Abstract
We have identified a point mutation in the promoter of the factor VII gene responsible for a severe bleeding disorder in a patient from a large French-Canadian family with known consanguinity. The proband has an extremely low plasma level of factor VII antigen and factor VII coagulant activity (<1 percent of normal) and suffers from hemarthroses and chronic arthropathy. Sequencing of the patient’s factor VII 5′ flanking region, intron/exon junctions, and coding regions showed a homozygous point mutation, a C to G transversion at position −94 relative to the translation start site. We show here that this mutation prevented binding of transcription factor Sp1 and of other nuclear proteins to this region of the factor VII promoter and resulted in a 20-fold reduction in reporter gene expression in HepG2 cells. These data underscore the importance of this region of the factor VII promoter for in vivo expression of the factor VII gene.
© 1998 by The American Society of Hematology.
HUMAN COAGULATION factor VII is a 406 amino acid, vitamin-K–dependent glycoprotein that is synthesized in the liver and circulates in blood as an inactive zymogen.1,2 Factor VII, in its activated form (factor VIIa), participates in the initiation of coagulation via the extrinsic pathway in association with tissue factor, an integral membrane protein that is exposed to the circulation upon vascular injury or stimulation of monocytes and endothelial cells.3-5 The factor VIIa–tissue factor complex initiates coagulation by activating both factor IX and factor X, leading to the localized generation of thrombin.6 7
A reduced plasma level of factor VII coagulant activity (VII:C) leads to factor VII deficiency, a bleeding diathesis of variable severity. It is inherited as an autosomal recessive disorder, and its incidence is estimated to be 1 per 500,000 in the general population.8,9In recent years, at least 30 different mutations in the factor VII structural gene have been identified in patients with factor VII deficiency.10-15 Although the majority of the mutations described are missense mutations, several nonsense mutations, small deletions, and splice site abnormalities have been identified as well.
Recognition of defects in the promoter of the human factor VII gene has also become possible with the identification and analysis of its 5′ regulatory region by several groups.16-18 Unlike the promoters of many eukaryotic genes, that of the factor VII gene has neither a CAAT nor a TATA box. However, it does include binding sites for both ubiquitously distributed and liver-enriched transcription factors upstream of the major transcriptional start site at position −51 (all numbering is with respect to the translational start site at position +1.) Among these, the sites for Sp1, a ubiquitous transcription factor of the zinc-finger family, and for hepatocyte nuclear factor-4 (HNF-4), a tissue-restricted orphan receptor, were shown to be important for expression of the factor VII gene.17 18
In contrast to the large number of mutations identified within the structural factor VII gene, there are few reports of alterations within regulatory regions that influence its expression. We have recently identified and characterized a point mutation within the 5′ regulatory region of the factor VII gene of a patient with severe factor VII deficiency.19 This mutation is a T to G substitution at position −61 that markedly reduces promoter activity by preventing HNF-4 binding and transactivation. In addition, there is a polymorphism in the 5′ regulatory region, a decanucleotide insertion at position −323, which is associated with modest reductions in plasma levels of factor VII antigen (VII:Ag) and VII:C.20,21 Although the decanucleotide insert is often found in allelic association with the Arg353Gln substitution, a polymorphism within exon 8, which itself reduces plasma levels of VII:Ag and VII:C,22 in vitro analysis of the insert alone indicated that it causes an approximately 33% reduction in factor VII promoter activity in human hepatoma cells.18
In this report, we describe a new mutation in the factor VII promoter that is responsible for factor VII deficiency in a large French-Canadian kindred with known consanguinity. The mutation, a C to G transversion at position −94, is within the core binding site of transcription factor Sp1. It disrupted binding of both Sp1 and other nuclear proteins to the factor VII promoter, thereby severely compromising the expression of the factor VII gene in reporter gene assays.
MATERIALS AND METHODS
Clinical history.
The patient is a member of a large kindred from a small town in Quebec province. Members of his extended family have been described previously in the medical literature. Two sisters with factor VII deficiency identified in 1955,23 shortly after the original description of the disorder,24 were found in 1971 to be related to 11 additional individuals (eight women and three men) with factor VII deficiency.25 These 13 patients exhibited plasma levels of VII:C ranging from 1.5% to 6% of normal with variable severities of bleeding. The female patients experienced menorrhagia; several patients had postsurgical hemorrhage, bruising, and epistaxis; and one patient suffered a cerebral hemorrhage. The parents of affected individuals were themselves free of clinical symptoms but had plasma levels of VII:Ag and VII:C that were approximately 50% of normal, suggesting that the trait was inherited in an autosomal recessive manner. All 13 patients were descendants of a couple who emigrated to Canada from France at the end of the 17th century. Although the incidence of severe factor VII deficiency is quite rare, it was calculated to be approximately 1 in 335 individuals in the descendants of this French couple.25
Informed consent was obtained from the patient and his family. This study was approved by the Human Studies Committee of the Brockton-West Roxbury Department of Veterans Affairs Medical Center.
Factor VII assays.
Plasma levels of VII:C and VII:Ag were measured on citrated plasma with a one-stage clotting assay using Automated Simplastin (Organon Teknika Corp, Durham, NC) and an enzyme-linked immunoabsorbent assay (American Bioproducts Co, Parsippany, NJ), respectively. The levels of VII:Ag and VII:C were expressed as a percentage of the level in a normal pool constructed from equal volumes of plasma from 30 healthy individuals.
DNA isolation and polymerase chain reaction (PCR).
Genomic DNA was purified from leukocytes isolated from whole blood of the patient and his parents by standard methods.26 The immediate 5′ flanking region of the factor VII promoter (spanning nucleotides −404 to +98) was amplified by PCR and subcloned into the vector pT7blue (Novagen, Madison, WI) for sequencing. The inserts were sequenced on a 373A DNA Sequencer (Applied Biosystems, Foster City, CA) by the dideoxy chain termination method.27 A clone carrying the patient’s mutation provided the template for the subsequent PCR reaction spanning nucleotides −185 to +1 (with base +1 designated as the translational start site), which was subcloned into the promoterless pOGH reporter plasmid containing the human growth hormone structural gene (Nichols Diagnostics Institute, San Juan Capistrano, CA); the presence of the mutation was confirmed by sequencing. A plasmid containing the wild-type sequence of the −185 to +1 fragment of the factor VII promoter was prepared similarly and used as a positive control. The exons of the factor VII gene were amplified by PCR from the patient’s genomic DNA as described previously.12 The PCR reactions of exons 1a, 5, 7, and 8 were then subcloned and sequenced, whereas those of exons 2, 3, 4, and 6 were analyzed by direct sequencing. All PCR amplifications were done with Taq DNA polymerase in a Perkin-Elmer-Cetus DNA Thermal Cycler (Norwalk, CT). Primers for PCR and sequencing were obtained from Integrated DNA Technologies (Coralville, IA).
Detection of polymorphic alleles in genomic studies.
Restriction analyses were performed as previously described22 to determine the genotype of the patient and his parents with respect to two polymorphisms that affect plasma levels of factor VII, the decanucleotide insert in the 5′ regulatory region and the Arg353Gln substitution in exon 8. Briefly, to detect the presence of the decanucleotide insert, a 431-bp fragment spanning position −323 (the insertion site for the decanucleotide) was amplified by PCR from the genomic DNA of the patient and his parents. Approximately 200 ng of PCR product was digested with 20 U ofEcoR I (New England Biolabs, Beverly, MA) and then subjected to electrophoresis on an 8% (wt/vol) polyacrylamide gel. The fragments generated were 328 and 103 bp in the absence of the insert, or 328 and 113 bp in its presence. To detect the Arg353Gln substitution, a 239-bp fragment of exon 8 surrounding the codon for amino acid 353 was analyzed similarly. In this instance, the presence of the mutation eliminates the only Msp I restriction site within the fragment. In the absence of the mutation, Msp I digestion produced fragments of 186 and 53 bp.
Cell culture and transient transfection analysis.
The human hepatoma cell line, HepG2 (ATCC HB 8065), was grown in minimal essential medium supplemented with 10% fetal bovine serum, 1 mmol/L sodium pyruvate, 10 mmol/L HEPES pH 7.4, 2 mmol/L glutamine, 100 U/mL penicillin G, and 100 mg/mL streptomycin, in a 5% CO2humidified atmosphere at 37°C. Twenty hours before the start of transfection, 1 × 106 HepG2 cells were seeded on replicate 60-mm culture dishes. Cells were transfected with mixtures of reporter plasmid DNA and an internal plasmid control to monitor transfection efficiency, complexed with the LipofectAmine reagent (GIBCO-BRL, Gaithersburg, MD) according to the manufacturer’s recommendations. Each transfection mixture contained 3 μg of the reporter plasmid and 1.5 μg of pSV-β-galactosidase control plasmid (Promega Corp, Madison, WI). After 16 hours of transfection, full growth media was given to the cells; 48 hours later, the culture media and cell lysates were assayed for expression of human growth hormone (hGH) and β-galactosidase, respectively. The media were assayed with the hGH assay kit (Nichols Diagnostics Institute) according to the manufacturer’s directions. Concentrations of hGH were calculated from a standard curve fitted with a four parameter logistic model. The cell lysates were analyzed with a colorimetric β-galactosidase assay kit (Promega Corp). Levels of β-galactosidase in the samples were calculated by comparison with a standard curve fitted by linear regression using the least-squares method. For each dish, the level of hGH expressed was normalized using the corresponding level of β-galactosidase to correct for differences in transfection efficiencies. In each experiment, the pOGH plasmid was also transfected into cells on control dishes; when normalized for expression of β-galactosidase, the values for hGH produced from the promoterless plasmid were subtracted from the experimental values reported.
Electrophoretic mobility shift assays.
HeLa cells were cultured as described above, and nuclear extracts were prepared according to the method of Dignam et al.28 The method of Bradford et al29 was used to determine protein concentrations in the nuclear extracts. Complementary oligonucleotides extending from position −108 to −84 of the factor VII 5′ flanking region were annealed and end-labeled with [γ-32P]–adenosine triphosphate (ATP) using T4 polynucleotide kinase, then purified using Sephadex G-50 spin columns (Boehringer Mannheim, Indianapolis, IN).19 For gel mobility shift assays, the procedure of Chodosh et al30 was used with minor modifications. Ten micrograms of HeLa nuclear extract, or 1 footprint unit of human recombinant transcription factor Sp1 (Promega Corp), was incubated on ice for 10 minutes with 0.5 μg bovine serum albumin (BSA), 2.5 μg poly (dI.dC), and 1 μg salmon sperm DNA (Sigma, St Louis, MO) in 20 μl of binding buffer (25 mmol/L HEPES pH 7.6, 14 mmol/L KCl, 10% glycerol, 0.1 mmol/L EDTA, 0.75 mmol/L dithiothreitol, 5 mmol/L MgCl2). After addition of 0.1 ng of radiolabeled double-stranded oligonucleotide (approximately 100,000 cpm/ng) to the mixture, the reaction was incubated on ice for an additional 20 minutes. In competition studies, the unlabeled competitor oligonucleotide was added during the initial incubation on ice. Following incubation, the reaction mixtures were electrophoresed on a 5% (wt/vol) polyacrylamide gel and autoradiographed. The oligonucleotide sequences used in the reactions were as follows: wild-type, 5′ −108 GTGTCCTCCCCTCCCCCATCCCTCT 3′ −84 and mutant 5′ −108 GTGTCCTCCCCTCCGCCATCCCTCT 3′ −84; Sp1 consensus sequence (Promega Corp), 5′ATTCGATCGGGGCGGGGCGAGC 3′.
RESULTS
Patient.
The patient is a 24-year-old Canadian man with a bleeding diathesis, having plasma levels of both VII:Ag and VII:C that are less than 1% of normal. He has a history of easy bruising and was diagnosed with factor VII deficiency at age 11, when he developed hemarthrosis of the left knee. Between the ages of 12 and 15 years, he required infusions of fresh frozen plasma at 2- to 3-month intervals for spontaneous bleeds within his elbow and ankle joints. He was then placed on chronic prophylaxis with factor VII concentrate (Immuno AG, Vienna, Austria) three times per week with amelioration of the bleeding diathesis. The patient’s parents do not have a bleeding disorder, although both exhibit reduced plasma levels of VII:Ag and VII:C (Table 1). His two sisters exhibit normal hemostasis and have normal factor VII levels (data not shown).
Identification of mutations in the factor VII gene.
Analysis of the entire coding sequence and the intron/exon boundaries of the patient’s factor VII gene revealed only the presence of two previously described polymorphisms but no alterations sufficient to account for his bleeding disorder. In exon 5, a C to T transition was observed at position 7880, which produces a neutral dimorphism in the codon for His115.31 The only other change in the coding sequence was a G to A substitution at position 10976, which produces the Arg353Gln polymorphism in exon 8. Subsequently, a 404-bp fragment of the 5′ flanking region of the patient’s factor VII gene was examined and several changes were found. These were the decanucleotide insert at position −323, a C to T substitution at position −122, and a C to G transversion at position −94. As mentioned, the decanucleotide insert frequently occurs in conjunction with the Arg353Gln polymorphism; these polymorphisms either alone or together induce modest reductions in the plasma levels of factor VII.18,20-22,32 The substitution at position −122 has been observed in an asymptomatic individual also carrying the decanucleotide insert18 and therefore is not expected to contribute to the patient’s phenotype.
The inheritance of these alterations in the factor VII gene was confirmed by sequence analysis of the parents (Table 1). Both parents were found to be heterozygous for the transversion at position −94. The mother was heterozygous for the decanucleotide insert, the −122 transition, and the Arg353Gln polymorphism. The father, like the patient, was homozygous for all three changes, which may account for the observation that his levels of VII:Ag and VII:C were slightly lower than the mother’s.
Characterization of the −94 C to G mutation.
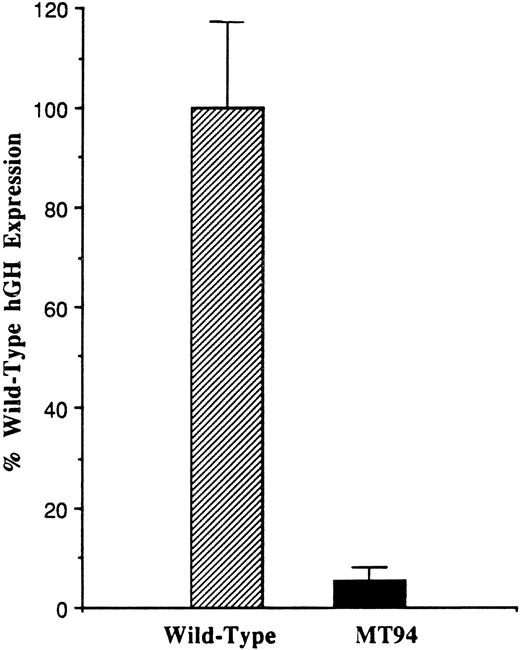
To analyze the influence of the −94 C to G mutation on promoter function, a fragment of the 5′ flanking region of the factor VII gene extending from position −185 to position +1 (the translational start site) was prepared from the patient’s DNA and inserted into the promoterless pOGH reporter plasmid as described in Materials and Methods. A reporter plasmid containing the wild-type sequence was similarly constructed. These plasmids were then used to direct the expression of the hGH structural gene in transient transfection experiments in HepG2 cells, a human liver hepatoma cell line expressing factor VII and other coagulation proteins.33 Figure 1 shows that the plasmid containing the mutant promoter fragment exhibited only 5.8 ± 2.2% (1 SD) of the activity observed with the plasmid containing the wild-type promoter fragment (n = 14).
Functional analysis of the wild-type and mutant (−94 C to G) factor VII promoters. One hundred eighty-six base pair fragments of wild-type or mutant (MT94) human factor VII promoter sequence were inserted into the hGH reporter vector and used to transiently transfect HepG2 cells. The data were corrected as described in Materials and Methods, and the level of hGH expression from reporter plasmid containing the wild-type sequence was considered 100%. Relative to this, the level of hGH expression from the MT94 sequence was 5.8 ± 2.2% (1 SD). The data shown were obtained from three experiments in which a total of 14 dishes were transfected with each expression vector.
Functional analysis of the wild-type and mutant (−94 C to G) factor VII promoters. One hundred eighty-six base pair fragments of wild-type or mutant (MT94) human factor VII promoter sequence were inserted into the hGH reporter vector and used to transiently transfect HepG2 cells. The data were corrected as described in Materials and Methods, and the level of hGH expression from reporter plasmid containing the wild-type sequence was considered 100%. Relative to this, the level of hGH expression from the MT94 sequence was 5.8 ± 2.2% (1 SD). The data shown were obtained from three experiments in which a total of 14 dishes were transfected with each expression vector.
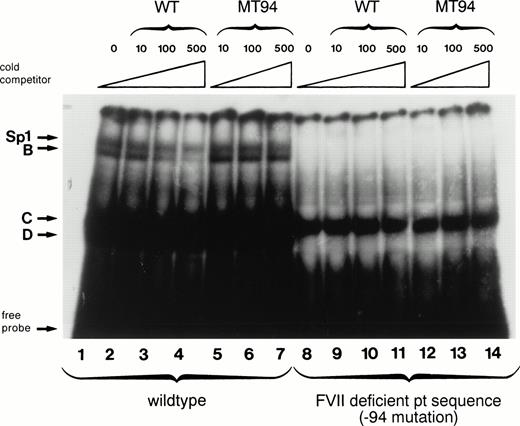
The −94 mutation occurs within the central hexanucleotide core of an Sp1 binding site (CCTCCC to CCTCCG) in an area shown by in vitro mutagenesis experiments to be important for factor VII expression.17,18 Therefore, we examined by electrophoretic mobility shift assays the behavior of an oligonucleotide containing the −94 C to G mutation with nuclear extracts from HeLa cells (which contain abundant amounts of Sp1) and compared it to that obtained with an oligonucleotide having the wild-type sequence (Fig 2). The labeled wild-type oligonucleotide formed four complexes with HeLa nuclear proteins designated Sp1, B, C, and D (lane 1). The presence of the ubiquitous transcription factor Sp1 in the slowest-mobility complex has been shown previously by the formation of a complex of identical mobility between recombinant human Sp1 and the wild-type factor VII sequence and by supershift experiments with anti-Sp1 antibody.18 Binding between HeLa nuclear proteins and the Sp1, B, and D complexes was specific as it could be competed away by incubation with increasing amounts of unlabeled wild-type oligonucleotide (lanes 2 through 4) but not by equivalent amounts of unlabeled mutant oligonucleotide (lanes 5 through 7). With the labeled mutant oligonucleotide, in contrast, only complex C was formed (lane 8). This complex is likely to be nonspecific because it cannot be competed away by either the mutant or the wild-type oligonucleotide (lanes 9 through 14).
Gel mobility shift assays show loss of nuclear protein binding with a mutant factor VII promoter sequence (MT94) from a patient with severe factor VII deficiency. A radiolabeled wild-type (WT) oligonucleotide including the Sp1 binding site (−108 to −84 bp before the translation start site) in the human factor VII promoter region showed binding to proteins (arrows) from HeLa nuclear extracts (lane 1). Specificity of binding is shown by competition with unlabeled WT oligonucleotide sequence at 10×, 100×, and 500× concentrations (lanes 2, 3, and 4) but not with an oligonucleotide having the mutant sequence (lanes 5, 6, and 7). Lanes 8 through 14 show the lack of binding of nuclear proteins by complexes labeled Sp1, B, and D to radiolabeled probe containing the MT94 sequence.
Gel mobility shift assays show loss of nuclear protein binding with a mutant factor VII promoter sequence (MT94) from a patient with severe factor VII deficiency. A radiolabeled wild-type (WT) oligonucleotide including the Sp1 binding site (−108 to −84 bp before the translation start site) in the human factor VII promoter region showed binding to proteins (arrows) from HeLa nuclear extracts (lane 1). Specificity of binding is shown by competition with unlabeled WT oligonucleotide sequence at 10×, 100×, and 500× concentrations (lanes 2, 3, and 4) but not with an oligonucleotide having the mutant sequence (lanes 5, 6, and 7). Lanes 8 through 14 show the lack of binding of nuclear proteins by complexes labeled Sp1, B, and D to radiolabeled probe containing the MT94 sequence.
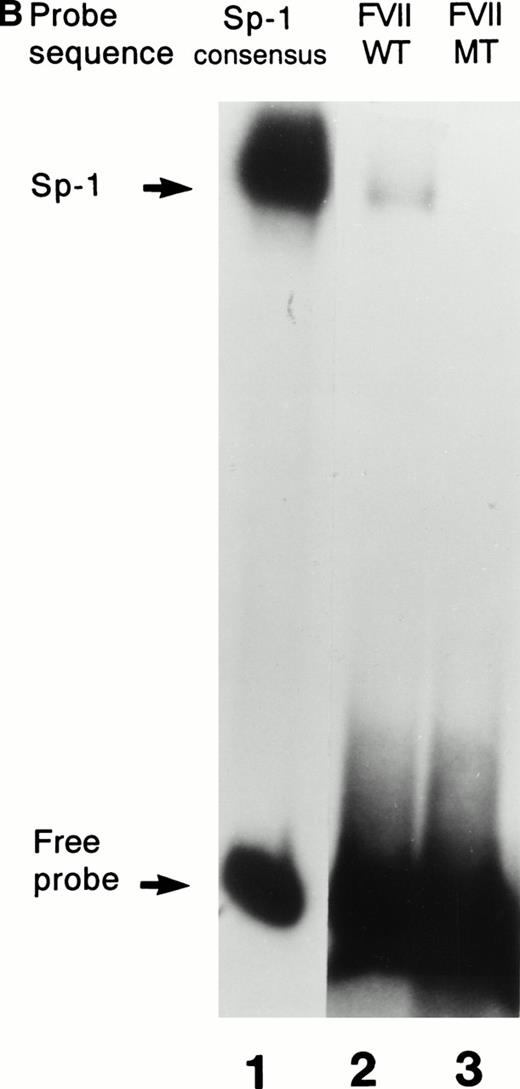
Further confirmation that the patient’s mutation disrupted the binding of Sp1 to the factor VII sequence is provided in Fig 3. As shown in Fig 3A, a labeled consensus Sp1 oligonucleotide bound a protein from HeLa nuclear extracts in the absence (lane 1) but not the presence (lane 2) of unlabeled Sp1 competitor oligonucleotide. The wild-type factor VII oligonucleotide (lane 3) competed effectively for binding to this protein, whereas at the same concentration the mutant factor VII oligonucleotide (lane 4) did not. Additionally, as shown in Fig 3B, purified recombinant human Sp1 bound directly not only to its consensus Sp1 oligonucleotide (lane 1) but also to the wild-type factor VII sequence (lane 2). However, the purified Sp1 did not bind detectably to the mutant factor VII oligonucleotide (lane 3). These data suggest that the −94 mutation prevents transcription of the factor VII gene through disruption of binding by Sp1 and other DNA binding proteins to this region of the promoter.
Interactions with an Sp1 consensus sequence and human Sp1 protein. (A) Unlabeled wild-type (WT) but not mutant (MT94) human factor VII sequence competes for Sp1 binding with radiolabeled Sp1 consensus sequence. Lane 1 shows binding of radiolabeled Sp1 consensus probe by HeLa nuclear extract in the absence of unlabeled competitor sequences. Lanes 2 through 4 show Sp1 consensus probe binding with HeLa nuclear extract in the presence of 1,000× concentrations of unlabeled competitor Sp1 consensus sequence (lane 2), WT factor VII sequence (−108 to −84; lane 3), and MT factor VII–deficient patient sequence (−108 to −84; lane 4). (B) WT but not MT94 factor VII–deficient patient sequence binds to the transcription factor Sp1. Binding of radiolabeled oligonucleotides with recombinant human Sp1 is shown with radiolabeled Sp1 consensus sequence (lane 1) and WT factor VII sequence (lane 2) but not with MT factor VII–deficient patient sequence (lane 3).
Interactions with an Sp1 consensus sequence and human Sp1 protein. (A) Unlabeled wild-type (WT) but not mutant (MT94) human factor VII sequence competes for Sp1 binding with radiolabeled Sp1 consensus sequence. Lane 1 shows binding of radiolabeled Sp1 consensus probe by HeLa nuclear extract in the absence of unlabeled competitor sequences. Lanes 2 through 4 show Sp1 consensus probe binding with HeLa nuclear extract in the presence of 1,000× concentrations of unlabeled competitor Sp1 consensus sequence (lane 2), WT factor VII sequence (−108 to −84; lane 3), and MT factor VII–deficient patient sequence (−108 to −84; lane 4). (B) WT but not MT94 factor VII–deficient patient sequence binds to the transcription factor Sp1. Binding of radiolabeled oligonucleotides with recombinant human Sp1 is shown with radiolabeled Sp1 consensus sequence (lane 1) and WT factor VII sequence (lane 2) but not with MT factor VII–deficient patient sequence (lane 3).
DISCUSSION
We have identified a naturally occurring point mutation, a C to G transversion at position −94 in the 5′ flanking region of the human factor VII gene, which is responsible for factor VII deficiency in a severely affected homozygous patient. The patient also has two polymorphisms, the decanucleotide insert at position −323 and Arg353Gln, each of which diminishes factor VII expression to a modest extent. However, these polymorphisms cannot account for his phenotype, although it is possible they contribute to a more severe bleeding diathesis than might have been observed in their absence. The patient is from a large French-Canadian family in which numerous members have factor VII deficiency. In view of the known consanguinity within the family,23 25 it is likely that homozygosity for the C to G transversion at position −94 is responsible for the factor VII deficiency observed in the other individuals of this kindred. Indeed, we have investigated a second severely affected individual from the same region, presumably a distant relative of the proband, and confirmed that he was homozygous for the mutation at position −94 (data not shown).
In reporter gene assays performed with HepG2 cells, a fragment of the mutant promoter extending from position −185 to +1 exhibited very low activity compared with the corresponding wild-type promoter fragment. In gel mobility shift assays, introduction of the mutant sequence into an oligonucleotide extending from position −108 to −84 disrupted its binding to several HeLa cell nuclear proteins including Sp1. Thus, the reduced activity observed in reporter gene assays may be due to disruption of binding not only to Sp1 but also to other transcription factors, as yet unidentified. Numerous examples of transcription factors that bind either in association with Sp1 or at overlapping binding sites have been reported, including Ets,34 GATA-1,35 SREB-1,36 and H4TF1.37 Other factors such as Sp3 have been reported to compete with Sp1,38 whereas the specific ratio of Sp1 to other transcription factors may also be important for determination of promoter activity.39
Binding of oligonucleotides with the factor VII promoter sequence to recombinant Sp1 protein was adversely affected by the presence of the mutation. Additionally, an excess (1,000×) of wild-type promoter sequence competed away binding of a radiolabeled Sp1 consensus oligonucleotide to Sp1 protein from HeLa cell extracts. However, excess mutant factor VII sequence did not, suggesting that the mutant oligonucleotide did not bind Sp1 at all under these in vitro conditions. It should be noted that direct binding of purified Sp1 to even the wild-type factor VII sequence was weak compared with its binding to the consensus Sp1 oligonucleotide. Within the 6-bp core Sp1 binding region (CCGCCC), the only difference is the central residue, a G in the consensus but a T in the factor VII sequence. A previous study investigating the effect of base changes within the core binding region on the interaction with purified Sp1 found that altering the central residue from G to T reduces Sp1 binding approximately threefold.40 Additional reductions in binding may be due to the effect of factor VII sequence flanking the core binding site. Greenberg et al 17 have also noted that the factor VII Sp1 site has lower affinity for purified Sp1 than does an Sp1 binding site identified in the human metallothionein promoter. Weak affinity of even the wild-type factor VII sequence for Sp1 may explain in part the low plasma concentration of factor VII that, at 500 ng/mL, is the lowest of the vitamin-K–dependent coagulation proteins.
Prior studies have shown that the region surrounding position −94 is essential for normal expression of the factor VII gene in hepatoma cells.17,18 Greenberg et al17 performed mutational analysis and showed that alteration of the nucleotides at positions −98 and −100 (CCCCTCCCCC to CACATCCCCC) or of residues −92 to −96 inclusive (CCCCTCCCCC to CCCCTAAAAA) decreased activity in reporter gene assays to approximately 12% and 7% that of the wild-type sequence, respectively. Pollak et al18 made point mutations at position −100 (CCCCTCCCCC to CACCTCCCCC), position −94 (CCCCTCCCCC to CCCCTCCACC), and at both positions. The −100 alteration diminished specific binding of an oligonucleotide spanning the region to both HeLa nuclear proteins and purified recombinant Sp1 and reduced the level of expression in reporter gene assays to 35% of that observed with the wild-type sequence. The −94 C to A mutation and the combined mutations abolished nuclear protein binding and reduced reporter gene expression to 2% of wild-type. Interestingly, our patient had nucleotide −94 mutated from C to G, confirming the importance of wild-type sequence in this region to factor VII expression.
Naturally occurring mutations involving Sp1 binding sites have recently been described in the promoters of several other genes with variable effects on expression. For example, the promoter of 5-lipoxygenase, a crucial enzyme in the synthesis of leukotrienes, contains five tandem repeats of a strong Sp1/Egr-1 consensus sequence. Deletions of one or two repeats, or addition of a sixth, significantly decreases activity in reporter gene assays.41 Point mutations within the proximal Sp1 binding site of the human low-density lipoprotein receptor promoter decrease both Sp1 binding and receptor expression, inducing moderate42 or severe43 forms of heterozygous familial hypercholesterolemia, depending on the particular base substitution. Point mutations were also associated with increases in both Sp1 binding and protein expression in the γ-hemoglobin gene, resulting in persistence of fetal hemoglobin.44 Naturally occurring mutations such as these, and the one described in this report, validate in vitro analyses of gene regulation and provide insight into mechanisms of human disease.
ACKNOWLEDGMENT
We thank Dr Georges Rivard (Hôpital Sainte-Justine, Montreal, Canada) for referring the patient and his family for investigation.
Supported by the Medical Research Service of the Department of Veterans Affairs (K.A.B.), the National Institutes of Health Grant No. RO1 HL48322 (K.A.H.), and a fellowship from the Southeastern Pennsylvania Affiliate of the American Heart Association (E.S.P.).
Address reprint requests to Kenneth A. Bauer, MD, Department of Veterans Affairs Medical Center, 1400 VFW Parkway, West Roxbury, MA 02132.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.