Abstract
Recurrent translocation t(10;11) has been reported to be associated with acute myeloid leukemia (AML). Recently, two types of chimeric transcripts, MLL-AF10 in t(10;11)(p12;q23) andCALM-AF10 in t(10;11)(p13;q14), were isolated. t(10;11) is strongly associated with complex translocations, including invins(10;11) and inv(11)t(10;11), because the direction of transcription of AF10 is telomere to centromere. We analyzed a patient of AML with t(10;11)(p11.2;q23) and identified ABI-1 on chromosome 10p11.2, a human homolog to mouse Abl-interactor 1 (Abi-1), fused with MLL. Whereas the ABI-1 gene bears no homology with the partner genes of MLL previously described, the ABI-1 protein exhibits sequence similarity to protein of homeotic genes, contains several polyproline stretches, and includes asrc homology 3 (SH3) domain at the C-terminus that is required for binding to Abl proteins in mouse Abi-1 protein. Recently, e3B1, an eps8 SH3 binding protein 1, was also isolated as a human homolog to mouse Abi-1. Three types of transcripts of ABI-1 gene were expressed in normal peripheral blood. Although e3B1 was considered to be a full-length ABI-1, the MLL-ABI-1fusion transcript in this patient was formed by an alternatively spliced ABI-1. Others have shown that mouse Abi-1 suppresses v-ABL transforming activity and that e3B1, full-length ABI-1, regulates cell growth. In-frame MLL-ABI-1 fusion transcripts combine the MLL AT-hook motifs and DNA methyltransferase homology region with the homeodomain homologous region, polyproline stretches, and SH3 domain of alternatively spliced transcript of ABI-1. Our results suggest that the ABI-1 gene plays a role in leukemogenesis by translocating to MLL.
© 1998 by The American Society of Hematology.
VARIOUS CHROMOSOMAL translocations associated with human cancers have been identified and characterized.1,2 Recurrent translocations involving chromosome 11 band q23 (11q23) observed in acute leukemia or myelodysplastic syndrome (MDS) are characterized by the presence of a variety of partner chromosomes.3 At least 20 chromosomal regions for partners of 11q23 have been observed, such as t(4;11), t(9;11) and t(11;19). The MLL gene4 (also calledALL-1, HRX, and HTRX-1) has been identified in 11q23 translocations,5-7 and its rearrangement was found in the majority of infant8-10 and secondary leukemias.11 12 Up to now, 14 partner genes for MLLhave been cloned from leukemia cells with various types of reciprocal translocations, and they formed fusion transcripts with MLL. However, it remains to be elucidated whether the MLL gene, the partner genes, or the fusion transcripts play a critical role in leukemogenesis.
There has been a series of reports of t(10;11) translocation in acute leukemia.13-15 Recently, two types of chimeric transcripts,MLL-AF10 in t(10;11)(p12;q23)16 andCALM-AF10 in t(10;11)(p13;q14),17 were isolated. Cytogenetic evidence suggests that there is heterogeneity in the breakpoints on 10p, with 10p11, 10p12, 10p13, 10p14, and 10p15. However, molecular analysis of these 10;11 translocations has shown that the AF10 gene at 10p12 is common in these abnormalities. In addition, as compared with other 11q23 translocations involvingMLL, t(10;11) is strongly associated with complex translocations, such as invins(10;11) and inv(11)t(10;11), because the direction of transcription of AF10 is telomere to centromere.15,18 19 Because of this opposite orientation, the t(10;11)(p12;q23) cannot form the regular head to tail fusion transcripts found in other 11q23 translocations. This may explain why the t(10;11)(p12;q23) is often complex and associated with 11q insertions.
Despite the heterogeneity of 10;11 translocations, previous papers reported only AF10 is involved in various 10;11 translocations. We identified here another gene, ABI-1, which is fused toMLL, in a patient of t(10;11)(p11.2;q23)-acute myeloid leukemia (AML). The ABI-1 is a human homolog to mouse Abl-interactor 1 (Abi-1), encoding an Abl-binding protein.20 TheABI-1 is the second partner gene located on chromosome 10p, suggesting that heterogeneity of 10;11 translocation is due to the presence of ABI-1 gene. These findings may help to clarify the mechanisms underlying chromosomal aberrations and leukemogenesis in leukemias with 11q23 abnormalities.
MATERIALS AND METHODS
Patient.
An 8-month-old boy was diagnosed as having acute myelomonocytic leukemia (AMMoL; French-American-British [FAB] M4), which was cytogenetically characterized as t(10;11)(p11;q23). He was treated by intensive chemotherapy followed by autologous bone marrow transplantation and has been in complete remission for 5 years.
Southern blot analysis.
High molecular weight DNA was extracted from bone marrow cells from the patient by proteinase K digestion and phenol/chloroform extraction. Ten micrograms of DNA was digested with appropriate restriction enzymes, subjected to electrophoresis on 0.8% agarose gels, transferred to charged nylon filters (Amersham, Buckinghamshire, UK), and hybridized to DNA probes labeled by the random hexamer method.21 A 0.9-kb BamHI fragment (designated probe x) derived from MLL cDNA22 was used as a probe.
Preparation of mRNA and cDNA libraries.
Poly(A)+ RNA from frozen cells was extracted with a Fast Track mRNA Isolation Kit (Invitrogen, San Diego, CA). A cDNA library was constructed with poly(A) selected mRNA from patient cells and BALM14 cell line following established procedures.23Briefly, random hexanucleotide-primed synthesized cDNAs were ligated with EcoRI adaptors, and cloned into the EcoRI-digested λgt10 cloning vector (Promega, Madison, WI). After packaging with commercial packaging kits (Epicentre Technologies, Madison, WI), phage plaques were screened with probes labeled using a random primer synthesis kit (Stratagene, La Jolla, CA). The probe x for the patient cDNA library and the 320-bp ABI-1 cDNA probe derived fromMLL-ABI-1 chimeric clone for the BALM 14 cDNA library were used for screening.
Reverse transcriptase-polymerase chain reaction (RT-PCR).
Total cellular RNA was extracted from bone marrow cells of the patient by the acid guanidine isothiocyanate-phenol-chloroform method.24 Four micrograms of total RNA was reverse transcribed to cDNA in a total volume of 20 μL with random hexamers and 20 U of reverse transcriptase (AMV; Boehringer Mannheim, Mannheim, Germany). One twentieth of the cDNA was amplified by PCR in a total volume of 100 μL with 50 mmol/L KCl, 1.5 mmol/L MgCl2, 10 mmol/L Tris-HCl (pH 9.0 at room temperature), 25 pmol of each primer, 75 μmol/L of each dNTP, and 2.5 U of Taq polymerase (Applied Biosystems, Urayasu, Japan). After 30 rounds of PCR (30 seconds at 94°C, 30 seconds at 55°C, and 1 minute at 72°C), 5 μL of PCR product was electrophoresed in a 3% agarose gel.25Primers used were as follows: MLL-7S, 5′-TCCTCAGCACTCTCTCCAAT-3′; m-1S, 5′-CAGCATAGTCCAGGCAGGA-3′; m-1A, 5′-GGAGAGTCATCAAACATGGG-3′.
Nucleotide sequencing.
PCR products were cloned into the TA cloning vector (Invitrogen). Nucleotide sequences of phage clones and PCR products were determined by the fluorometric method (Dye Terminator Cycle Sequencing Kit; Applied Biosystems).
Northern blot analysis.
Multiple human tissue Northern blots (Clontech, Palo Alto, CA) were hybridized with 32P-labeled clone 48 as a probe.23
Fluorescence in situ hybridization (FISH) analysis.
Chromosomal mapping of the genomic clones (H4, H5-2, H7-2, H18, and H20) was performed by the FISH method.26 The phage clones were labeled by the standard nick translation method using biotin-16-dUTP (Boehringer Mannheim). To confirm the origin of cloned DNAs, we also mapped these genomic clones to leukemic cells together with whole chromosome painting probe for chromosome 10 (WCP10; Coatasome 10, digoxigenin-labeled; Oncor, Gaithersburg, MD). We also performed a series of FISH studies on leukemic samples to further characterize t(10;11) using WCP11 (Coatasome 11, digoxigenin-labeled; Oncor).
RESULTS
Rearrangement of MLL and noninvolvement of AF10.
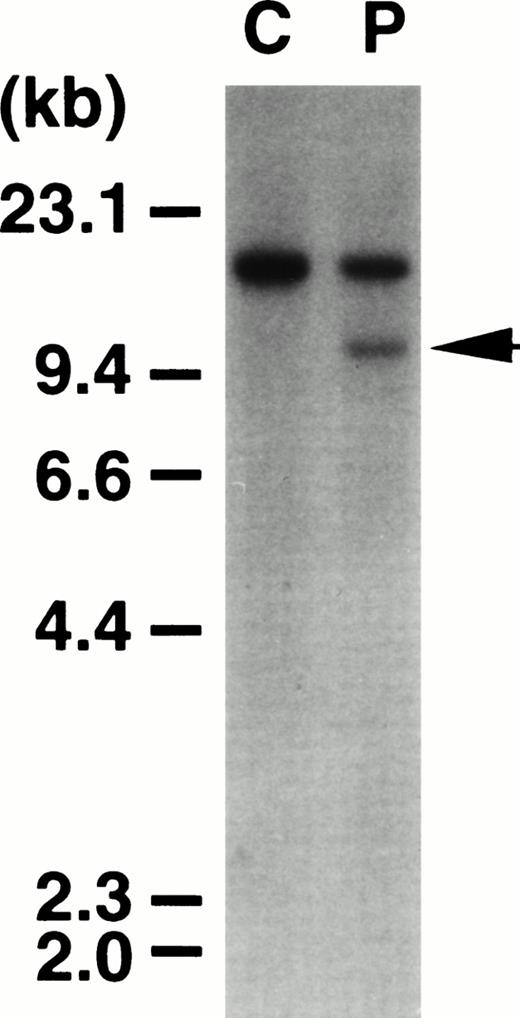
Southern blot analysis of DNA prepared from the leukemic cells of the patient using probe x showed a chromosomal breakpoint within the breakpoint cluster region of MLL gene at 11q23 (Fig 1) that spans exons 5 through 11 in the MLL locus.22 However, no PCR products were amplified from the cDNA prepared from these cells when MLL-AF10specific primer pair was used. We inferred that the MLL in this patient is fused to a novel partner gene.
Southern blot of DNA digested with HindIII and probed with the 0.9-kb fragment of the MLL gene. C, normal peripheral lymphocytes. P, leukemic cells from the patient. The patient exhibited a rearranged band (arrowhead) with this probe.
Southern blot of DNA digested with HindIII and probed with the 0.9-kb fragment of the MLL gene. C, normal peripheral lymphocytes. P, leukemic cells from the patient. The patient exhibited a rearranged band (arrowhead) with this probe.
Isolation of the MLL fusion cDNAs in t(10;11)(p11;q23).
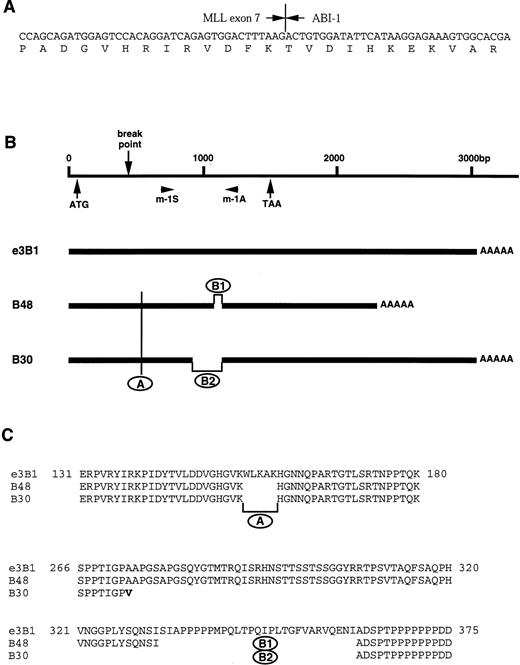
To isolate fusion transcripts of MLL, we prepared a cDNA library from mRNA of the patient’s leukemic cells. Four cDNA clones were isolated by screening with probe x, and one (clone 15-21) of them was found to represent a fusion transcript of MLL. Clone 15-21, 682 bp in size, contained a 362-bp sequence corresponding to exons 5 to 7 in the MLL gene at the 5′ region, and the remaining 320-bp sequence did not match the MLL gene or the partner genes of MLL previously cloned (Fig 2A).
(A) Partial sequences of MLL-ABI-1 chimeric transcript. Vertical lines indicate the exon-exon junctions of each gene. Arrowheads indicate the fusion points of each cDNA. (B) ABI-1 cDNA clones (B48, B30) and e3B1. AAAAA, poly(A) tail. (C) Comparison among three types of ABI-1 (e3B1, B48, and B30) predicted amino acid sequence.
(A) Partial sequences of MLL-ABI-1 chimeric transcript. Vertical lines indicate the exon-exon junctions of each gene. Arrowheads indicate the fusion points of each cDNA. (B) ABI-1 cDNA clones (B48, B30) and e3B1. AAAAA, poly(A) tail. (C) Comparison among three types of ABI-1 (e3B1, B48, and B30) predicted amino acid sequence.
Isolation of the ABI-1 gene.
The 320-bp sequence identified in the chimeric clone was used as a probe to screen a cDNA library from the BALM14 cell line. We isolated two kinds of clones. Clones B30 and B48 contained sequences of 2,920 and 2,195 nucleotides with an open reading frame, encoding a protein of 387 (42.7 kD) and 447 (48.8 kD) amino acids, respectively (Fig 2B). Both clones include a poly(A) tail. Between these two clones, there are two sequence differences (Fig 2B and C). Based on the sequence analysis, a 177-bp stretch was inserted in the middle of the coding region in clone B48, whereas a 1,121-bp sequence in the 3′ untranslated region was shortened in clone B48. The entire amino acid sequence exhibits high similarity (81.5%) to mouse Abi-1, a SH3 protein that suppresses v-abl transforming activity by binding to the Abl protein,20 suggesting that the protein is a human homolog to mouse Abi-1. Therefore, we designated these clones as ABI-1.ABI-1 has no significant similarity to other MLLpartner genes reported previously, except for the SH3 domain of EEN. Recently, another group isolated an eps827 SH3 binding protein 1, e3B1,28 as a human homolog to mouse Abi-1. The amino acid (nucleotide) sequence of ABI-1 completely matched that of e3B1 except in two regions, an additional 5 amino acids stretch (region A in Fig 2B and C) and 29 amino acids stretch (region B1 in Fig 2B and C). Region A (residues 155 to 159 in e3B1) occurs after the homeodomain homologous region, which is homologous to the DNA-binding sequence of homeodomain proteins. Region B1 (residues 333 to 361 in e3B1) occurs just before the polyproline stretches, but both regions are located after the chromosomal breakpoint of t(10;11). Region B2 (residues 274 to 361 in e3B1) lacking in clone 30 included a part of the homeodomain homologous region (the second PEST domain in e3B1) and region B1.
Expression of the ABI-1 gene in normal tissues.
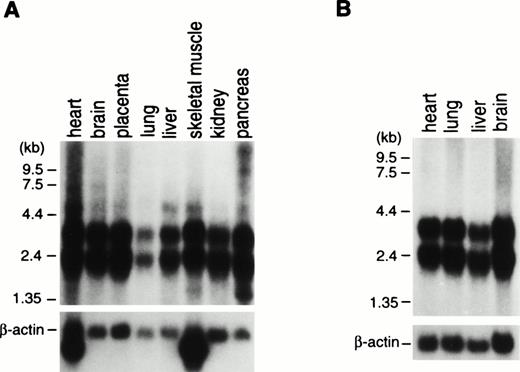
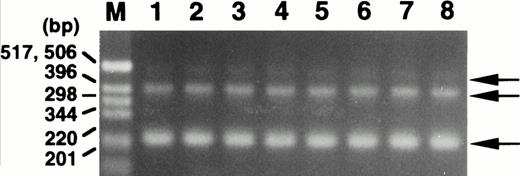
To examine for expression of the ABI-1 gene, we performed Northern blot analysis on poly(A)-selected RNA from various human tissues. The expressions of the 2.4- and 3.2-kb transcripts appeared constant in most tissues examined (Fig 3). Clone B48 of the ABI-1 cDNA obtained encompasses approximately 2,195 nucleotides and most likely corresponds to the 2.4-kb transcript. On the other hand, the 3.2-kb transcript may be the product of clone B30 and full-length ABI-1, e3B1. Furthermore, we performed RT-PCR analysis for RNAs from normal peripheral blood using primers m-1S and m-1A and detected two major transcripts and an extremely weak expression of the largest sized transcript in all cells (Fig 4). Sequence analysis showed that the two major products correspond to clones B48 and B30, respectively, and that the largest sized band corresponds to e3B1, suggesting that the alternatively spliced transcripts of ABI-1, clones B48 and B30, are major transcripts of ABI-1 in normal peripheral blood.
Northern blot analysis of RNAs from adult (A) and fetal (B) human tissues. ABI-1 cDNA fragment was used as a probe for the Northern blots in the upper figures. Membranes were rehybridized to the β-actin probe for the lower figures. Names of the organs are indicated on top of the figures.
Northern blot analysis of RNAs from adult (A) and fetal (B) human tissues. ABI-1 cDNA fragment was used as a probe for the Northern blots in the upper figures. Membranes were rehybridized to the β-actin probe for the lower figures. Names of the organs are indicated on top of the figures.
Expression of the ABI-1 in normal peripheral blood by RT-PCR analysis. Total RNAs isolated from peripheral blood of 8 healthy volunteers were used as the cDNA template. M, size marker (1-kb DNA ladder; GIBCO BRL, Gaithersburg, MD). Arrows indicate the amplified bands.
Expression of the ABI-1 in normal peripheral blood by RT-PCR analysis. Total RNAs isolated from peripheral blood of 8 healthy volunteers were used as the cDNA template. M, size marker (1-kb DNA ladder; GIBCO BRL, Gaithersburg, MD). Arrows indicate the amplified bands.
MLL-ABI-1 fusion transcript is formed by alternatively spliced transcript of ABI-1.
To clarify the expression of MLL-ABI-1 fusion transcript in the patient’s leukemic cells, we performed RT-PCR analysis for RNA from leukemic cells using primers MLL-7S and m-1A that could detect regions A, B1, and B2, absent in clones B48 or B30, and detected only about a 1.0-kb band (data not shown). The amplified fragment was cloned into TA cloning vector and sequence analysis showed that it was formed of the fusion transcript of ABI-1 that lacked regions A and B1, as observed in clone B48. In clone 15-21 of chimeric MLL-ABI-1transcript isolated from the patient’s leukemic cells, a portion ofABI-1 with 320 bp in the fusion cDNA fragment also lacked region A. These findings suggest that MLL-ABI-1 fusion transcript is formed by alternatively spliced transcript ofABI-1, corresponding to clone B48. On the other hand, when we used reciprocal primers for identifying an ABI-1/MLL fusion transcript, no PCR products were obtained.
Assignment of the ABI-1 gene.
To assign a chromosomal localization for the ABI-1 gene, we obtained 6 phage clones (H4, H5-2, H7-2, H18, H20, and H7-3) after screening of a genomic library from human placental DNA using clone B48 as a probe. All phage clones were assigned to band 10p11.2 on normal metaphase chromosomes by FISH analysis (data not shown). Because multiple phage clones mapped to only one chromosomal locus, it is also suggested that ABI-1 (B30 and B48) and e3BI are alternatively spliced transcripts of a single gene.
Precise chromosome analysis of leukemic cells by FISH.
FISH analysis on patient’s metaphase chromosomes using human genomic DNA clones spanning the ABI-1 gene showed that five (H4, H5-2, H7-2, H18, and H20) of the clones containing the 5′ end of the breakpoint of the ABI-1 gene hybridized to der(10) and one (H7-3) containing the 3′ end to der(11) in leukemic cells (data not shown). Furthermore, we performed FISH analysis on U937 cell line, in which AF10 on 10p12 translocated to CALM on 11q14,17 and showed that all phage clones of ABI-1hybridized to der(10) but not der(11) (data not shown). These results suggest that the ABI-1 gene is disrupted in t(10;11)(p11.2;q23)-AMMoL and that the breakpoint is located centromeric to the AF10 gene.
DISCUSSION
Two types of t(10;11) have been identified: t(10;11)(p12;q23), which creates the MLL-AF10 fusion gene,16 and t(10;11)(p13;q14), which creates CALM-AF10 fusion gene.17 In the present study, we isolated a novel chimeric transcript, MLL-ABI-1, in a patient of AMMoL with t(10;11)(p11.2;q23). FISH analysis shows that ABI-1 gene is located centromeric to the AF10 gene. At the cytogenetic level, it may be difficult to distinguish MLL-ABI-1 andMLL-AF10 fusions, and it is likely that other cases of t(10;11) may result in this gene fusion. Recent molecular analysis showed thatENL,5ELL29/MEN,30 andEEN31 genes in the same chromosomal region, 19p13, were partner genes of MLL with t(11;19)(q23;p13), respectively. That no amplified product was detected by RT-PCR may be due not only to the heterogeneity of the breakpoints of the same gene, but also to the different partner genes.
ABI-1, a human homolog to mouse Abi-1,20 has high homology to ABI-2.32 These proteins exhibit sequence similarity to homeotic genes and contain several polyproline stretches and an SH3 domain at the C-terminus. Although EEN, a partner gene for MLL, also has an SH3 domain, the remaining region of EEN does not have any homology to ABI-1.31 The e3B1, which showed full-length ABI-1, was isolated as an eps8 binding protein.28 eps8 is a substrate of receptor tyrosine kinases and is an SH3 domain containing protein that plays an important role in mitogenic signaling.27 e3B1 is considered to be a downstream target for eps8. Overexpression of e3B1 inhibits cell growth, suggesting that e3B1 is associated with regulation of cell proliferation. On the other hand, the mouse Abi-1 protein was isolated as an ABL-binding protein that suppresses v-ABL transforming activity.20 Similarly, ABI-2 was isolated as another ABL-binding protein that interacts with c-ABL through both the SH3 domain and C-terminus of the c-ABL and modulates c-ABL transforming activity.32 The c-ABL protein, originally identified as the cellular homolog of the v-abl oncogene product of Abelson murine leukemia virus, is a tyrosine kinase. c-ABL is closely associated with leukemogenesis of chronic myelogenous leukemia and acute lymphoblastic leukemia (ALL) creating BCR-ABL fusion transcript in t(9;22)(q34;q11). Although it was suggested that both Abi-1 and ABI-2 are regulators of ABL function in transformation or in signal transduction, the functional correlation between Abi-1, ABI-2, and BCR-ABL is still unknown. Furthermore, it is not known whether the function of alternatively spliced transcripts of ABI-1, clones B30 and B48, is the same as that of e3B1 or Abi-1.
The MLL gene is fused to various partner genes by 11q23 chromosomal translocations. Up to now, 14 partner genes for MLLhave been cloned from leukemia cells with various types of reciprocal translocations, including t(4;11), t(9;11), and t(11;19). They include the putative transcriptional factors (AF4, AF9, AF10, AF17, and ENL), a target gene for Ras (AF6),33 and an RNA polymerase II elongation factor (ELL).34 Furthermore, transcriptional coactivators and/or histone acetyltransferases, CBP and p300, were shown to be novel partner genes for MLL in therapy-related AML (MDS) with t(11;16) and t(11;22), respectively by us35,36and others.37,38 The functions of the normal MLLgene and the fusion transcripts have remained unknown. Two studies reported that chimeric mice carrying the mouse Mll-AF9 fusion gene developed AML39 and that hematopoietic progenitor cells transduced with HRX(MLL)-ENL induced AML in vitro,40respectively. Rearrangements of the MLL gene found in ALL, AML, and MDS suggest that this gene fused to various partner genes plays a causative role in the dysregulation of differentiation along both lymphoid and myeloid pathways.
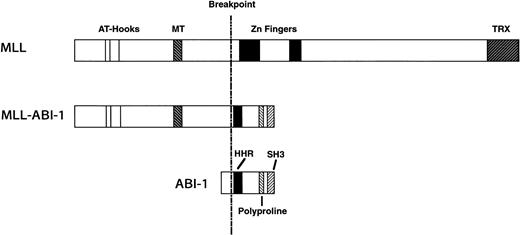
A schematic representation of the predicted MLL-ABI-1 fusion proteins is shown in Fig 5, with the predicted MLL-ABI-1 fusion transcripts encoding a protein of 1,727 aa containing 1,406 aa from MLL in the N-terminus and 321 aa from ABI-1 in the C-terminus. The MLL-ABI-1 chimeric protein retains the AT-hook domain, DNA methyltransferase homology region, and a transcriptional repression domain of MLL, and the homeodomain homologous region and the SH3 domain of ABI-1. Although e3B1 was reported to be a cytosolic protein,28 many partner genes of MLL encode nuclear proteins, suggesting that ABI-1 is a cytosolic protein; MLL-ABI-1 chimeric product may localize in the nuclei similar to MLL-AF6 despite the fact that AF6 itself localizes in the cytosol.41 Because ABI-1 is suggested to function as a tumor suppressor, one of the mechanisms of leukemogenesis by the MLL-ABI-1 fusion may be that the function of normal ABI-1 is dominantly suppressed by the fusion protein. One report suggested that gain-of-function by creating chimeric transcripts involving MLLinduced transforming activity.42 These findings suggest that not only dysfunction of normal ABI-1, but also gain-of-function by fusion of MLL and ABI-1 is necessary to induce leukemogenesis. Further functional analysis of the MLL-ABI-1fusion gene will provide new insights into leukemogenesis by 11q23 chromosomal translocations.
Schematic representation of the MLL, ABI-1, and MLL-ABI-1 fusion proteins. MT, DNA methyltransferase homology region; TRX,Drosophila trithorax. HHR, homeodomain homologous region.
Schematic representation of the MLL, ABI-1, and MLL-ABI-1 fusion proteins. MT, DNA methyltransferase homology region; TRX,Drosophila trithorax. HHR, homeodomain homologous region.
ACKNOWLEDGMENT
The authors thank M. Seto (Aichi Cancer Center Research Institute) for providing the MLL cDNA probe (probe x). We also express appreciation to S. Sohma for technical assistance.
Supported by a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare of Japan, a Grant-in-Aid for Scientific Research on Priority Areas, and Grant-in-Aid for Scientific Research (B) and (C) from the Ministry of Education, Science, Sports and Culture of Japan.
Address reprint requests to Yasuhide Hayashi, MD, Department of Pediatrics, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.