Abstract
Transcripts for the retinoic acid receptors (RARs) α1, α2, γ1, and γ2 were found in the granulocytic lineage (Gr-1+cells) through semiquantitative polymerase chain reaction (PCR) analysis. The screening of single cell cDNA libraries derived from hematopoietic progenitors also showed the presence of RARα and, to a lesser extent, RARγ transcripts in committed granulocyte (colony-forming unit-granulocyte [CFU-G]) or granulocyte-macrophage (CFU-GM) colony-forming cells. The contribution of RARα1 and γ to hematopoietic cell differentiation was therefore investigated in mice bearing targeted disruption of either one or both of these loci. Because RARγ and RARα1γ compound null mutants die shortly after birth, bone marrow cells were collected from fetuses at 18.5 days postcoitum (dpc) and evaluated for growth and differentiation in culture in the presence of Steel factor (SF), interleukin-3 (IL-3), and erythropoietin (Epo). The frequency of colony-forming cells from bone marrow populations derived from RARα1/γ double null mice was not significantly different from that of RARγ or RARα1 single nulls or from wild-type controls. In addition, the distribution of erythroid, granulocyte, and macrophage colonies was comparable between hematopoietic cells from all groups, suggesting that lineage commitment was not affected by the lack of RARα1 and/or RARγ. Colony cells were then harvested individually and evaluated by morphologic criteria. While terminal granulocyte differentiation was evident in wild-type cells and colonies from either single null mutant, colonies derived from RARα1−/−γ−/− bone marrow populations were blocked at the myelocyte and, to a lesser extent, at the metamyelocyte stages, whereas erythroid and macrophage differentiation was not affected. Together, these results indicate that both RARα1 and γ are required for terminal maturation in the granulocytic lineage in vitro, but appear to be dispensable for the early stages of hematopoietic cell development. Our results raise the possibility that in acute promyelocytic leukemia (APL), the different RARα fusion proteins cause differentiation arrest at a stage when further maturation requires not only RARα, but also RARγ. Finally, bone marrow cells appear to differentiate normally in vivo, suggesting an effective compensation mechanism in the RARα1/γ double null mice.
THE GENERATION OF hematopoietic cells proceeds through complex yet discrete differentiation events that include an irreversible commitment step from a multipotent precursor, followed by terminal maturation events resulting in functional end cells (ie, granulocytes, monocytes, erythrocytes, platelets, and lymphocytes). There is increasing evidence that differentiation along a given pathway is driven by lineage-restricted transcription factors that act cell autonomously to activate a defined set of target genes, thus specifying cell types in normal hematopoiesis (reviewed by Orkin1). Leukemic cells, however, are arrested at specific stages of differentiation and leukemias have thus been classified into distinct subtypes according to their morphologic appearance. Several of the French-American-British (FAB) classification subtypes correlate with consistent chromosomal translocations, which frequently involve loci encoding transcription factors. Not surprisingly, these transcription factors are also often crucial for normal hematopoiesis (reviewed by Wolff2).
Retinoids are essential regulators of complex differentiation processes (reviewed by Lohnes et al3). The retinoid signal is transduced by two families of nuclear receptors, the retinoic acid (RA) receptors (RARα, β, and γ and their isoforms) and the retinoid X receptors (RXRα, β, and γ) (reviewed by Kastner4). RARs function as ligand-inducible transcription regulators by binding, together with an RXR partner,5 to specificcis-acting sequences (RA response elements; RAREs). RARs can be activated by both all-trans and 9-cis RA, whereas RXRs are activated only by 9-cis RA. RXRs are also essential heterodimeric partners for a number of other nuclear receptor signalling pathways.6 7
Targeted gene disruption has been used to evaluate the function of RAR types and isoforms in vivo. Mice deficient for RARα1, the major RARα isoform, and RARγ2 are apparently unaffected.3,8,9However, ablation of all RARα or γ isoforms results in a low frequency of skeletal malformations, as well as a subset of defects related to postpartum vitamin A deficiency.3,8 In contrast to the relatively minor consequence of disruption of a single receptor, defects evoked by loss of various combinations of RARs recapitulate essentially all of the pathologies associated with congenital vitamin A deficiency, together with the appearance of malformations not previously seen in such dietary manipulation studies.3,10,11 Thus, gene targeting suggests some degree of functional redundancy between RARs5,12,13 (reviewed by Perlmann and Evans14).
A role for RARα in hematopoiesis was first suggested by its locating at the breakpoint of several chromosomal translocations associated with acute promyelocytic leukemia (APL; M3 in the FAB classification). The predominant t(15;17) translocation and the variant t(11;17) and t(5;17) translocations result in the creation of PML-RARα, PLZF-RARα, and NPM-RARα chimeras, respectively.15-20 In APL, granulocytic differentiation is blocked at the promyelocyte stage. RA treatment in vivo relieves this block and favors terminal granulocyte differentiation in patients with the t(15;17) translocation,21-24 albeit resulting in the appearance of polymorphonuclear cells with abnormal granulations.25Together, these observations indicate an important role for RARα in terminal granulocyte differentiation.
Both PML-RARα and PLZF-RARα inhibit RA-dependent transactivation of wild-type RARs.26-28 Moreover, a dominant negative C-terminal truncated RARα (RARα Δ403) can also elicit differentiation arrest in the granulocytic lineage at the promyelocyte stage.29 These observations further suggest that attenuation of RARα signaling plays an important role in the pathogenesis of APL. Dominant negative mutants, however, can interfere with the function of all RAR types, and could conceivably affect other RXR-dependent pathways, such that the contribution of each RAR to hematopoiesis has not been clearly established. To directly address the role of RARα1 and RARγ in hematopoietic cell differentiation, we evaluated the growth and differentiation potential of bone marrow cells from mice in which either one or both loci were inactivated.
MATERIALS AND METHODS
Generation of RAR mutants.
The RARα1, RARγ, and RARα1/γ knockout animals used in these studies have been described previously.8-10 To generate wild-type and RAR null fetuses, the appropriate matings were established and females surveyed daily for a vaginal plug; noon of the day of plug was considered 0.5 dpc. Females were killed at 18.5 dpc, fetuses obtained by caesarean section, and genotype assigned by polymerase chain reaction (PCR) using placental DNA as previously described.30
Isolation of bone marrow cells and methylcellulose cultures.
Long bones and the sternum were freed of surrounding tissues and cut into small pieces. Bone marrow cells were isolated by repeated aspiration through a Pasteur pipette and bone particles eliminated through differential deposition in Iscove's medium (IMDM; GIBCO, Grand Island, NJ) supplemented with 10% fetal calf serum (FCS).
Bone marrow cells were counted and either cytosmeared for evaluation by morphologic criteria or cultured in methylcellulose, as previously described.31-33 For methylcellulose cultures, 5 × 104 mononuclear cells were plated in 1 mL of IMDM supplemented with 10 % FCS, bovine serum albumin (20 mg/mL; Sigma, St Louis, MO), iron saturated transferrin (160 μg/mL; Sigma), recombinant murine SF, WEHI-3 conditioned medium (2.5%, as a source of IL-3) and recombinant human erythropoietin (hu Epo; 1 U/mL), and viscified with 1% methylcellulose (Fluka, Rokonka, NY). Colonies were scored on day 7 of culture with an inverted microscope as described.31 After counting, colonies were harvested individually and cells evaluated by morphologic criteria using Giemsa-stained cytosmears.
Isolation of Gr-1+ cells.
Gr-1+ and Gr-1− cells were purified from wild-type adult mouse bone marrow by immunomagnetic selection. Briefly, after erythrocyte lysis by osmotic shock, nucleated bone marrow cells were depleted of B cells through incubation with magnetic beads coupled to a polyclonal goat antimouse antibody at a bead to cell ratio of 8. Negative cells were coated with human Ig (Sigma) in ice cold IMDM/10% FCS for 30 minutes to block Fc binding. The cells (5 × 106) were then labeled with purified Gr-1 (1 μg/100 μL) for 30 minutes, washed, followed by a second labeling with MAR18.5 (specific for rat κ chains). After washing, the cells were resuspended in medium containing goat antimouse coupled magnetic beads as above. Both positive and negative cells were resorted twice to eliminate nonspecific cosedimenting cells and cell pellets (5 × 106) frozen for RNA extraction.
Semiquantitative reverse transcription-PCR (RT-PCR) analysis.
RNA was purified from approximately 5 × 106 Gr-1+ or Gr-1− cells or from 100 μg of newborn mouse skin by Trizol extraction (GIBCO-BRL). Approximately 2 μg of RNA was reverse transcribed with 300 U of Moloney murine leukemia virus (MMLV) reverse transcriptase (GIBCO-BRL) using hexanucleotide primers under standard reaction conditions. One tenth (2 μL) of the first strand cDNA was then used as substrate for PCR amplification using primers specific for the transcripts of interest. Amplification reactions were terminated every second cycle from 25 to 35 cycles or after 50 cycles. Reaction products were resolved on a 1.5% agarose gel, transferred to Gene Screen plus membranes (DuPont) according to the manufacture's instructions, and hybridized with end-labeled oligonucleotides corresponding to internal sequences specific to each predicted PCR product. Oligonucleotides used for PCR were as follows; for amplification of RARα isoforms, the primer CCGGATGATTTGTCTTGACA was used in combination with either GTTGGGCTGACCACCCAACC or AGTGACCTGCAGACTTAGGC for amplification of RARα1 or RARα2, respectively; RARβ transcripts (all isoforms) were amplified using the oligonucleotides ACATGAACCCTTGACCCCAAG and TTTAAACTAGATTCTGGTGG; RARγ isoforms were amplified using the primer CTTCACAGGAGCTGACCCCAT and either AGCCTGGCCCAGTATGTAGG or ATCCCTTACCCCCCATGC for RARγ1 or RARγ2, respectively; β-actin was amplified using the primers GAGGGCATACCCCTCGTAGAT and CAGAAGGATTCCTATGTGGGC. End-labeled oligonucleotides used for Southern blot analysis to confirm the fidelity of amplification were as follows; RARα isoforms, ATCGAGACCCAGAGCAGCAG; RARβ isoforms, CCATCGAGACACAGA; RARγ isoforms, CAGAGCACCAGCTCGGAGGA; β-actin, AGAGAGGTATCCTGACC.
Analysis of single cell cDNA.
3′ murine RARα or γ cDNA probes corresponding to sequences immediately upstream of the polyadenylation signal was generated by 3′ rapid amplification of cDNA ends (RACE) using TTGGACACTCTAAGCGGA and TTGAGGACGACTCCTCGA as 5′ primers for RARα and γ, respectively. The identity of the amplified product was confirmed by restriction analysis and by specific hybridization to internal oligonucleotides. Southern blots of single cell cDNA libraries identified to specific progenitors by sib analysis as detailed previously,34 35 were hybridized to the probes, exposed to a PhosphorImager screen, and quantitated.
RESULTS
Gr-1 is a cell surface marker of the Ly-6 family of adhesion molecules and its expression is restricted to more differentiated granulocytes,36 whereas granulocyte precursors (colony-forming unit-granulocyte [CFU-G]) and other hematopoietic lineage precursors are Gr-1−.37 Because of differentiation arrest at the promyelocyte stage in APL and its association with expression of dominant negative RARα chimeras, we chose to study RAR gene expression in bone marrow populations separated on the basis of Gr-1 expression. After cell sorting, the purity of the Gr-1+ population was confirmed through morphologic examination of cytosmears and was found to consist of 97% differentiated granulocytes (metamyelocytes, bands and polymorphonuclear [PMN]) and 1.5% myelocytes. Myeloblasts and promyelocytes were found in the Gr-1−population, together with cells of all other lineages.
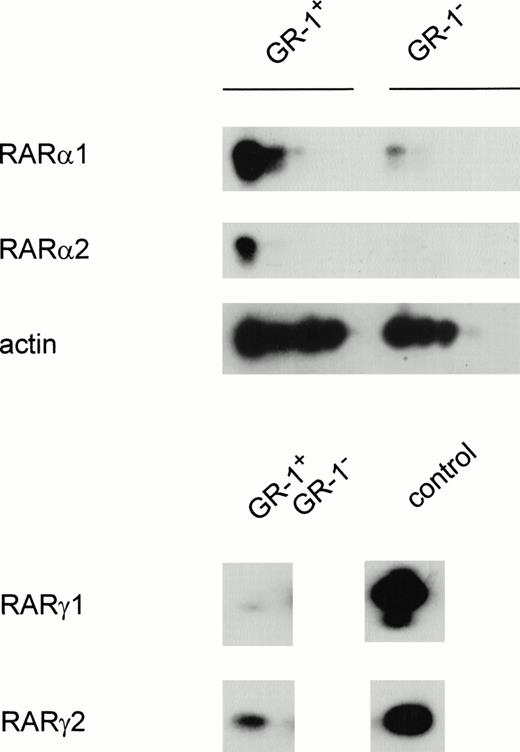
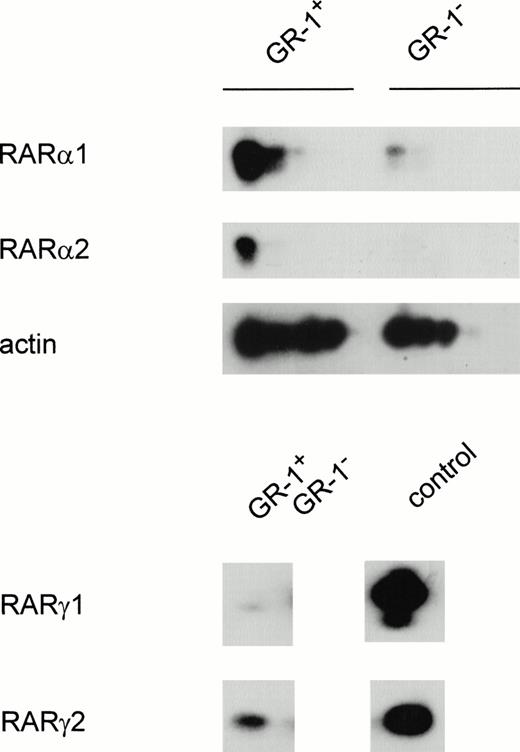
RT-PCR analysis of Gr-1+ and Gr-1− cells indicated that the RARα1 message was present in both cell populations, although Gr-1− cells appeared to express much less of this transcript compared with the Gr-1+ pool (Fig 1). In contrast, RARα2 transcripts were undetectable in Gr-1− cells under conditions where it was readily observed in Gr-1+ isolates. Transcripts encoding RARγ isoforms were undetectable after 35 cycles of PCR, irrespective of Gr-1 status. However, RARγ2 transcripts were detected in Gr-1+, and to a lesser extent in Gr-1−, cells after 50 cycles of amplification. RARγ1 transcripts were barely detectable in the former cell population and were undetectable in Gr-1− cells. RARβ transcripts were not detectable in either cell population despite extensive PCR amplification (data not shown). Thus, RARα1 appears to be the major RAR transcript in Gr-1+ cells, while RARα2, RARγ1 and RARγ2 are much less abundant. In contrast, only RARα1 and γ2 were detected at low levels in GR-1− cells. Similar results were observed with cells that were sorted by flow cytometry, suggesting that these low levels of RARα1 and RARγ2 in GR-1− cells were not due to contaminating Gr-1+ cells (data not shown).
Semiquantitative PCR analysis of RAR expression in differentiated granulocytes (Gr-1+) and other cell types (Gr-1−). Cells were separated as described in Materials and Methods. The PCR reaction was performed for 25 to 35 cycles for RARα1 and RARα2 and 50 cycles for RARγ and RARβ. Shown is a Southern blot analysis of the PCR product hybridized with an internal oligonucleotide probe specific for each amplification product, as described in Materials and Methods.
Semiquantitative PCR analysis of RAR expression in differentiated granulocytes (Gr-1+) and other cell types (Gr-1−). Cells were separated as described in Materials and Methods. The PCR reaction was performed for 25 to 35 cycles for RARα1 and RARα2 and 50 cycles for RARγ and RARβ. Shown is a Southern blot analysis of the PCR product hybridized with an internal oligonucleotide probe specific for each amplification product, as described in Materials and Methods.
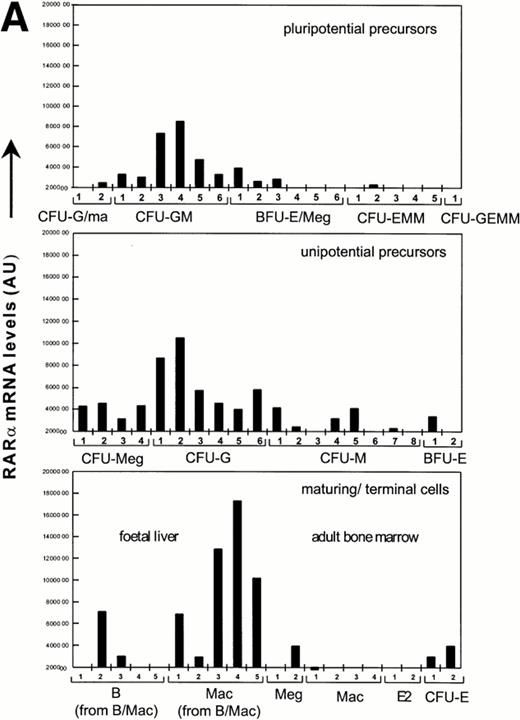
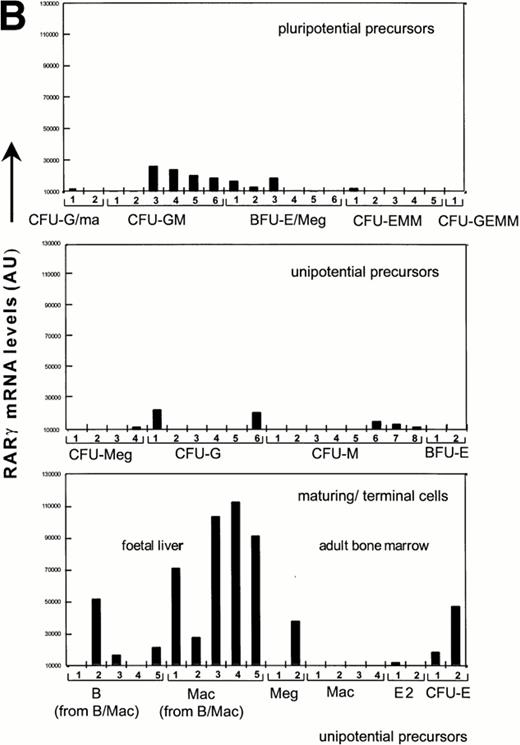
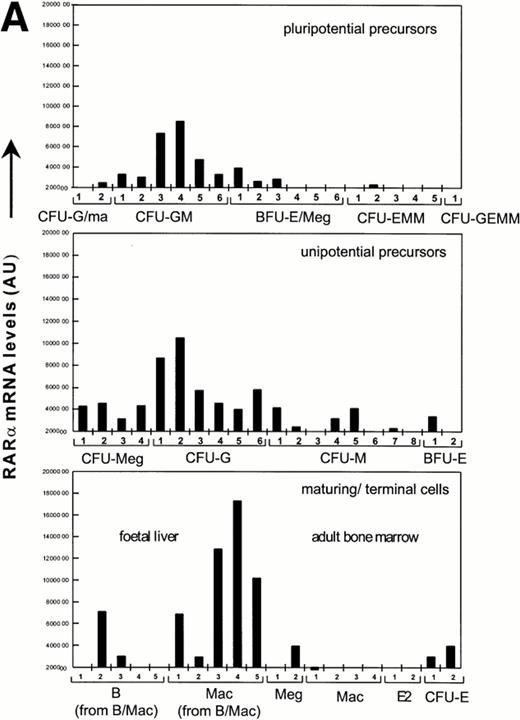
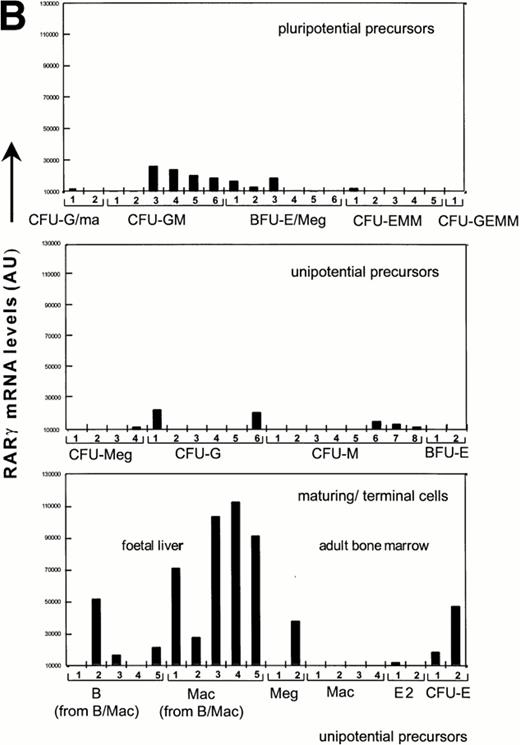
To further address the possibility of RAR expression in a rare subpopulation of GR-1−cells, we derived probes from the 3′untranslated region (UTR) of RARα and RARγ cDNAs for the screening of single cell cDNAs from hematopoietic precursors that were identified through sib analysis as detailed previously.34 Previous studies have validated these sample sets through hybrization with a ribosomal L32 probe, which showed a uniform level in all cDNA libraries.34 In contrast, RARα and RARγ expression was observed mainly in myeloid cells, with highest levels in maturing cell populations, specifically in all macrophage populations from fetal liver and in two of five B-cell populations from fetal liver grown in the presence of colony-stimulating factor (CSF)-1 and/or IL-7, respectively (Fig 2). In contrast, hybridization to cDNA samples from adult macrophages, shown previously to be highly positive for c-fms and lysozyme,34 was not observed. In this sample set, the cDNA libraries obtained from maturing granulocytes were not sufficiently representative34 for accurate assessment of RAR expression. Strikingly, both RARα and RARγ transcripts in hematopoietic progenitors were mostly confined to committed CFU-G and CFU-GM. This pattern of expression is in marked contrast to that previously reported for the hematopoietic transcription factors, SCL and GATA-1, which are coexpressed in multipotent progenitors and committed erythroid progenitors.35 Consistent with RT-PCR analysis, hybridization signals for RARα were much stronger than for RARγ, again suggesting that RARα is the major RAR transcript in hematopoietic progenitors. RARβ expression was not analyzed.
Quantitation of relative RARα and RARγ transcript levels in cDNA samples from diverse differentiation stages of the hematopoietic hierarchy. Southern blots of cDNA samples from the different cell types as shown were sequentially hybridized with probes for RARγ and α, and exposed to a PhosphorImage screen for quantitation.
Quantitation of relative RARα and RARγ transcript levels in cDNA samples from diverse differentiation stages of the hematopoietic hierarchy. Southern blots of cDNA samples from the different cell types as shown were sequentially hybridized with probes for RARγ and α, and exposed to a PhosphorImage screen for quantitation.
Given the above patterns of expression for RARα and RARγ messages, we analyzed the role of RARα1 and RARγ in hematopoietic cell differentiation through the study of the corresponding single or double knockout mice. Because bone marrow cells may be affected by developmental deficiencies in the bone mass itself, we compared the carcasses from RAR null mutants, heterozygote littermates, or wild-type controls. There was no significant difference in bone weight or in the number of bone marrow cells recovered, indicating that hematopoietic cell development within the bone marrow is not grossly affected (data not shown). Similarly, there was no detectable difference in cell type or lineage distribution in bone marrow populations elicited by receptor knockout, as judged by analysis of cytospins from either wild-type, RARα1, and/or RARγ null mice (data not shown).
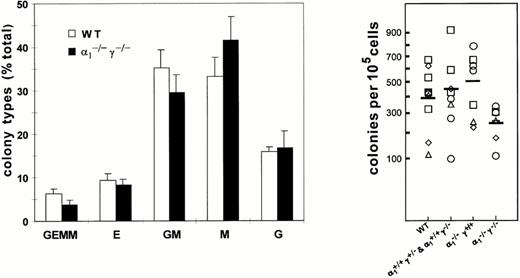
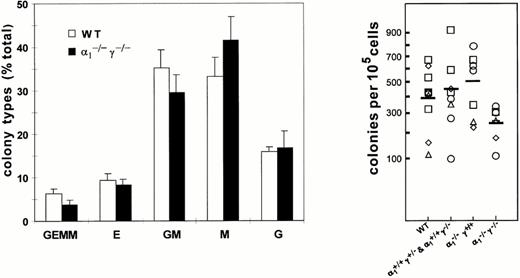
Consistent with the normal distribution of bone marrow cell populations, quantitative assessment by methylcellulose culture indicated that the frequency of colony-forming cells from bone marrow isolates did not differ significantly between wild-type controls and RARα1, RARγ, or RARα1/γ null mutants or heterozygous littermates (Fig 3, right panel). Moreover, the distribution of multilineage and unilineage colony-forming cells was comparable between all genotypes (Fig 3, left panel, data shown for wild-type and compound null mutants). Together, these observations suggest that the lack of either or both RARα1 and RARγ does not affect the generation of committed precursors per se, which may be taken as an indication of lineage commitment in vivo.
Distribution of the different types of colonies in methylcellulose cultures of bone marrow cells from compound RARα1−/−γ−/− null mice and littermate or wild-type controls. Colonies were scored on day 7 and the results expressed as percent total colony count (left panel). Total number of nucleated bone marrow cells recovered from the long bones of RARα1−/−γ−/− mice and wild-type controls or heterozygous littermates. There was no significant difference among the four groups (right panel).
Distribution of the different types of colonies in methylcellulose cultures of bone marrow cells from compound RARα1−/−γ−/− null mice and littermate or wild-type controls. Colonies were scored on day 7 and the results expressed as percent total colony count (left panel). Total number of nucleated bone marrow cells recovered from the long bones of RARα1−/−γ−/− mice and wild-type controls or heterozygous littermates. There was no significant difference among the four groups (right panel).
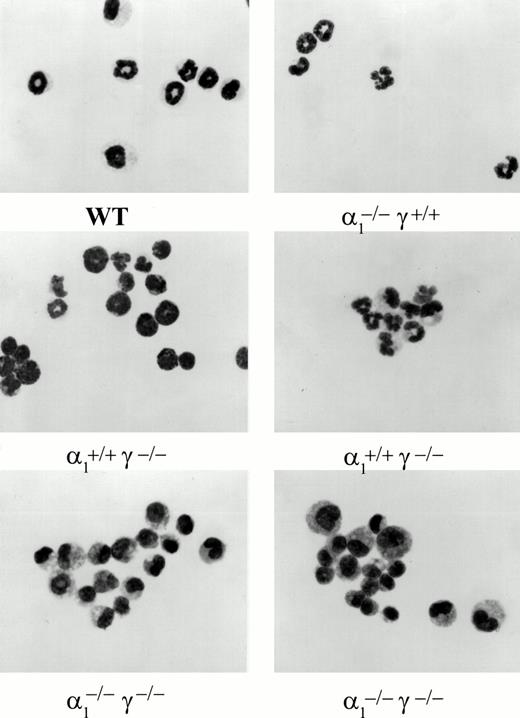
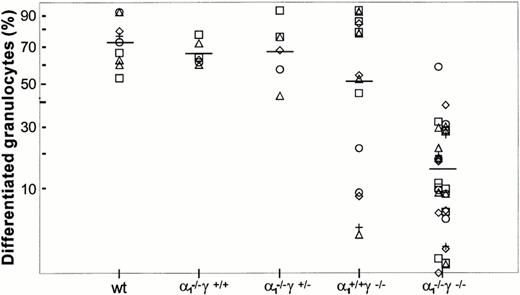
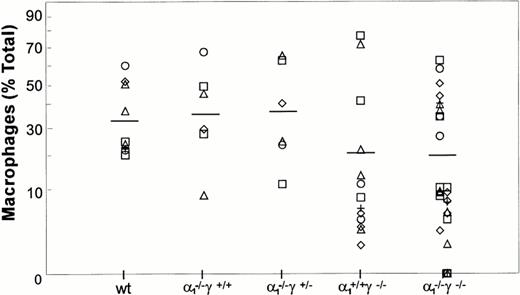
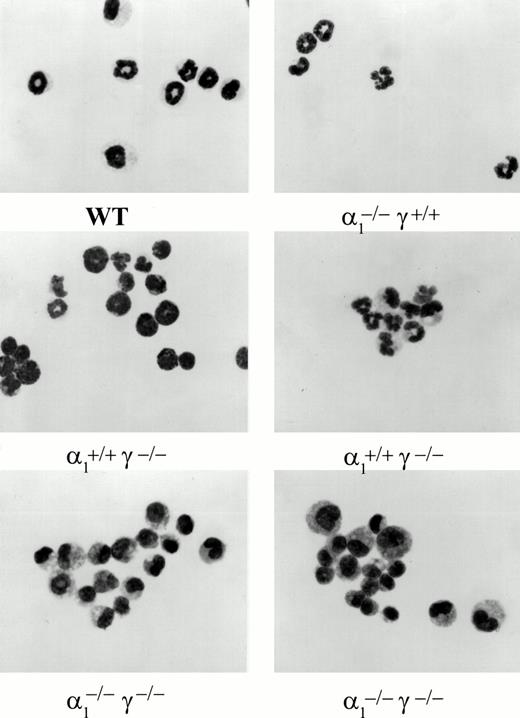
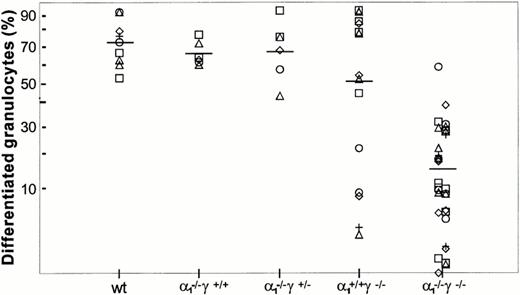
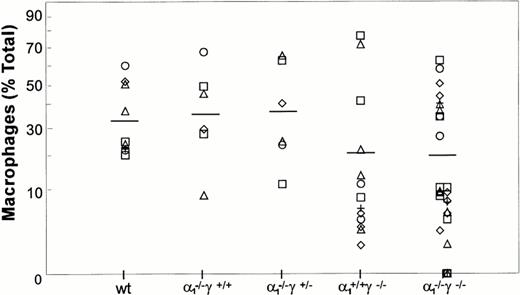
Maturation along the granulocytic and monocytic lineages was further examined in cytospins from individual colonies isolated from methylcellulose cultures. Terminal maturation into bands or polymorphonuclear granulocytes was evident in wild-type colonies (Fig 4). Similarly, terminal granulocyte differentiation, as judged by nuclear segmentation, was normal in RARγ−/− or RARα1−/−cells. In contrast, granulocyte differentiation was largely arrested at the myelocyte stage in RARα1−/−γ−/− cells. When the proportion of differentiated granulocytes (metamyelocytes, bands, and PMN) was compiled as percent total cells in the granulocytic lineage, including myeloblasts and promyelocytes, a significant difference between the RARα1−/−γ−/− group and wild-type controls was observed (P < .05) (Fig 5). In contrast, RARα1−/− or RARα1−/−γ+/-littermates were not significantly different from wild-types. The phenotype of RARγ−/− isolates, although not statistically different from wild-type, exhibited a higher degree of variability with some highly differentiated and some less differentiated colonies. In contrast to the maturation arrest in the granulotype lineage, the proportion of macrophages per colony was comparable between all genotypes (Fig 6). Finally, there was no correlation between the percentage of monocytes/macrophages and the degree of granulocytic differentiation observed. Together, our observations suggest that the lack of RARα1 and RARγ significantly impaired granulocytic differentiation in vitro without affecting macrophage or erythroid differentiation.
Light microscopic photographs of colony cells harvested from individual granulocyte macrophage colonies. Colony cells were cytosmeared and stained with Wright-Giemsa. Genotypes are shown underneath.
Light microscopic photographs of colony cells harvested from individual granulocyte macrophage colonies. Colony cells were cytosmeared and stained with Wright-Giemsa. Genotypes are shown underneath.
Granulocyte differentiation in methylcellulose culture. GM colonies were harvested individually and stained as in Fig 4. Results are shown as percent differentiated granulocytes (metamyelocytes, bands, and PMN) within the granulocytic lineage. The difference between wild-type controls and RARα1−/−γ−/− was significantly different (P < .05), whereas the difference with other groups was not significant.
Granulocyte differentiation in methylcellulose culture. GM colonies were harvested individually and stained as in Fig 4. Results are shown as percent differentiated granulocytes (metamyelocytes, bands, and PMN) within the granulocytic lineage. The difference between wild-type controls and RARα1−/−γ−/− was significantly different (P < .05), whereas the difference with other groups was not significant.
Macrophage differentiation in methylcellulose culture. The macrophage content of each GM colony is shown as a percentage of total cells. There was no significant difference among the various groups.
Macrophage differentiation in methylcellulose culture. The macrophage content of each GM colony is shown as a percentage of total cells. There was no significant difference among the various groups.
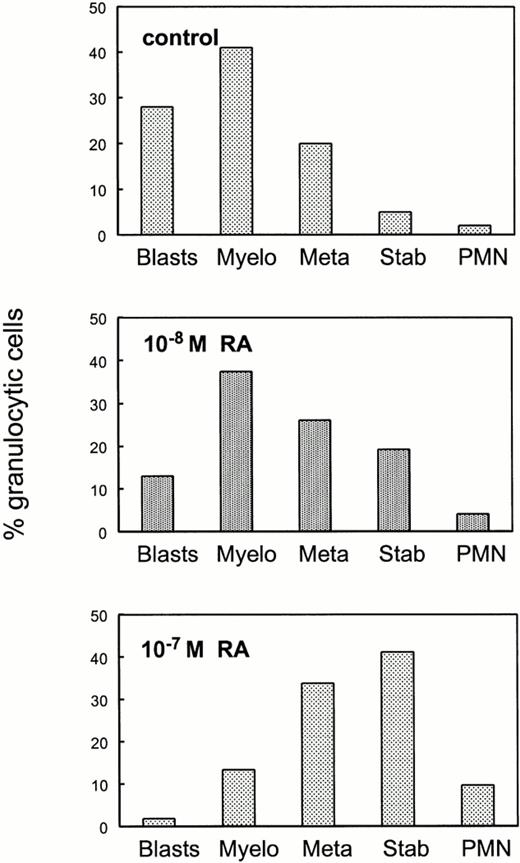
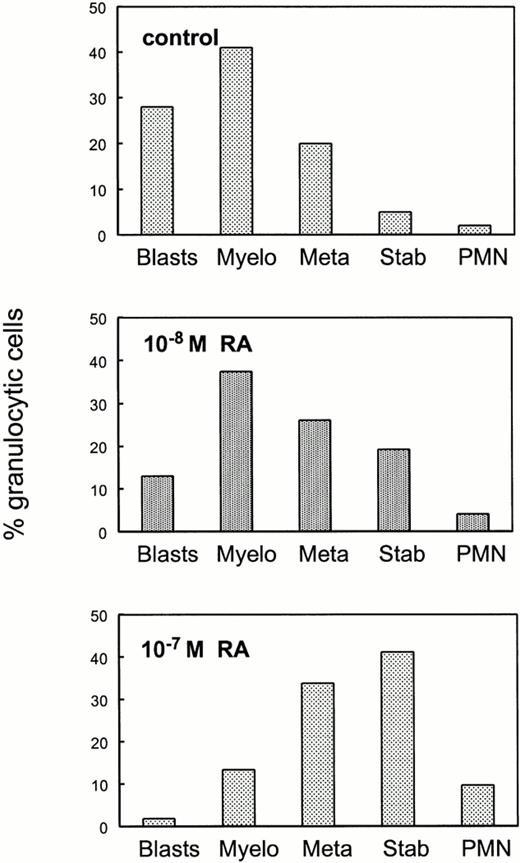
To determine whether the in vitro differentiation block could be relieved by excess retinoids, bone marrow cells from RARα1−/−γ−/− mice were plated in the presence of RA at two different concentrations (Fig 7). Analysis of individual granulocyte macrophage colonies indicate that pharmacologic concentrations of RA overcame the maturation block imposed by the lack of RARα1 and γ.
Pharmacologic doses of RA overcome the maturation block in granulopoietic cells lacking RARα1 and RARγ. Bone marrow cells from RARα1−/−γ−/− mice were plated in methycellulose cultures in the presence or in the absence of RA at the indicated concentrations. GM colonies were harvested individually and stained as in Fig 4. Results shown are the average of differential counts of 30 independent colonies in the control group, three colonies for the lower concentration of RA (10−8 mol/L) and 15 colonies for the higher concentration of RA (10−7 mol/L) and are expressed as percent total cells within the granulocytic lineage.
Pharmacologic doses of RA overcome the maturation block in granulopoietic cells lacking RARα1 and RARγ. Bone marrow cells from RARα1−/−γ−/− mice were plated in methycellulose cultures in the presence or in the absence of RA at the indicated concentrations. GM colonies were harvested individually and stained as in Fig 4. Results shown are the average of differential counts of 30 independent colonies in the control group, three colonies for the lower concentration of RA (10−8 mol/L) and 15 colonies for the higher concentration of RA (10−7 mol/L) and are expressed as percent total cells within the granulocytic lineage.
DISCUSSION
Results from semiquantitative PCR and expression analysis of representative cDNAs from clonal hematopoietic isolates are consistent with our finding of a role for RARα1, together with RARγ, in terminal maturation along the granulocyte pathway. Both RARα1 and RARα2 isoforms were detected in Gr-1+ cell populations with the former appearing more abundant. Likewise, transcripts for both RARγ isoforms were also detected in differentiated granulocytes, although only after extensive amplification. Although it remains to be seen whether message abundance correlates directly with protein levels,38 the functional assays described here are direct indicators of the importance of RARs in hematopoietic cell differentiation. Our data clearly show that, despite its low apparent abundance, RARγ must play a role in granulocyte differentiation, as disruption of this gene, in combination with RARα1 knockout, was required to affect cell differentiation in vitro. However, analysis of RARα2 null mice, or studies of the impact of other RAR knockouts on hematopoiesis, have not been reported. We cannot, therefore, exclude a role for RARα2 alone or in combination with other receptors, in this process.
Tissue culture models and the phenotype of RAR single versus double null mutants indicate specific, as well as overlapping, functions for the RARs.3,39,40 In the present study, it would appear that RARα1 and RARγ play redundant roles in granulocyte differentiation. However, as discussed previously,12,13,39 we cannot exclude that gene disruption itself evokes functional redundancy, which does not ordinarily exist, and that granulopoiesis normally operates under the control of a specific RAR type. Consistent with a degree of functional specificity, we found that the differentiation block could be relieved by RA, albeit at pharmacologic concentrations. This suggests that RARα2 can fulfill the role(s) of the disrupted receptors, but only in a situation of retinoid excess. It may be possible to further address this possibility through the use of RAR type-specific agonists or antagonists.12
In vivo, disruption of RARα1 and/or RARγ appeared to have no observable consequence regarding granulocyte differentiation. One possible mechanism to explain this observation is that granulocyte differentiation proceeds via non-cell autonomous cues that are supported by the intact RARs in nongranulocytic cells, which promote normal granulocyte differentiation. Once removed from this environment (in isolated culture conditions), this essential factor(s) is not sufficiently expressed in the knockout granulocyte precursors, resulting in the observed differentiation block in vitro. Alternatively, it is conceivable that the RARα chimeras associated with APL may exert effects in addition to attenuation of retinoid signaling, and that both mechanisms are required to elicit a differentiaiton block in vivo. In support of the first possibility, we provide evidence that high doses of RA in vitro overcome the maturation block in RARα1−/−γ−/− cells, suggesting that granulocytic differentiation in the compound null mice occurs in vivo through a compensation mechanism, which is non-cell autonomous. These observations, however, do not rule out the possibility that maturation arrest in APL may also be due to contributions that are specific to the different RARα fusion partners.41-46
The lack of RARα1 and RARγ did not affect the distribution of the different types of colonies (ie, multilineage and committed unilineage colonies) despite the presence of both transcripts in committed granulocyte and macrophage precursors. Because the distribution of colony types is believed to reflect the process of lineage commitment in vivo, it is most likely that RARα1 and RARγ do not drive early decisional events in multipotent stem cells. In contrast, both genes are essential for terminal granulocyte maturation in vitro, consistent with their pattern of expression in Gr-1+ populations. Our observations therefore suggest that in APL, the various translocations involving RARα and the resulting dominant negative RAR mutants cause differentiation arrest at the promyelocyte stage, when further maturation and nuclear segmentation require signaling via both RARα1 and γ.
An outstanding question remains the nature of retinoid target genes that are important for terminal granulocyte maturation. A cell surface marker, RIG-E, was recently identified as a downstream RAR target through differential display analysis of RA-treated NB4 cells, a promyelocytic leukemic cell line.47 RIG-E is a member of the Ly-6 family of adhesion molecules that include Gr-136and may be involved in neutrophil function. It is not presently clear, however, whether RIG-E (or Gr-1) is a direct target of RAR function. Recently, homeobox (hox) gene products have been implicated in several hematopoietic differentiation pathways.48-50 Several members of this gene family are direct RA targets40 and, in some cases, exhibit RAR type-specific regulation in cell culture models.13,39,40,51,52 Moreover, the skeletal defects inherent to the RARγ and RARα1/γ knockout mice phenocopy certain of the defects seen in some Hox null mice10 (and references therein), further supporting a direct relationship. It is therefore possible that normal granulocyte differentiation occurs, in part, through retinoid-dependent regulation of certain Hox genes, a possibility we are currently addressing.
Supported by funds from the Cancer Research Society Inc (Montreal, Quebec, Canada), the National Cancer Institute with funds from the National Cancer Society of Canada, and a fellowship from the Leukemia Research Fund (Toronto, Ontario, Canada; to J.L.). D.L. is a scholar of the Medical Research Council of Canada and T.H. a senior scientist of the Fonds de la Recherche en Santé du Québec.
Address reprint requests to Trang Hoang, PhD, Director, Laboratory of Hemopoiesis and Leukemia, Clinical Research Institute of Montréal, 110 Pine Ave West, Montréal, Quebec, Canada H2W 1R7.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.