Abstract
We recently determined that erythropoietin (EPO) activates 3 members of the signal transducer and activator of transcription (STAT) family, Stat1α, Stat3, and Stat5, in the human EPO-dependent cell lines, UT-7 and UT-7/EPO (Kirito et al, J Biol Chem 272:16507, 1997). In addition, we have shown that Stat1α, but not Stat3, is involved in EPO-induced cellular proliferation. In this study, we examined the roles of Stat1α and Stat3 in EPO-induced erythroid differentiation. UT-7/GM was used as a model system, because this cell line can differentiate into erythroid-lineage cells with EPO treatment (Komatsu et al, Blood 89:4021, 1997). We found that EPO did not activate Stat1α or Stat3 in UT-7/GM cells. Transfection experiments showed that both Stat1α and Stat3 inhibited the induction by EPO of γ-globin and erythroid-specific 5-aminolevulinate synthetase transcripts, resulting in a reduction of the percentage of hemoglobin-positive cells. Dominant negative forms of Stat1α or Stat3 promoted the EPO-induced erythroid differentiation of UT-7/GM cells, even in the presence of granulocyte-macrophage colony-stimulating factor, although this cytokine never induced erythroid differentiation of the parent UT-7/GM cells with or without EPO. A cell cycle analysis showed that the constitutive activation of Stat1α, but not Stat3, shortened the period of G0/G1 prolongation caused by EPO stimulation. Taken together, our data suggest that Stat1α and Stat3 act as negative regulators in EPO-induced erythroid differentiation. Specifically, Stat1α may activate a cell cycle-associated gene(s), leading to the entry of cells into the cell cycle.
ERYTHROPOIETIN (EPO) plays an important role in the proliferation and differentiation of erythroid progenitor cells.1 The binding of EPO to its specific receptor on the cell surface induces tyrosine phosphorylation and the activation of several proteins, including Janus kinase 2 (JAK2),2 signal transducer and activator of transcription 5 (Stat5),3-5mitogen-associated protein kinases,6 and phospholipase C-γ1.7
We recently demonstrated that EPO induces the activation of Stat1α and Stat3 in the EPO-dependent cell lines UT-7, its subline UT-7/EPO, and F36E.8-13 By contrast, EPO did not activate Stat1α and Stat3 in UT-7/GM, another subline of UT-7, or in TF-1 cells, which grow only minimally in the presence of EPO.13-15 Transfection experiments with Stat1α and/or Stat3 showed that the overexpression of Stat1α, but not Stat3, promoted the EPO-induced proliferation of UT-7/GM cells. In addition, the introduction of EPO receptor (EPOR) cDNA into UT-7/GM cells restored not only the growth response to EPO, but also the activation of Stat1α and Stat3.13 These results suggested that Stat1α is at least partly involved in EPO-induced cell growth.
In the present study, we examined the roles of Stat1α and Stat3 in EPO-induced erythroid differentiation, because STAT proteins appear to play some role in hematopoiesis.16-18 For example, Stat3 plays a critical role in interleukin-6 (IL-6)–induced macrophage differentiation and granulocyte colony-stimulating factor (G-CSF)–induced granulocytic differentiation.16-18Although the concept is controversial at present, Stat5 may be involved in EPO-induced erythroid differentiation.19,20 Thus, STAT proteins appear to be involved in the differentiation of hematopoietic cells. Because UT-7/GM and TF-1 cells can differentiate into erythroid cells after exposure to EPO,14,15 the loss of activation of Stat1α and Stat3 by EPO may be involved in the EPO-induced erythroid differentiation of these cell lines. To test this possibility, we used UT-7/GM transfectant cells exogenously expressing Stat1α and/or Stat3 proteins13 and examined the biological functions of these Stat proteins in EPO-induced erythroid differentiation. We show here that both Stat1α and Stat3 proteins inhibit the EPO-induced erythroid differentiation of UT-7/GM cells.
MATERIALS AND METHODS
Cells.
The UT-7 cell line was derived from the bone marrow of a patient with megakaryoblastic leukemia.10 UT-7 and UT-7/GM cells were maintained in liquid culture with Iscove's modified Dulbecco's medium (IMDM; GIBCO Laboratories, Grand Island, NY) containing 10% fetal calf serum (FCS; Hyclone Laboratories, Logan, UT) and 1 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF).
Hematopoietic growth factors and reagents.
Recombinant human EPO was a gift from the Life Science Research Institute of the Snow Brand Milk Company (Tochigi, Japan). Recombinant human GM-CSF was provided by Sumitomo Pharmaceutical Co (Osaka, Japan). Neomycin (G418) was purchased from GIBCO BRL Life Technologies (Gaithersburg, MD). Human erythroid-specific 5-aminolevulinate synthetase (ALAS-E) cDNA was provided by Dr M. Yamamoto (Tsukuba University, Tsukuba, Japan). Human γ-globin cDNA and a human ribosomal DNA probe were provided by the Japanese Cancer Research Resources Bank (Tokyo, Japan). The human EPOR cDNA was a gift from Dr H. Nakauchi (Tsukuba University, Tsukuba, Japan).
Dianisidine staining.
Hemoglobin concentrations were examined by incubating cells in serum-free IMDM medium containing 0.2% 3,3′-dimethoxybenzidine, fast blue B (dianisidine; Sigma Chemicals, St Louis, MO), 0.3% acetic acid, and 0.3% H2O2 for 30 minutes at room temperature (RT).21
Preparation of nuclear and cytoplasmic extracts.
After stimulation, cells were washed with ice-cold phosphate-buffered saline (PBS) containing 2 mmol/L Na3VO4, resuspended in a hypotonic buffer (20 mmol/L HEPES [pH 7.9], 10 mmol/L KCl, 1 mmol/L MgCl2, 10% glycerol, 0.5 mmol/L dithiothreitol [DTT], 1 mmol/L phenylmethylsulphonyl fluoride [PMSF], 15 μg/mL aprotinin, 3 μg/mL leupeptin, 3 μg/mL pepstatin, and 2 mmol/L Na3VO4) with 0.2% Nonidet P-40 (NP-40) and then homogenized. After centrifugation at 1,000g for 5 minutes, the supernatant was separated from the nuclear pellet and then centrifuged at 14,000g for 20 minutes at 4°C. The debris was removed, and the supernatants were collected as cytoplasmic extracts. The nuclear pellets were resuspended in hypotonic buffer with 300 mmol/L NaCl, debris was removed by centrifugation (14,000g for 20 minutes), and the supernatants were collected as nuclear extracts.
Electromobility shift assay (EMSA).
Nuclear or cytoplasmic extracts (5 μg of protein) were incubated for 15 minutes at 4°C in 10 mmol/L Tris-HCl, pH 7.5, 75 mmol/L KCl, 1 mmol/L DTT, 1.5 μg of poly dIC/dAT, 1 mmol/L EDTA, and 4% Ficoll type 400, with 2 ng of a 32 P-labeled oligonucleotide. Samples were loaded on a 5% nondenaturating polyacrylamide gel, run for 1.5 hours at 150 V, vacuum-dried, and exposed to x-ray film. For the supershift study, nuclear extracts were incubated with antibodies at RT for 10 minutes before the above reaction. The probes were double-stranded oligonucleotides corresponding to the sis-inducible element (SIE; upper strand, 5′-GTCGACATTTCCCGTAAATC-3′)22 and the β-casein promoter region (β-CAP; upper strand, 5′-AGATTTCTAGGAATTCAAATC-3′).23 In the study of competition, extracts were incubated with a 150 molar excess of the unlabeled probe.
Generation of stable transfectants.
UT-7/GM cells were transfected with mammalian expression vector (pCAGGSneo) alone or pCAGGSneo containing human Stat1α cDNA, murine Stat3 cDNA,14 or human EPOR cDNA by conventional electroporation (1,000 V, 25 μFD). We selected 3 independent clones resistant to neomycin (0.8 mg/mL). To make UT-7/GM cells that overexpressed both Stat1α and Stat3, we constructed a pCAGGS-BSD vector containing a blasticidin-resistant gene.24
The tyrosine residues at 701 of Stat1 and at 705 of Stat3 were replaced with phenylalanine to make Stat1F and Stat3F, respectively.16 Stat1F or Stat3F cDNA also was inserted into pCAGGS vector and introduced into UT-7/GM cells under the above conditions.
Luciferase assay.
UT-7/GM cells were transfected with DNA by the lipofectin method according to the manufacturer's instructions (Promega, Madison, WI). Typically, 1.0 μg of one of the reporter plasmids containing the firefly luciferase gene and 1 μg of pSV-β-galactosidase (Promega), an expression vector containing the LacZ gene encoding β-galactosidase as an internal control for transfection efficiency were used. Two micrograms of the pCAGGS-Neo expression vector system with a cDNA encoding HA-Stat1, HA-Stat3, or their derivatives were cotransfected. After transfection, cells were starved for 24 hours and stimulated with 10 ng/mL of GM-CSF for 6 hours. The cells were then collected in 100 μL lysis buffer and subjected to the assay for luciferase and β-galactosidase activity. The reporter genes contain 4 copies of the acute-phase response element (APRE), which is transactivated by Stat1α and Stat3, in front of the minimaljunB promoter linked to the luciferase gene (4XAPREluc).16
Colorimetric MTT assay for cell proliferation.
Cell growth was also examined by a colorimetric assay according to Mosmann,25 with some modification. Briefly, cells were incubated at a density of 1 × 104/0.1 mL in 96-well plates in IMDM containing 10% FCS in the absence or presence of various concentrations of GM-CSF or EPO. After 72 hours of culture at 37°C, 20 μL of sterilized 5 mg/mL MTT [3-(4.5-dimethylthiazol-2-yl)-2.5-diphenyltetrazolium bromide; Sigma] was added to each well. After 2 hours of incubation at 37°C, 100 μL of 10% sodium dodecyl sulfate (SDS) was added to each well to dissolve the dark-blue crystal product. The optical density (OD) was measured at a wavelength of 595 nm using a microplate reader (model 3550; Bio-Rad, Richmond, CA).
RNA extraction and Northern blotting.
Total RNA was isolated from cells according to the method of Chomczynski and Sacchi.26 RNA was resolved by electrophoresis on agarose formaldehyde gels, transferred to nylon membranes (Zeta-probe; Bio-Rad) in 10× standard sodium citrate (SSC), and hybridized to human cDNA fragments for ALAS-E or γ-globin. The fragment was labeled with 32P-αCTP by random-priming. After an overnight incubation at 43°C in the presence of 50% formamide, blots were washed 3 times with 2× SSC, 0.5× SSC, or 0.1× SSC containing 0.1% SDS for 15 minutes each. The membranes were autoradiographed using Kodak XAR-5 film (Eastman Kodak, Rochester, NY) with an intensifying screen at −70°C.
Cell cycle analysis.
A cell cycle analysis was performed by staining DNA with propidium iodide in preparation for flow cytometry with the FACScan/ModFitLT system (Becton Dickinson, San Jose, CA).
RESULTS
Correlation between the GM-CSF–induced activation of Stat1α and Stat3 and the GM-CSF–induced inhibition of EPO-induced erythroid differentiation.
We previously reported that Stat1α, Stat3, and Stat5 were commonly activated by EPO in UT-7 cells and that EPO induced the activation of Stat5 but not of Stat1α or Stat3 in UT-7/GM cells.13 In addition, we demonstrated that UT-7/GM cells can differentiate along the erythroid lineage in the presence of EPO and that GM-CSF inhibited the EPO-induced erythroid differentiation of UT-7/GM cells in a dose-dependent manner.14 Collectively, the activation of Stat1α and Stat3 may be closely involved in the inhibition by GM-CSF of EPO-induced erythroid differentiation.
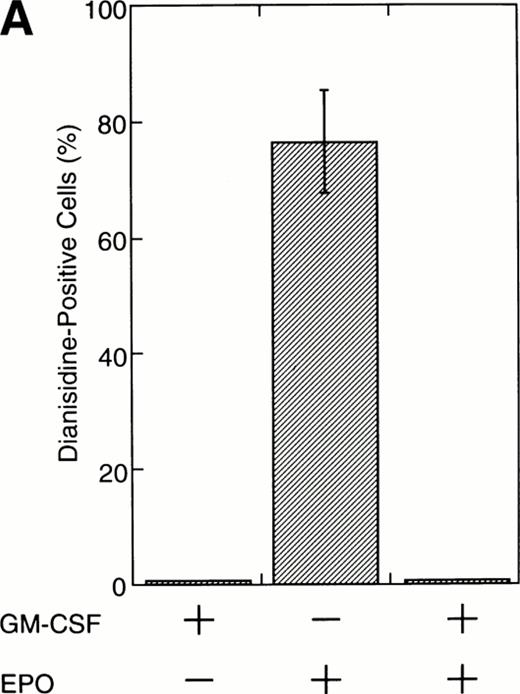
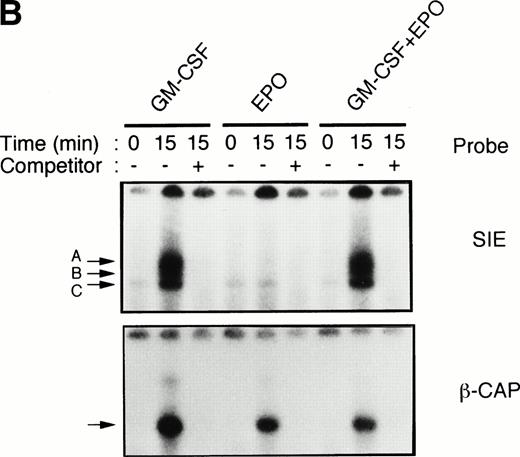
To test this notion, we examined whether SIE-binding complexes formed after stimulation with EPO (10 U/mL) plus GM-CSF (10 ng/mL). About 80% of the cells became dianisidine-positive after 7 days of exposure to EPO alone (Fig 1A). By contrast, GM-CSF strongly reduced the percentage of dianisidine-positive cells, even in the presence of high-dose EPO (Fig 1A). GM-CSF activated Stat1α and Stat3 at 10 ng/mL, at which concentration the dianisidine-positive cells completely disappeared (Fig 1B). However, Stat5 was commonly activated by GM-CSF and EPO (Fig 1B). Thus, there seems to be a positive correlation between the GM-CSF–induced activation of Stat1α and Stat3 and the GM-CSF–induced inhibition of EPO-dependent erythroid differentiation.
Activation by GM-CSF of Stat1α and Stat3 and suppression by GM-CSF of EPO-induced erythroid differentiation. (A) UT-7/GM cells were cultured with GM-CSF (10 ng/mL) and/or EPO (10 U/mL). Seven days later, cells were harvested for dianisidine staining. The data are the means ± standard deviation (SD) of triplicate cultures. (B) Growth factor-starved UT-7/GM cells were treated with EPO (10 U/mL) and/or GM-CSF (10 ng/mL) for 15 minutes. Nuclear extracts were then prepared for an EMSA with32P-labeled SIE or β-CAP probes. Arrows A, B, and C indicate a homodimer of Stat3, a heterodimer of Stat1α and Stat3, and a homodimer of Stat1α, respectively (see Kirito et al13and text).
Activation by GM-CSF of Stat1α and Stat3 and suppression by GM-CSF of EPO-induced erythroid differentiation. (A) UT-7/GM cells were cultured with GM-CSF (10 ng/mL) and/or EPO (10 U/mL). Seven days later, cells were harvested for dianisidine staining. The data are the means ± standard deviation (SD) of triplicate cultures. (B) Growth factor-starved UT-7/GM cells were treated with EPO (10 U/mL) and/or GM-CSF (10 ng/mL) for 15 minutes. Nuclear extracts were then prepared for an EMSA with32P-labeled SIE or β-CAP probes. Arrows A, B, and C indicate a homodimer of Stat3, a heterodimer of Stat1α and Stat3, and a homodimer of Stat1α, respectively (see Kirito et al13and text).
Overexpression of Stat1α or Stat3 proteins suppressed the EPO-induced erythroid differentiation.
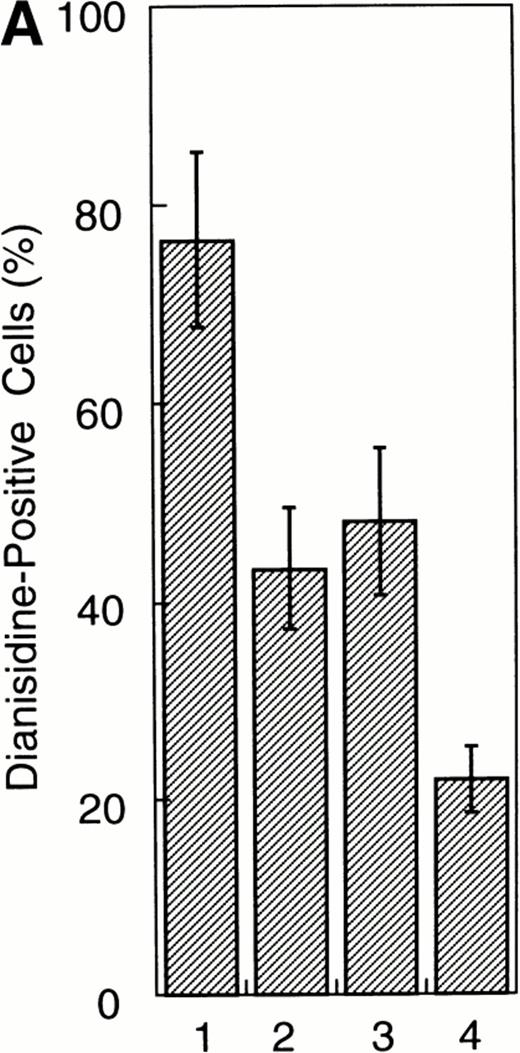
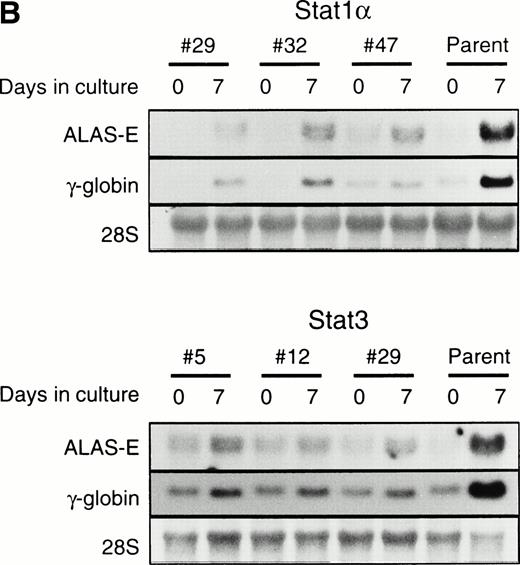
To examine whether the activation of Stat1α and Stat3 inhibits the EPO-induced erythroid differentiation, we used Stat1α- or Stat3-transfected UT-7/GM cells that we generated previously.13 We confirmed that Stat1α and Stat3 proteins are overexpressed and constitutively activated in each type of transfectant cells (Kirito et al13 and data not shown). UT-7/GM clones transfected with vector alone were used as controls. These transfectant cells were cultured with 10 U/mL EPO for 7 days and then harvested for dianisidine staining. Dianisidine-positive cells were not observed before exposure to EPO, but 70% to 80% of the control cells were positive for dianisidine after 7 days of exposure to EPO (Fig 2A). The percentage of dianisidine-positive cells decreased to 40% to 45% in the Stat1α or Stat3 transfectant clones (Fig 2A). We also evaluated the expression levels of γ-globin or ALAS-E mRNA, both of which are involved in hemoglobin synthesis. EPO enhanced the expression level of both genes in the control cells. Although the expression levels of these two genes were elevated by the EPO treatment of Stat1α or Stat3 transfectants, the degree of elevation was lower than that in the control cells (Fig2B). This finding is consistent with dianisidine-staining.
Effects of overexpression of Stat1α and/or Stat3 on the EPO-induced differentiation of UT-7/GM cells. (A) UT-7/GM clones transfected with vector alone (lane 1; n = 10), Stat1α-transfected clones (lane 2; n = 10), Stat3-transfected clones (lane 3; n = 10), and Stat1α and Stat3-cotransfected clones (lane 4; n = 3) were cultured with EPO (10 U/mL). Seven days later, cells were harvested for dianisidine-staining. The data are the mean ± SD. (B) Parent UT-7/GM cells, Stat1α-transfected clones (clones 29, 32, and 47), and Stat3-transfected clones (clones 5, 12, and 29) were cultured with EPO (10 U/mL) and harvested for the isolation of total cellular RNA. γ-Globin and ALAS-E transcripts were detected by Northern blotting. The membrane was rehybridized with a32P-labeled human ribosomal DNA probe to show the amounts of RNA loaded.
Effects of overexpression of Stat1α and/or Stat3 on the EPO-induced differentiation of UT-7/GM cells. (A) UT-7/GM clones transfected with vector alone (lane 1; n = 10), Stat1α-transfected clones (lane 2; n = 10), Stat3-transfected clones (lane 3; n = 10), and Stat1α and Stat3-cotransfected clones (lane 4; n = 3) were cultured with EPO (10 U/mL). Seven days later, cells were harvested for dianisidine-staining. The data are the mean ± SD. (B) Parent UT-7/GM cells, Stat1α-transfected clones (clones 29, 32, and 47), and Stat3-transfected clones (clones 5, 12, and 29) were cultured with EPO (10 U/mL) and harvested for the isolation of total cellular RNA. γ-Globin and ALAS-E transcripts were detected by Northern blotting. The membrane was rehybridized with a32P-labeled human ribosomal DNA probe to show the amounts of RNA loaded.
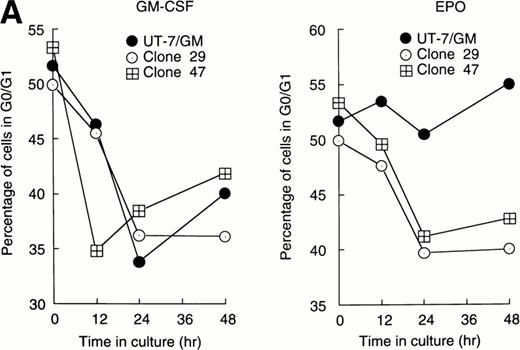
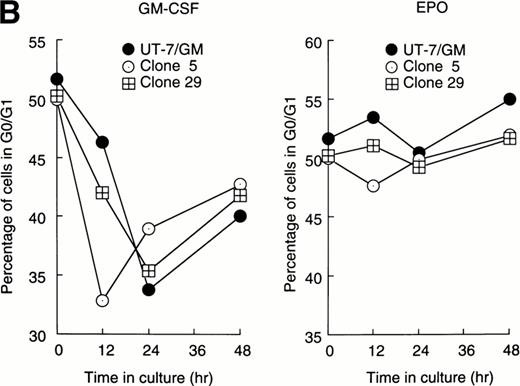
Stat1α suppressed the prolongation of the G0/G1 phase of the cell cycle after EPO stimulation.
Cellular proliferation and differentiation are closely associated with cell cycle events. For example, G0/G1 prolongation is a critical event for erythroid differentiation.27,28 We previously reported that prolongation of the G0/G1 phase of the cell cycle was required for the EPO-induced erythroid differentiation of UT-7/GM cells.14 This observation prompted us to examine whether Stat proteins inhibited the EPO-induced erythroid differentiation through an effect on cell cycle progression. After the 24-hour deprivation of growth factors, parent UT-7/GM or transfectant cells were exposed to EPO for various periods, and a cell cycle analysis was performed using flow cytometry. The percentage of G0/G1 cells at the start time was 50% to 55% in all clones. The G0/G1 percentage decreased after 12 to 24 hours of exposure to GM-CSF in both the parental and transfectant cells (Fig 3A and B). By contrast, there was a large difference in the cell cycle patterns after EPO treatment. In the parental UT-7/GM and Stat3 transfectant cells, the percentage of G0/G1 cells was unchanged even after 48 hours of culture with EPO (Fig 3B). However, when Stat1α-transfected cells were cultured with EPO, the percentage of G0/G1 cells was reduced to 35% to 40% after 24 to 48 hours of exposure to EPO (Fig 3A). These kinetics were almost identical to those produced by GM-CSF treatment (Fig 3A). These results may explain our previous observation that the cells transfected with Stat1α grew better than did the parent or Stat3-transfected cells.13
Cell cycle analysis. Parental UT-7/GM cells, Stat1α-transfected clones (A; clones 29 and 47), or Stat3-transfected clones (B; clones 5 and 29) were starved for 24 hours. The cells were subsequently stimulated with GM-CSF (10 ng/mL, left panel) or EPO (10 U/mL, right panel) and harvested at the indicated times for cell cycle analysis. The percentage of cells in G0/G1 was determined by a ModFitLT program. The data are the means of three independent experiments.
Cell cycle analysis. Parental UT-7/GM cells, Stat1α-transfected clones (A; clones 29 and 47), or Stat3-transfected clones (B; clones 5 and 29) were starved for 24 hours. The cells were subsequently stimulated with GM-CSF (10 ng/mL, left panel) or EPO (10 U/mL, right panel) and harvested at the indicated times for cell cycle analysis. The percentage of cells in G0/G1 was determined by a ModFitLT program. The data are the means of three independent experiments.
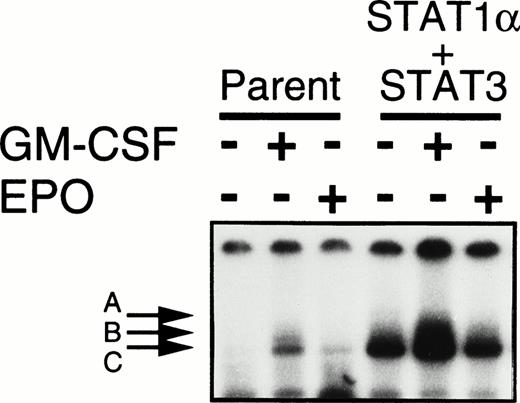
Activation of both Stat1α and Stat3 additively inhibited the EPO-induced erythroid differentiation.
Shortening of the G0/G1 phase after EPO stimulation occurred in the cells transfected with Stat1α but not in those transfected with Stat3 cDNA. These results imply that Stat1α and Stat3 inhibit EPO-induced erythroid differentiation through distinct pathways. If so, the activation of both Stat proteins could be expected to reduce the erythroid differentiation additively or synergistically. To test this notion, we introduced Stat3 cDNA into a Stat1α-transfected clone (clone 29) and selected 3 independent clones overexpressing both Stat1α and Stat3 by Western blotting analysis (data not shown). The constitutive activation of Stat1α and Stat3 was observed in these clones (Fig 4). However, the growth patterns in these clones were unchanged; the clone cells required GM-CSF or EPO for growth and survival (data not shown). Only 20% to 30% of these cells became dianisidine-positive after EPO treatment (Fig 2A). However, cell cycle patterns of the cells doubly transfected with Stat1α and Stat3 were similar to those of the cells transfected with Stat1α alone (data not shown).
Establishment of UT-7/GM cells coexpressing Stat1α and Stat3. UT-7/GM cells were cotransfected with Stat1α and Stat3 cDNA. UT-7/GM and transfectant cells were deprived of growth factors for 24 hours and then incubated with GM-CSF (10 ng/mL) or EPO (10 U/mL) for 15 minutes. Nuclear extracts were then prepared from the cells, and an EMSA was performed using 32P-labeled SIE probes.
Establishment of UT-7/GM cells coexpressing Stat1α and Stat3. UT-7/GM cells were cotransfected with Stat1α and Stat3 cDNA. UT-7/GM and transfectant cells were deprived of growth factors for 24 hours and then incubated with GM-CSF (10 ng/mL) or EPO (10 U/mL) for 15 minutes. Nuclear extracts were then prepared from the cells, and an EMSA was performed using 32P-labeled SIE probes.
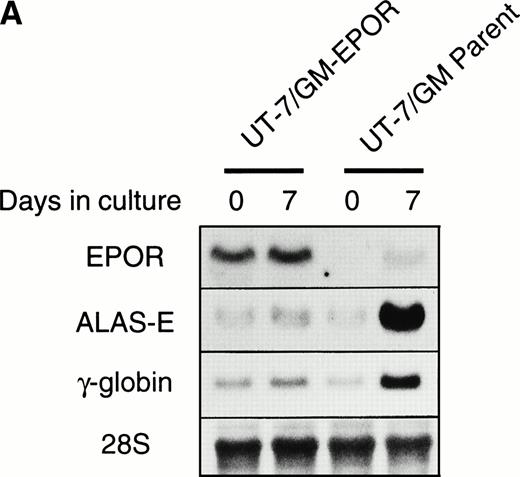
Overexpression of the full-length EPOR also decreased the EPO-induced erythroid differentiation.
Overexpression of the full-length EPOR (UT-7/GM-EPOR) restored the growth response to EPO in UT-7/GM cells (Kirito et al13 and data not shown). Moreover, we confirmed by EMSA that Stat1α and Stat3 proteins were activated by EPO in these transfectant cells (Kirito et al13 and data not shown).
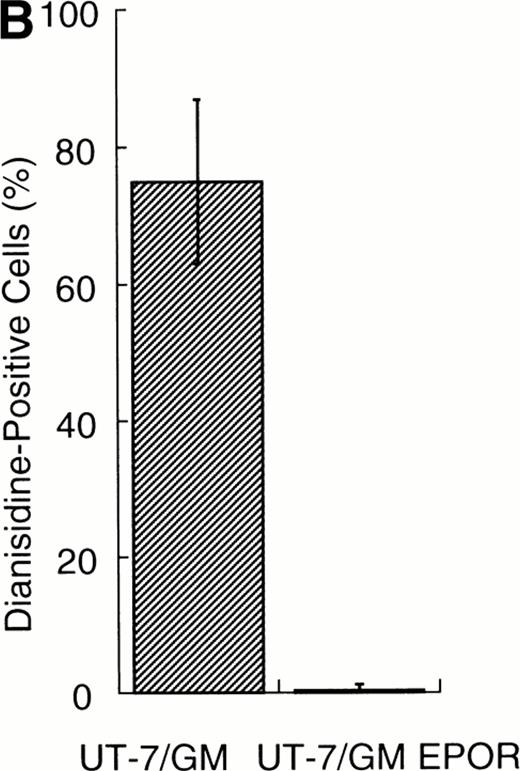
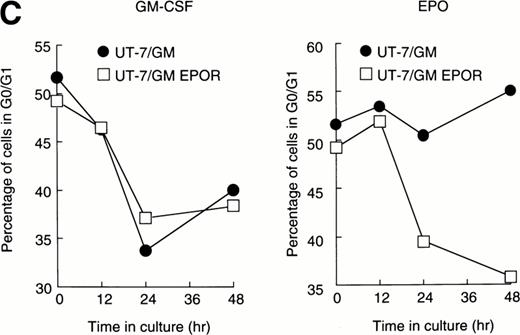
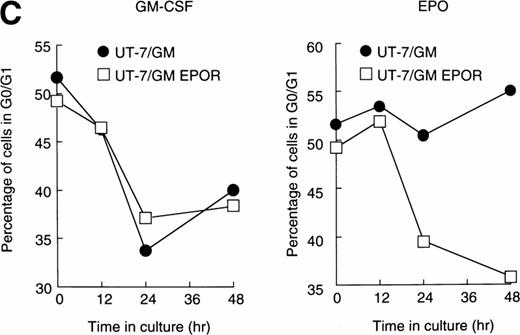
We examined whether EPOR overexpression affects the EPO-induced erythroid differentiation of UT-7/GM cells. We confirmed by Northern blotting that the parental UT-7/GM cells expressed EPOR at low levels, whereas the transfectant cells abundantly expressed EPORs (Fig 5A). Transfectant cells were cultured with EPO for 7 days and then harvested for dianisidine staining and Northern blot analysis. The percentages of dianisidine-positive cells in the EPOR-transfected cells were much lower than in the parental UT-7/GM cells (Fig 5B; 0% v 80%). EPO induced EPOR, ALAS-E, and γ-globin genes at the mRNA level in the parental UT-7/GM cells, whereas neither ALAS-E nor γ-globin gene was induced in the EPO-treated UT-7/GM-EPOR cells (Fig 5A). This result was consistent with dianisidine staining. The percentage of cells in G0/G1 was clearly reduced after EPO treatment in the UT-7/GM-EPOR cells but not in the parental UT-7/GM cells (Fig 5C), suggesting that G0/G1 prolongation did not occur in the UT-7/GM-EPOR cells.
Effects of overexpression of EPOR on the EPO-induced erythroid differentiation of UT-7/GM cells. (A) Parent UT-7/GM and UT-7/GM-EPOR cells were treated with EPO (10 U/mL) for 7 days, and total cellular RNA was isolated. The transcripts of EPOR, ALAS-E, and γ-globin were examined by Northern blotting. The membrane was rehybridized with a 32P-labeled human ribosomal DNA probe to show the amounts of RNA loaded. (B) Dianisidine-staining. UT-7/GM-EPOR cells were cultured with 10 U/mL of EPO for 7 days. The cells were harvested for dianisidine staining. The data are the mean ± SD of triplicate cultures. (C) Parent UT-7/GM cells and UT-7/GM-EPOR cells were starved for 24 hours. The cells were stimulated with GM-CSF (10 ng/mL, left panel) or EPO (10 U/mL, right panel) and harvested for cell cycle analysis. The percentage of cells in G0/G1 was determined. The data are the means of three independent experiments.
Effects of overexpression of EPOR on the EPO-induced erythroid differentiation of UT-7/GM cells. (A) Parent UT-7/GM and UT-7/GM-EPOR cells were treated with EPO (10 U/mL) for 7 days, and total cellular RNA was isolated. The transcripts of EPOR, ALAS-E, and γ-globin were examined by Northern blotting. The membrane was rehybridized with a 32P-labeled human ribosomal DNA probe to show the amounts of RNA loaded. (B) Dianisidine-staining. UT-7/GM-EPOR cells were cultured with 10 U/mL of EPO for 7 days. The cells were harvested for dianisidine staining. The data are the mean ± SD of triplicate cultures. (C) Parent UT-7/GM cells and UT-7/GM-EPOR cells were starved for 24 hours. The cells were stimulated with GM-CSF (10 ng/mL, left panel) or EPO (10 U/mL, right panel) and harvested for cell cycle analysis. The percentage of cells in G0/G1 was determined. The data are the means of three independent experiments.
Effect of dominant negative forms of Stat1α and Stat3 on the inhibition by GM-CSF of EPO-induced erythroid differentiation.
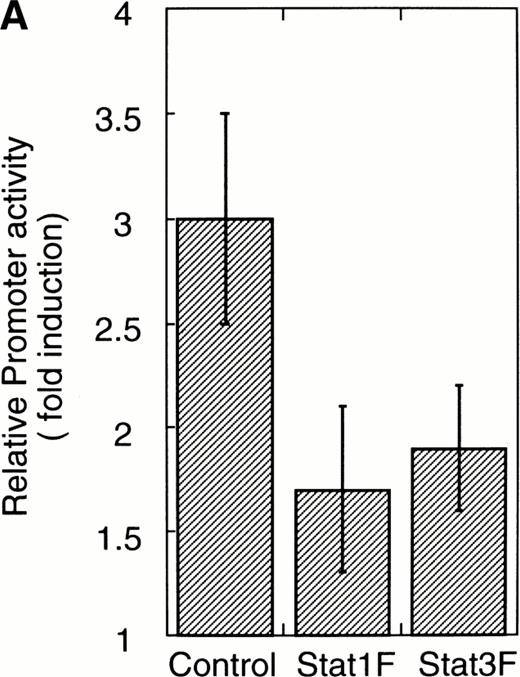
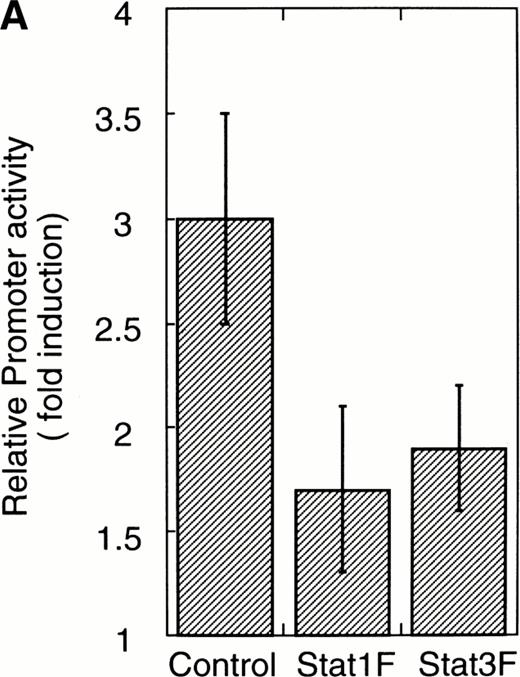
To confirm that Stat1F and Stat3F function as the dominant negative forms in our system,16,29 30 we cotransfected these cDNAs and 4XAPREluc into the parent UT-7/GM cells. After 24 hours of starvation, these transfectant cells were stimulated with GM-CSF, and the cells were harvested 6 hours later for a luciferase assay. As shown in Fig 6A, the luciferase activity was clearly reduced in the Stat1F and Stat3F transfectant cells, indicating that these dominant-negative forms work well in our system. In addition, we confirmed by EMSA that the dominant-negative Stats actually inhibited the DNA binding activities of the wild-type Stats in UT-7/GM cells (data not shown).
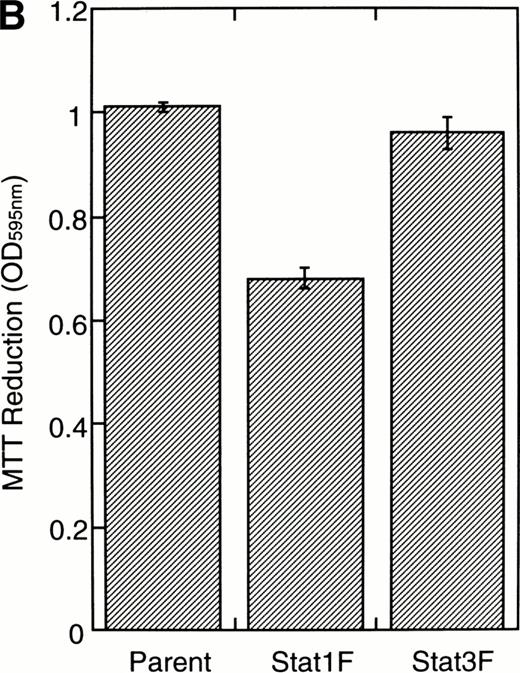
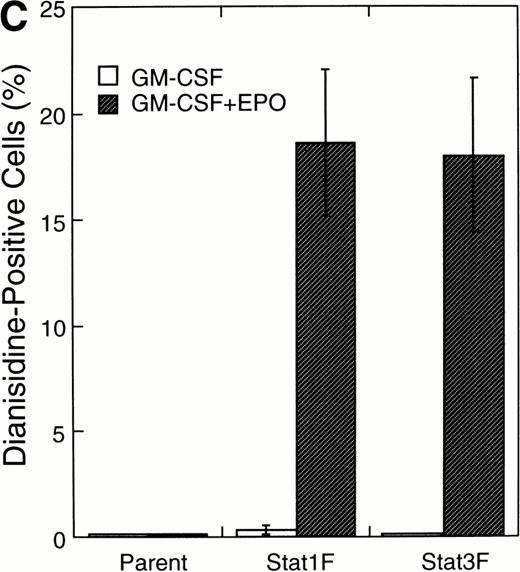
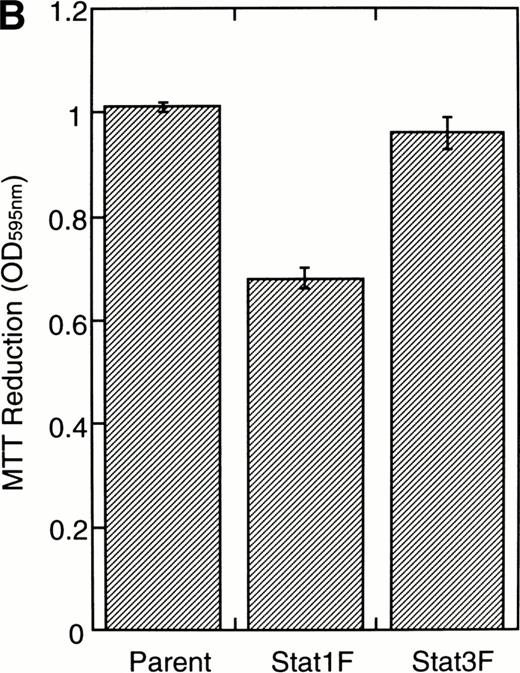
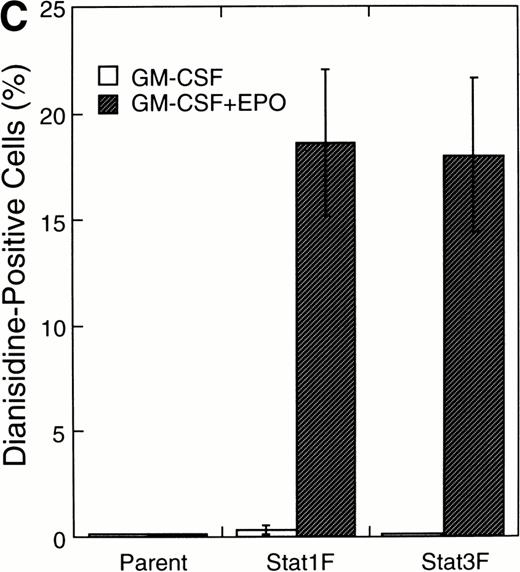
Effects of dominant-negative forms of Stat1α and Stat3 on the inhibition by GM-CSF of EPO-induced erythroid differentiation. (A) UT-7/GM cells were transfected with a plasmid DNA mixture (1 μg of reporter genes containing 4 copies of APRE in front of the minimaljunB promoter linked to the luciferase gene, 2 μg of either expression vector pCAGGS-Neo, with no insert or with an insert of Stat cDNA encoding either HA-Stat3F or HA-Stat1F, and 1 μg of pSV-β-galactosidase. After transfection, the cells were cultured without growth factors and then stimulated with GM-CSF (10 ng/mL) for 6 hours. Luciferase values were normalized for β-galactosidase activity and expressed relative to the normalized luciferase activity in the extracts from unstimulated cells transfected with the reporter plasmids and a control expression plasmid. The data are the mean ± SD of more than four independent experiments. (B) MTT reduction assay. Cells were plated at a density of 104/well in IMDM supplemented with 5% FCS and cultured with GM-CSF (10 ng/mL). MTT reduction was measured after 3 days of culture. The data are the mean ± SD of triplicate cultures. (C) Dianidisine staining. Cells were cultured in the presence of GM-CSF (10 ng/mL) or a combination of EPO (10 U/mL) and GM-CSF (10 ng/mL). Seven days later, cells were harvested for dianisidine staining. The data are the mean ± SD from three independent clones.
Effects of dominant-negative forms of Stat1α and Stat3 on the inhibition by GM-CSF of EPO-induced erythroid differentiation. (A) UT-7/GM cells were transfected with a plasmid DNA mixture (1 μg of reporter genes containing 4 copies of APRE in front of the minimaljunB promoter linked to the luciferase gene, 2 μg of either expression vector pCAGGS-Neo, with no insert or with an insert of Stat cDNA encoding either HA-Stat3F or HA-Stat1F, and 1 μg of pSV-β-galactosidase. After transfection, the cells were cultured without growth factors and then stimulated with GM-CSF (10 ng/mL) for 6 hours. Luciferase values were normalized for β-galactosidase activity and expressed relative to the normalized luciferase activity in the extracts from unstimulated cells transfected with the reporter plasmids and a control expression plasmid. The data are the mean ± SD of more than four independent experiments. (B) MTT reduction assay. Cells were plated at a density of 104/well in IMDM supplemented with 5% FCS and cultured with GM-CSF (10 ng/mL). MTT reduction was measured after 3 days of culture. The data are the mean ± SD of triplicate cultures. (C) Dianidisine staining. Cells were cultured in the presence of GM-CSF (10 ng/mL) or a combination of EPO (10 U/mL) and GM-CSF (10 ng/mL). Seven days later, cells were harvested for dianisidine staining. The data are the mean ± SD from three independent clones.
To investigate the functional roles of Stat1α and Stat3 in erythroid differentiation, we generated stable transfectant cells exogenously expressing Stat1F or Stat3F. An MTT assay showed that Stat1F but not Stat3F partially inhibited the GM-CSF–induced cell growth of UT-7/GM cells (Fig 6B). Then, these transfectant cells were cultured with GM-CSF alone (10 ng/mL), EPO (10 U/mL), or a combination of GM-CSF (10 ng/mL) and EPO (10 U/mL) for 7 days and then harvested for dianisidine staining. GM-CSF completely blocked the EPO-induced erythroid differentiation in the parental UT-7/GM cells. However, unlike the parental UT-7/GM cells, about 20% of the cells transfected with Stat1F or Stat3F showed the positivity for dianisidine staining after exposure to EPO, even in the presence of GM-CSF (Fig 6C). Approximately 80% of the cells treated with EPO became positive for dianidisine staining in these transfectant cells (Stat1F, 75.0% ± 2.7%; Stat3F, 75.4% ± 5.0%), as in the case of the parental UT-7/GM cells (76.5% ± 8.7%).
DISCUSSION
In this study, we established the functional role of Stat proteins in EPO-induced erythroid differentiation. The forced activation of Stat1α and/or Stat3 suppressed the induction of γ-globin and the erythroid-specific ALAS gene, resulting in decreased hemoglobin synthesis. In addition, UT-7/GM cells transfected with EPOR cDNA restored the EPO-induced activation of Stat1α and Stat3 and acquired high responsiveness to EPO for cell proliferation. However, these cells lost their capacity to become hemoglobin-positive in the presence of EPO. Dominant negative forms of Stat1α or Stat3 promoted EPO-induced erythroid differentiation even in the presence of GM-CSF. These results indicate that both Stat1α and Stat 3 act as negative regulators of EPO-induced erythroid differentiation.
The regulation of cell cycle progression appears to be involved in the cellular switch to the proliferation and differentiation of erythroid progenitor cells.27,28 Carroll et al27 reported that prolongation of the G0/G1 phase was required for EPO-induced erythroid differentiation. We recently found that the addition of GM-CSF reduced the G0/G1 prolongation, accompanied by a reduction in the ratio of dianisidine-positive cells.14 In addition, we showed here that the inhibition by GM-CSF of EPO-induced erythroid differentiation correlated with the activation of Stat1α and Stat3 (Fig 1A and B) and that the activation of Stat1α promoted the entry of UT-7/GM cells into the cell cycle. This notion was also supported by the finding that Stat1F but not Stat3F inhibited the GM-CSF–induced cell growth of UT-7/GM cells. Taken together with reports that Stat1α is also involved in the proliferative effects of platelet-derived growth factor (PDGF) and epidermal growth factor (EGF),31our data raise the possibility that Stat1α is directly or indirectly involved in the activation of cell cycle-associated gene transcription.
There are several lines of evidence that the activation of Stat proteins is involved in the cell cycle. It was demonstrated that a dominant-negative form of Stat3 inhibited growth arrest at the G0/G1 phase induced by IL-6 in a murine leukemic cell line, M1.16In addition, another group reported the EGF- or interferon-γ (IFN-γ)–induced growth arrest of A431 or HT29 cells by the upregulation of the cyclin-dependent kinase inhibitor, p21; this was mediated through the binding of activated Stat1α to the promoter of the p21 gene.32 Thus, Stat1α and Stat3 proteins appear to play a role in the blockade of cell cycle progression. However, this notion is inconsistent with our present finding that Stat1α proteins promoted entry into the cell cycle. This discrepancy may be explained partly by the evidence that p21 acts not only as a negative regulator, but also as a positive regulator of cell cycle progression.33 34
It is apparent that the inhibition by Stat3 of the EPO-induced erythroid differentiation of UT-7/GM cells is independent on the cell cycle. Indeed, the forced overexpression of Stat3 had no effect on the EPO-induced proliferation of UT-7/GM cells.13 Moreover, Stat3 promotes the differentiation of M1 cells to macrophages. Very recently, it was found that Stat3 inhibits the neural differentiation of PC12 cells.29 Whereas all of these data were obtained using immortalized cell lines, it was recently shown that Stat3 is involved in the maintenance of embryonic stem (ES) cells by leukemia inhibitory factor (LIF); the Stat3 dominant-negative form induced morphological differentiation of ES cells even in the presence of LIF.35 Thus, it is unlikely that the inhibitory effects of Stat3 on differentiation are limited to immortalized cells. Stat3 could also be expected to play an important role in normal development, presumably as an inhibitor of differentiation.
Although the dominant-negative forms of Stat1α and Stat3 certainly inhibited the GM-CSF–induced activation of Stat1α and Stat3, the majority of the transfectant cells did not become dianisidine-positive in the presence of GM-CSF alone, as was the case for the parental UT-7/GM cells. However, unlike the parent cells, about 20% of the transfectant cells became dianidisine-positive after exposure to EPO, even in the presence of GM-CSF (Fig 6C). These observations strongly suggest that the loss of activation of Stat1α and Stat3 is required, but is not sufficient by itself, for EPO-induced erythroid differentiation, and that the EPO signal(s) is a prerequisite for erythroid differentiation. In addition, the finding that the constitutive activation of Stat1α and Stat3 proteins partially but not completely inhibited EPO-induced erythroid differentiation of UT-7/GM cells suggests that other molecules such as Lyn36and SHP-137 may be in part involved in the suppression by GM-CSF of the EPO-induced erythroid differentiation of UT-7/GM cells.
The constitutive activation of Stat1α or Stat3 is frequently observed in leukemia cells.38-40 In addition, Stat1α and Stat3 are activated by Bcr-Abl products41 and src-kinase,42,43 respectively. The infection of Friend spleen focus-forming virus in the EPO-dependent murine cell line HCD 57 causes cytokine-independent cell growth and the constitutive activation of Stat1α and Stat3.44 A mutation in the Drosophilia homolog of the JAK kinase gene causes the hyperactivation of this signal transduction pathway and subsequent leukemia-like hematopoietic defects.45-47 Thus, aberrations in the JAK-STAT pathway could cause the development of hematologic malignancy. Although the constitutive activation of Stat1α and/or Stat3 blocked the EPO-induced erythroid differentiation of these transfectant cells, we did not observe autonomous cell growth (Kirito et al13 and data not shown). These results indicate that the constitutive activation of Stat1α and/or Stat3 is insufficient for the autonomous growth of leukemia cells in vitro. The mechanism by which exogenous expression of Stat1α and Stat3 in UT-7/GM cells resulted in constitutive activation of these molecules is unknown (Fig 4). Because mere overexpression of Stat molecules, even in COS cells, usually does not give rise to constitutive activation, the overexpressed Stat1α and Stat3 proteins might be showing the action of normally subthreshold amounts of an autocrine factor.
Because Stat3 knockout mice have a lethal defect, it is difficult to estimate the function of Stat3 on erythropoiesis.48Stat1α knockout mice studies demonstrated that the elimination of the Stat genes does not affect erythropoiesis.49 50 This seems to contradict our observation. This discrepancy may be explained by two possibilities. One is that other Stat proteins such as Stat3 compensate for the lack of Stat1α. The other is that Stat1α is involved in abnormal erythropoiesis, as shown by the results obtained using leukemic cell line cells.
It is also interesting that most of the UT-7/GM cells lost the capacity to differentiate into erythroid cells by overexpression of the full-length EPOR. By contrast, these transfectant cells had a restored growth response to EPO and activation of Stat1α and Stat3 by EPO.13 This indicates that Stat1α and Stat3 lie downstream of the EPO receptor in the EPO signaling pathway. However, Stat3-Stat3 homodimers were almost undetectable in the EPO-treated UT-7/GM-EPOR cells (data not shown). This may be explained by one possibility that the ratio of activated Stat3 to activated Stat1α by EPO is relatively small, resulting in the increased formation of Stat1α-Stat3 heterodimers and the decreased formation of Stat3-Stat3 homodimers.
It seems likely that the inability of UT-7/GM cells to proliferate in response to EPO is due to low levels of endogenous EPO receptor expression. Although we cannot completely exclude the possibility that decreased sensitivity to EPO is due to the presence of mutations in the endogenous EPO receptor, we and another group demonstrated that the original UT-7 cells expressed structurally normal EPOR mRNA.51 52
Whether Stat5 plays a critical role in EPO-induced erythroid differentiation is still controversial. Chretien et al19reported that the activation of Stat5 promotes cell growth and inhibits erythroid differentiation in a human erythroleukemia cell line. By contrast, Iwatsuki et al20 found that the activation of Stat5 leads to erythroid differentiation in a murine erythroleukemia cell line. We observed that EPO induced erythroid differentiation in UT-7/GM cells but not in UT-7 cells. However, there was no significant difference in the degree of Stat5 activation by EPO between these cell lines (Kirito et al13 and data not shown). Although the possibility that this discrepancy is caused by differences in the cell lines cannot be excluded completely, our data suggest that Stat5 activation is insufficient for EPO-induced erythroid differentiation. A separate factor(s) is required for the terminal maturation of erythroid cells.
It is still controversial as to whether EPO induces the activation of Stat1α and/or Stat3 as well as Stat5. Some investigators reported that Stat5, but not Stat1α or Stat3, is activated by EPO in Ba/F3 cells transfected with EPOR cDNA.53,54 To test this, we also generated Ba/F3 cells exogenously expressing human EPOR and examined whether EPO induces the activation of Stat1α and Stat3 proteins in these transfectant cells. As reported previously, neither of these Stat proteins was activated by EPO stimulation, although Stat5 protein was strongly activated by EPO treatment (data not shown). Thus, the data obtained from Ba/F3 transfectant cells were in contrast with those from the UT-7 cells expressing endogenous human EPOR. Although we cannot completely exclude the possibility that this discrepancy may be due to the different cell lines used in these experiments or the different EPOR number on the surface of the cells, it is likely that EPO induces the activation of Stat1α and/or Stat3 proteins in primary erythroid cells and cell lines abundantly expressing endogenous EPOR.13,44,55 56
In summary, we propose a novel function of Stat1α and Stat3 in EPO-induced erythroid differentiation of a leukemia cell line; Stat1α may inhibit EPO-induced erythroid differentiation, presumably mediated through the activation of cell cycle-associated gene(s). Although the roles of Stat1α and Stat3 in normal erythropoiesis remains to be undetermined, the constitutive activation of these Stat proteins may be involved in the erythroid differentiation arrest in erythroleukemia.
ACKNOWLEDGMENT
The authors thank the Life Science Research Institute of Snow Brand Milk Co for providing recombinant human EPO. We are also grateful to Tomoko Ando for technical assistance and Motoko Yoshida for manuscript preparation.
Supported by Grants-in-Aid for Cancer Research and Scientific Research from the Ministry of Education, Science and Culture of Japan and by a grant from the Yamanouchi Foundation for Research on Metabolic Disorders.
Address reprint requests to Norio Komatsu, MD, PhD, Division of Hematology, Department of Medicine, Jichi Medical School Minamikawachi-machi, Tochigi-ken 329-04, Japan; e-mail:nkomatsu@ms.jichi.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.