that can compensate for loss of proteasome function (AAFcmk)37 is very similar in sequence to our NS protease inhibitor (APFcmk), both of which contain a phenylalanine (F) residue at the critical P1 position. In addition, although MG132 failed to promote substantial accumulation of p53 in CLL cells, zAPFcmk consistently did (D.J. McConkey, unpublished observations, April 1998), and the effects of the inhibitors on NFκB activity are similar (Fig 6). We are currently isolating the NS protease complex, and a detailed analysis of its structure and biochemical regulation will reveal how it is related (if at all) to the proteasome.
The proteasome is central to normal cell physiology and hence complete inhibition of its activity is ultimately cytotoxic. However, appropriate titration of proteasome activity can elicit significant efficacy with limited side effects (Peter Elliot, Julian Adams, Proscript Inc, personal communication, April 1998). Indeed, the present report shows that proteasome inhibitors induce marked apoptosis in mobilized hematopoietic progenitor cells with more modest effects on normal lymphocytes. Moreover, extensive preclinical profiling of such compounds has clearly shown that maximum tolerated doses have only modest myelosuppressive activity (P. Elliot, J. Adams, personal communication). It is likely that progenitor cell mobilization and probably other manipulations that induce cell cycling will in general sensitize cells to proteasome inhibitor-induced apoptosis, as previous work suggests that proliferating cells are more sensitive to their effects than are postmitotic cells.48 Additionally, the proteasome inhibitor, PS-341, has been shown to possess antitumor activity of its own and this effect is enhanced when combined with other chemotherapeutics (P. Elliot, J. Adams, personal communication). With the advent of acceptable phase I safety data, the present results would strongly argue that PS-341 should be evaluated in patients with refractory CLL.
ACKNOWLEDGMENT
The authors thank Virginia Snell for providing the purified hematopoietic stem cells, Yuko Miyamoto for purified peripheral lymphocytes, and Julian Adams and Peter Elliot (Proscript Inc, Cambridge, MA) for sharing preliminary data on PS-341.
Supported by grants from the National Institutes of Health (CA16672, CA55164, CA49639) (to M.A.), Physicians’ Referral Service, MDACC, the American Cancer Society (RPG-97-169-01-CDD) (to D.J.M.), and an American Legion Auxiliary Fellowship (to J.C.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to David J. McConkey, PhD, Department of Cell Biology - 173, U.T. M.D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; email: dmcconke@notes.mdacc.tmc.edu.