Abstract
The t(12;21)(p13;q22) translocation, fusing the ETV6 andAML1 genes, is the most frequent chromosomal translocation associated with pediatric B-cell precursor acute lymphoblastic leukemia. Although the genomic organization of the ETV6 gene and a breakpoint cluster region (bcr) in ETV6 intron 5 has been described, mapping of AML1 breakpoints has been hampered because of the large, hitherto unknown size of AML1 intron 1. Here, we report the mapping of the AML1 gene between exons 1 and 3, cloning of ETV6-AML1 breakpoints from different patients, and localization of the AML1 breakpoints withinAML1 intron 1. In contrast to the tightly clustered ETV6breakpoints, the AML1 breakpoints were found to be dispersed throughout AML1 intron 1. Although nucleotide sequence analysis of the breakpoint junctions showed several 5/7 matches for the V(D)J consensus heptamer recognition sequence, these matches were present only on the ETV6 alleles and not on the AML1 alleles, making it unlikely that the translocations were mediated by a simple V(D)J recombination mistake. Interestingly, several breakpoints as well as a stable insertion polymorphism mapped close to a polymorphic, alternating purine-pyrimidine tract in the ETV6 gene, suggesting that this region may be prone to DNA recombination events such as insertions or translocations. Finally, the presence of an insertional polymorphism within the ETV6 bcr must be recognized to avoid incorrect genotype designation based on Southern blot analysis.
CHROMOSOMAL TRANSLOCATIONS occur in a recurrent and nonrandom fashion in many patients with acute leukemia. The biological relevance of these translocations is underscored by the fact that a number of known and putative proto-oncogenes have been identified through the study of genes located at these translocation breakpoints.1-3 Further characterization of these translocations on a molecular level has led to insights into the mechanism(s) which generate these translocations, as well as the role(s) of the genes involved in producing the malignant phenotype.1-3 Additionally, clinical studies of patients with specific nonrandom translocations have aided oncologists in devising risk-specific therapy, and, in some instances, identifying molecular markers for tracking minimal residual disease.4-6
The ETV6 gene (also known as TEL) was first identified in a patient with chronic myelomonocytic leukemia and a t(5;12)(q33;p13) which involved ETV6 andPDGFRβ.7 Since that time, additional translocations involving ETV6, namely t(3;12)(q26;p13),8,9 t(9;12)(q34;p13),10t(12;22)(p13;q11),11 and t(12;21)(p13;q22)12-15translocations which fuse ETV6 with MDS1/EVI1, ABL, MN1, and AML1/CBFA2, respectively, have been described in both myeloid and lymphoid malignancies.
Although both an ETV6-AML1 fusion and an AML1-ETV6fusion may potentially be produced by the t(12;21) translocation, reverse transcriptase polymerase chain reaction (RT-PCR) analysis has consistently shown the presence of the ETV6-AML1transcript, transcribed from the der(21), in all cases with the t(12;21),16 whereas the reciprocal AML1-ETV6transcript is not universally found in these patients. The former protein is therefore thought to play a role in leukemogenesis.17 The ETV6-AML1 transcript fusesETV6 exons 1 through 5 with AML1 exons 2 through 8 and encodes a protein that contains the amino-terminal portion of theETV6 protein, including the putative helix loop helix (HLH) domain, fused to amino acids 20-480 of the AML1cprotein.17 The ETV6-AML1 fusion transcript is detected in about 17% to 25% of pediatric B-cell precursor acute lymphoblastic leukemia (ALL) patients, making it the most common nonrandom, recurrent translocation associated with childhood ALL.18,19 Furthermore, a frequent loss of the normal, untranslocated ETV6 allele has been described by several authors, raising the possibility that ETV6 may function as a tumor suppressor gene.19
The precise molecular mechanisms leading to chromosomal translocations remain largely unknown. In some cases there is compelling evidence that the translocation is caused by mistakes in normal V(D)J recombinase activity as evidenced by the presence of cryptic heptamer/nonamer sequences, the addition of nontemplated “N-region” nucleotides at the breakpoints, and by exonucleolytic deletion of germline nucleotides at these breakpoint junctions.20,21 In other translocations, homologous recombination between Alu repeats has been implicated; this mechanism is almost certainly involved in producing the partial tandem duplication of MLL.22 In addition, a number of epidemiological studies have implicated the therapeutic use of topoisomerase II inhibitors, especially the epipodophyllotoxins etoposide and teniposide, in the generation ofMLL and AML1 translocations associated with therapy-related acute myeloid leukemia.23 Finally, a handful of chromosomal translocations have been found near regions of DNA containing alternating purine and pyrimidine nucleotides (pu/py tracts), leading some investigators to speculate that these pu/py tracts may be regions of the genome that are predisposed to chromosomal rearrangements.24-26
Although a number of investigators have clearly shown that the t(12;21) is the most frequent chromosomal translocation associated with B-cell precursor ALL, until very recently,27 none of these translocation breakpoints had been cloned and characterized at the nucleotide sequence level. Characterization of ETV6-AML1genomic breakpoints has been hampered because of the large (estimated at >100 kb) first intron of the AML1 gene.17,28Because most ETV6-AML1 fusion mRNAs join ETV6 exon 5 toAML1 exon 2, it has been presumed that the AML1breakpoints occur within AML1 intron 1.17,28However, it is not known whether the AML1 breakpoints are clustered within this large intron; indeed, the singleETV6-AML1 breakpoint reported was not localized with respect to the AML1 intron-exon structure.27 To gain some insight into the potential mechanism that generated these translocations, and to determine if the breakpoints are tightly clustered, we cloned and sequenced the ETV6-AML1 breakpoints from several different patients.
MATERIALS AND METHODS
Preparation and analysis of genomic DNA.
Informed consent to participate in research studies was obtained in accordance with institutional guidelines. Leukemic blasts from bone marrow of patients with acute leukemia were isolated by Ficoll-Hypaque (Sigma, St Louis, MO) density centrifugation. Genomic DNA was isolated by using a salting out procedure as described.29 Ten micrograms of genomic DNA was digested with the indicated restriction enzyme (GIBCO-BRL, Gaithersburg, MD), size fractionated on 0.8% agarose gels containing 1 μg of ethidium bromide/mL, photographed, denatured, neutralized, and transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH) by the Southern technique.30 31 DNA was immobilized by ultraviolet cross-linking.
P1-derived artificial chromosome (PAC) clone isolation and analysis.
The RPCI 5 human PAC library was screened with a 0.7-kbEcoRI-SmaI AML1 cDNA probe (nucleotides 1 to 704 of GenBank accession no. D43969) encompassing AML1 exons 1, 2, and 3.32 Single colonies of the hybridizing PAC clones were grown in 3 mL Luria-Bertani (LB) broth with 25 μg/mL of kanamycin and PAC DNA was isolated by a modification of Qiagen protocol.33 Restriction enzyme digests and Southern blot transfer were performed as described above for genomic DNA. Field inversion gel electrophoresis was performed to separate large molecular weight fragments (25 to 150 kb), using 1% agarose, 0.5× TBE (45 mmol/L Tris, 45 mmol/L Borate, 1.0 mmol/L EDTA, pH 8.3), with a forward vector of 180 V, a reverse vector of 120 V, switch time of 0.4 to 3.5 seconds, and linear shape, for 20 hours at room temperature.
Hybridization and probes.
Probes used to screen patient samples were a 0.9-kb SstI-BamHI ETV6cDNA (nucleotides 673 to 1580 of GenBank accession no.U11732) encompassing ETV6 exons 5 through 8, a 0.3-kbMluI-BamHI ETV6 cDNA (nucleotides 813 to 1142 of GenBank accession no. U11732) encompassing ETV6 exons 5 and 6, several single copy AML1 genomic fragments mapping to AML1 intron 1 (0.3 BR, 0.3 BSca, 0.6 BS, and 0.9 BR), and ETV6-AML1genomic fusion fragments from each patient. When necessary, reiterated sequences were suppressed by prehybridization to human placental DNA (Sigma) at 65°C for 2 to 4 hours. Southern blots were hybridized to32P probes labeled by the random priming technique, using a Prime-It II kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. Oligonucleotides were labeled using a terminal deoxynucleotidyl transferase end-labeling technique,30 and hybridization performed as previously described30; the blots were washed twice with 0.1× SSC (1× SSC is 0.15 mol/L sodium chloride and 0.015 mol/L sodium citrate) and 0.1% sodium dodecyl sulfate (SDS) at 52°C for random primed DNA fragments or with 6× SSC and 0.1% SDS at 10°C below the melting temperature (Tm) for oligonucleotide probes. Autoradiography of the blots was performed at −70°C with an intensifying screen.
Fluorescence in situ hybridization (FISH) analyses.
Bone marrow cells stored at −20°C in 3:1 methanol:glacial acetic acid were dropped on microscope slides and air-dried. FISH was performed using the Hybrite hybridization system (Vysis, Downers Grove, IL), which combines denaturation of the probe solution and specimen. Briefly, a mixture of LSI TEL/AML1 ES probe (Vysis), specimen hybridization buffer, and water were applied to the slide target area, coverslipped, and denatured at 75°C (1 minute), and hybridized overnight at 37°C in a Hybrite unit. After hybridization, the coverslips were removed and slides were washed in successive washes of 0.4× SSC/0.3% NP-40 (73°C) and 2× SSC/0.1% NP-40 (room temperature). Slides were air-dried in darkness. Cells were counterstained with 4′-6′-diamine-2-phenylindole dihydrochloride (DAPI) and analyzed by fluorescence microscopy using a triple-band pass filter for DAPI, SpectrumGreen, SpectrumOrange. Representative cells were photographed with a computer-based imaging system (Perceptive Scientific Instruments, Inc, League City, TX).
Genomic library construction.
Genomic DNA (10 μg) was digested to completion with BamHI, extracted with phenol-chloroform, ethanol precipitated, and used without further size fractionation. In a total volume of 5 μL at 14°C for 16 hours, 0.2 to 0.6 μg of BamHI-digested genomic DNA was ligated to 1 μg of BamHI-digested λ DASH II (Stratagene) phage arms. Recombinant phage DNA was packaged using Gigapack III Gold (Stratagene) extracts and protocols. Five to 10 × 105 recombinant phages were plated with XL-1Blue MRA (Stratagene) cells and incubated at 37°C. Filter lifts, hybridization, and plaque purification were performed as described.30
DNA sequence analysis.
Relevant genomic fragments were subcloned into plasmid vectors (pBluescript II; Stratagene) and sequenced using either an automated sequencer (ABI PRISM model no. 373A Stretch; Perkin Elmer, Norwalk, CT) or manual sequencing with Sequenase reagents and protocols. When necessary, oligonucleotide primers were synthesized on an oligonucleotide synthesizer (model no. 394; Applied Biosystems, Foster City, CA).
RESULTS
Screening of patient samples for ETV-6 rearrangements.
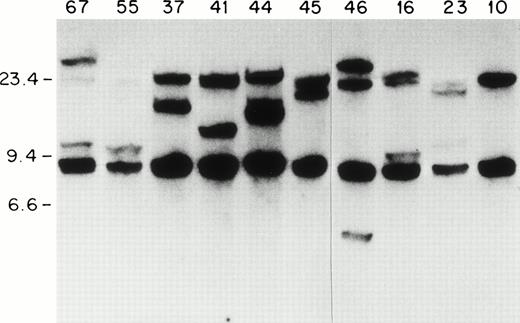
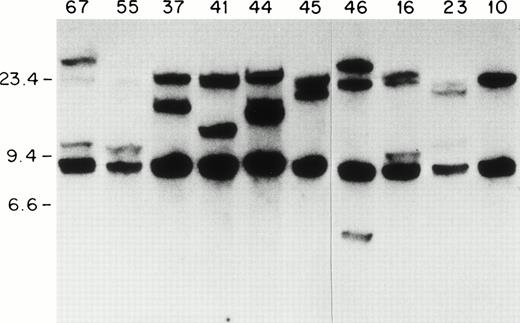
Because most of the ETV6-AML1 rearrangements seem to occur within a 20-kb genomic fragment of the ETV6 gene,14we screened 92 patients with childhood acute leukemia for ETV6gene rearrangements using an ETV6 cDNA probe. Southern blots of genomic DNA extracted from leukemic blasts were screened with a 0.9-kbSstI-BamH1 ETV-6 cDNA probe containing sequences from exons 5 to 8 (nucleotides 673 to 1580 of GenBank accession no. U11732). With the above probe, two germline fragments of 20 kb and 7 kb were identified on a BamHI digest. As shown in Fig 1, one or two rearranged fragments can be detected in lymphoblasts from patients 67, 55, 44, 46, 16, 23, 37, 41, and 45. In patient 41 (as we show later) there were two 20-kb fragments that represented the germline ETV6 fragment, and a rearranged fragment that comigrated with the germline fragment. We hybridized Southern blots from the same patients to a 329-bp MluI-BamHI ETV6 cDNA fragment that contained exon 5 and 6 sequences (nucleotides 813 to 1142). This probe identified the 20-kb germlineETV6 fragment and the same rearranged fragments identified by the previous probe, indicating that all the ETV6 rearrangements identified with the exon 5 to 8 probe were located within this 20-kbBamHI fragment. In total, 13 patients with ETV6 gene rearrangements were identified; all of these patients had B-cell precursor ALL. Therefore, 13/92 or 14% of the entire sample, and 13/68 or 20% of the patients with B-cell precursor ALL had ETV6 gene rearrangements. This frequency (20%) is similar to that found in other series of childhood B-cell precursor ALL.34
ETV6 gene rearrangements in patient samples. Southern blot of BamHI-digested genomic DNA hybridized to the 0.9-kb ETV6 cDNA probe. Size standards are in kb. The unique patient numbers are indicated above each lane. Lane 10 contains DNA from a leukemic patient without an ETV6 gene rearrangement. Twenty-kb and 7-kb bands represent germline ETV6 fragments; rearranged fragments representing ETV6 translocations can be identified in all samples except no. 10.
ETV6 gene rearrangements in patient samples. Southern blot of BamHI-digested genomic DNA hybridized to the 0.9-kb ETV6 cDNA probe. Size standards are in kb. The unique patient numbers are indicated above each lane. Lane 10 contains DNA from a leukemic patient without an ETV6 gene rearrangement. Twenty-kb and 7-kb bands represent germline ETV6 fragments; rearranged fragments representing ETV6 translocations can be identified in all samples except no. 10.
FISH analyses of patient samples.
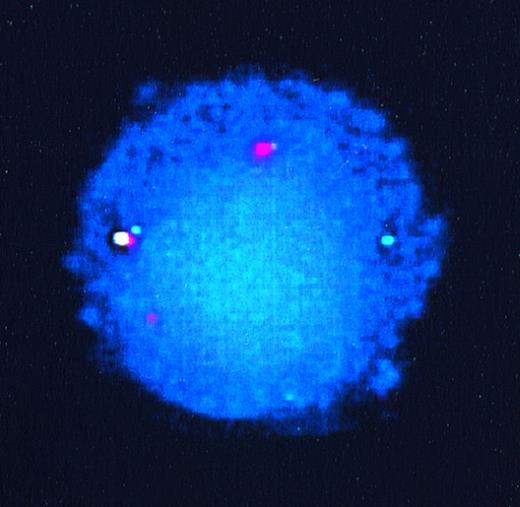
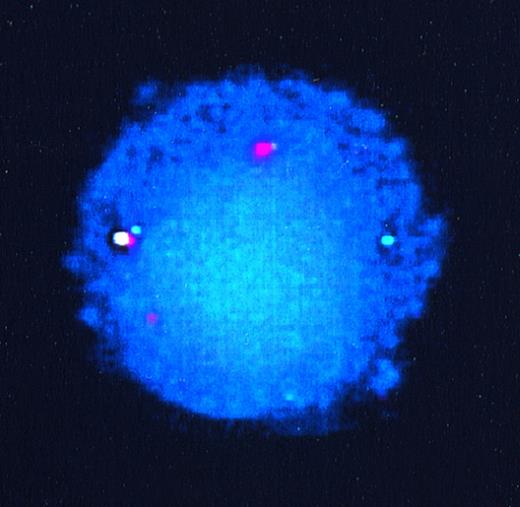
To confirm that the other partner in these translocations wasAML1, we performed FISH analyses on interphase nuclei from patient samples for which interphase preparations were available. The probes used consisted of a 350-kb ETV6 genomic probe encompassing exons 1A to 3, labeled with SpectrumGreen, and a 500-kbAML1 probe encompassing the entire AML1 gene, labeled with SpectrumOrange. Figure 2 shows hybridization signals from the germline ETV6 allele (green), the germline AML1 allele (orange), the derivative 21 containing the ETV6-AML1 fusion (yellow), and the derivative 12, also as orange. The derivative 12 does not appear yellow because the fusion occurs centromeric to the ETV6 region (exons 1A to 3) encompassed by the ETV6 probe.
FISH of interphase nuclei from lymphoblasts of patient 55. The germline ETV6 allele is seen as a green signal, germline AML1 as orange, derivative 21 (ETV6-AML1fusion) as yellow, and the derivative 12 (AML1-ETV6 fusion) also as orange.
FISH of interphase nuclei from lymphoblasts of patient 55. The germline ETV6 allele is seen as a green signal, germline AML1 as orange, derivative 21 (ETV6-AML1fusion) as yellow, and the derivative 12 (AML1-ETV6 fusion) also as orange.
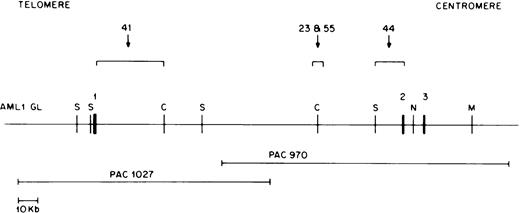
Construction of an AML1 germline map in the region of AML1 exons 1 to 3.
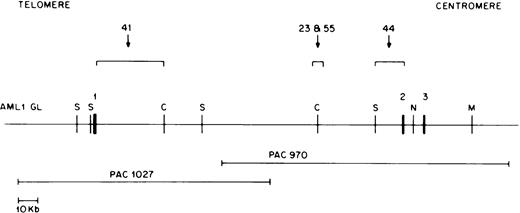
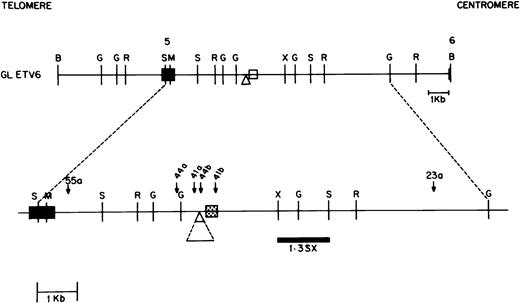
RT-PCR experiments have shown that ETV6-AML1 fusion cDNAs usually result from ETV6 exon 5 splicing to AML1 exon 2, or, less commonly, to AML1 exon 3.17 18 TheAML1 breakpoints in the t(12;21) translocation are therefore thought to lie in the 5′ end of the AML1 gene, most likely between exons 1 and 2. Because the genomic organization of theAML1 gene has not been completely characterized, we developed an AML1 PAC clone contig of this region. We screened the RPCI 5 human genomic PAC library with an AML1 cDNA probe containing exon 1, 2, and 3 sequences (nucleotides 1 to 718 of GenBank accession no. D43969). Using multiple restriction enzyme digests and Southern blot hybridization to selected probes, a PAC contig of six overlapping clones was constructed. Of these, four contained exon 1 sequences but not exon 2 sequences, and two contained exon 2 and 3 sequences but not exon 1 sequences; no single clone contained both exon 1 and exon 2 sequences. Two of these clones, PAC 1027, containing exon 1 sequences, and PAC 970, containing exon 2 and 3 sequences, overlapped as shown in Fig 3 and were chosen for construction of an AML1 germline map. Using multiple restriction enzyme digests of the above two PAC clones and a combination of pulsed field gel and conventional agarose gel electrophoresis as well as hybridization to selected probes, we constructed a map of the AML1 gene as shown in Fig 3. Exons were localized using an AML1 exon 1 cDNA (0.5-kb EcoRI-NsiI) fragment, an exon 2 oligonucleotide (5′TGCATACTTGGAATGAATCCTTCTAG AGAC 3′), and an exon 3 oligonucleotide (5′ TGAGCCCAGGCAAGATGAGC 3′) as probes. The large intron between AML1 exon 1 and exon 2 was determined to be approximately 165 kb.
Restriction map of the AML1 gene. Chromosomal orientation from telomere to centromere is as indicated. Exons 1, 2, and 3 are shown as solid boxes (▪). The location of the breakpoints in different patients is indicated by a bracket. Restriction enzyme sites are C, ClaI; M, MluI ; N,NotI; and S, SalI. The two overlapping P1 artificial chromosome clones containing either AML1 exon 1 or exon 2 and 3 are depicted below the AML1 restriction map.
Restriction map of the AML1 gene. Chromosomal orientation from telomere to centromere is as indicated. Exons 1, 2, and 3 are shown as solid boxes (▪). The location of the breakpoints in different patients is indicated by a bracket. Restriction enzyme sites are C, ClaI; M, MluI ; N,NotI; and S, SalI. The two overlapping P1 artificial chromosome clones containing either AML1 exon 1 or exon 2 and 3 are depicted below the AML1 restriction map.
Cloning and localization of the ETV6-AML1 breakpoints.
We chose to clone and characterize the ETV6-AML1 genomic breakpoints from 4 of the 13 patients identified with ETV6 gene rearrangements. Patient-specific genomic DNA libraries were constructed by cloning BamH1-digested genomic DNA into the λ DASH II vector. Five to 10 × 105 recombinant phages were screened with the 329-bp MluI-BamHI ETV6 cDNA probe. Independent hybridizing plaques were identified and purified. The phage inserts were subcloned into plasmids, restriction mapped, and sequenced from both ends to identify plasmids with either the ETV6germline allele or rearranged inserts representing either the derivative 12 or derivative 21 alleles.
To rapidly identify and subclone the germline AML1 fragment involved in each translocation, we hybridized the cloned, rearrangedETV6-AML1 genomic fragments to Southern blots ofBamHI-digested AML1 PAC clones. Reiterated sequences in the probes were suppressed by prehybridization with human placental DNA. Using this technique, a single genomic AML1 BamHI fragment was identified from each patient. The fragment was gel purified, subcloned into plasmid vectors, and mapped with restriction enzymes. The regions that corresponded to the breakpoints were then sequenced.
In this manner, we were able to clone and sequence the derivative 21 breakpoint from all four selected patients. In addition, we were able to clone and sequence the derivative 12 breakpoint from two patients; we were not able to recover the derivative 12 allele from the remaining two patients. Figure 3 shows the location of the AML1breakpoints in these four patients with respect to AML1 exons 1, 2, and 3. The AML1 breakpoints in patients 23 and 55 were in a 6-kb BamHI fragment within intron 1 located approximately 40 kb upstream of exon 2 (Fig 3). In patient 44, the AML1breakpoint occurred within a 7-kb germline BamHI fragment containing AML1 exon 2 sequences, whereas patient 41 had anAML1 breakpoint directly downstream of exon 1 as indicated in Fig 3. Therefore, three out of four patients studied had AML1breakpoints within a 40-kb region of intron 1.
To determine if the AML1 breakpoints from our 13 patients were tightly clustered near any of these four breakpoints, we hybridized single-copy AML1 genomic probes (0.3 BSca near breakpoint of patient 41, 0.6 BS and 0.3 BR near breakpoints of patients 23 and 55, and 0.9 BR near breakpoint of patient 44; Fig 3) derived from these four breakpoints to Southern blots of BamHI- andHindIII-digested genomic DNA from our 13 patients. Of the four probes used, only the 0.3 BR probe was able to identify rearranged fragments in more than one sample; this AML1 genomic probe identified rearranged fragments in two patients (patients 23 and 55).
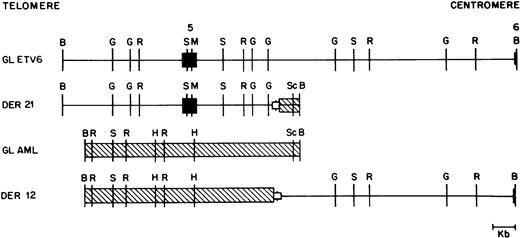
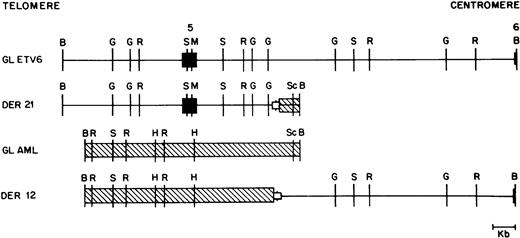
As mentioned above, we were able to clone both germline alleles and both derivative alleles from two patients (patients 41 and 44). The four relevant alleles from patient 41 are shown in Fig 4, and show that this translocation was actually a translocation-deletion with portions of both ETV6and AML1 being deleted (see below).
Restriction map of germline ETV6 (GL ETV6), germline AML1 (GL AML), and the derivative 12 (DER 12) and derivative 21 (DER 21) alleles from patient 41. ETV6 exons are shown as solid boxes (▪). ETV6 is depicted as a straight line; AML1 is crosshatched (▧). The open boxed area (□) depicted at the breakpoint junction in the two derivative alleles represents a deletion of 540 bp of the ETV6 gene and 165 bp of the AML1 gene observed in this patient. Restriction enzyme sites are B, BamHI; G, BglII; R, EcoRI; S,SstI; M, MluI; H, HindIII; and Sc, ScaI .
Restriction map of germline ETV6 (GL ETV6), germline AML1 (GL AML), and the derivative 12 (DER 12) and derivative 21 (DER 21) alleles from patient 41. ETV6 exons are shown as solid boxes (▪). ETV6 is depicted as a straight line; AML1 is crosshatched (▧). The open boxed area (□) depicted at the breakpoint junction in the two derivative alleles represents a deletion of 540 bp of the ETV6 gene and 165 bp of the AML1 gene observed in this patient. Restriction enzyme sites are B, BamHI; G, BglII; R, EcoRI; S,SstI; M, MluI; H, HindIII; and Sc, ScaI .
ETV6 breakpoints have previously been reported to cluster in a 5.6-kb EcoRI fragment within ETV6 intron 5.8 In two of our four patients, at least one breakpoint occurred within this fragment. However, in the other two patients, theETV6 breakpoint was either 2 kb centromeric (patient 23) or 1.7 kb telomeric (patient 55) to this region.
Sequence characteristics in the vicinity of the breakpoints.
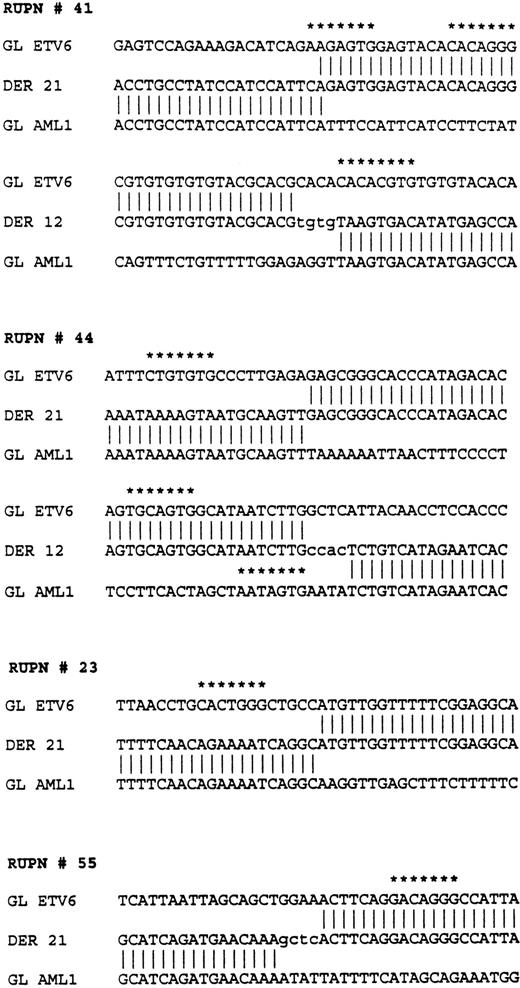
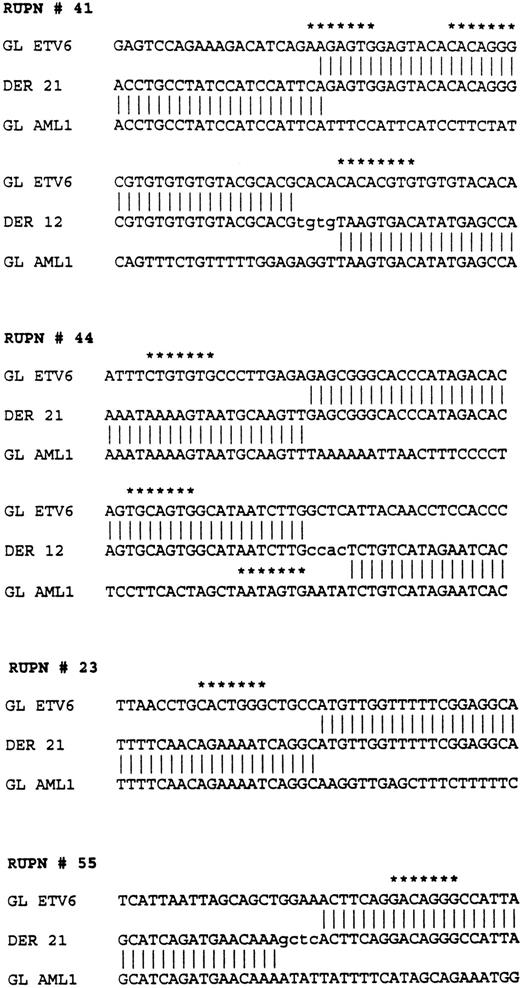
The nucleotide sequences flanking each of the breakpoints was determined on each derivative and germline allele through sequence analysis of relevant portions of plasmid subclones. In patients 44 and 41, both derivative alleles were cloned and sequenced. Close analysis of the nucleotide sequence showed that these were not perfectly reciprocal translocations, and that both patients had a variable number of nucleotides deleted at the breakpoints from both the ETV6and the AML1 genes. Patient 41 had 540 nucleotides ofETV6 deleted and 165 nucleotides of AML1 deleted; patient 44 had 659 nucleotides of ETV6 deleted and 205 nucleotides of AML1 deleted.
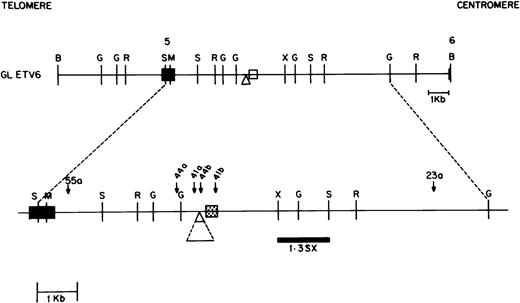
Several unique sequence characteristics found in the ETV6 gene near the breakpoints are depicted in the map in Fig 5; sequences flanking the breakpoints in the four different patients studied are shown in Fig 6. A nucleotide sequence consisting of alternating purine and pyridimine nucleotides (a pu/py tract) was found in ETV6 intron 5 within the breakpoint cluster region. Of six alleles that were sequenced through this region, the length of this tract varied from 206 to 233 nucleotides, indicating that this tract undergoes expansions and contractions. The nucleotide sequence of the pu/py tract was identical in three of these six alleles and single nucleotide polymorphisms in the nucleotide sequence was observed in the remaining three. However, in the latter three, the single nucleotide polymorphisms were all G ↔ A or T ↔ C changes that maintained the integrity of the alternating pu-py tract.
Map of the ETV6 gene between exon 5 and 6 containing the ETV6 breakpoint cluster region (bcr). Chromosomal orientation from telomere to centromere is as indicated. Exons 5 and 6 are shown as solid boxes. The breakpoints in the different patients are shown as downward arrows with the patient identification numbers marked above. Patients 41 and 44 have two breakpoints each, designated 41a and 41b, and 44a and 44b, respectively. The portion of the gene between these breakpoints is deleted in each patient. Only the derivative 21 was cloned from patients 23 and 55. The 0.3-kb area in the ETV6 gene, shown as a speckled box, represents a pu/py repeat region. The 0.7-kb region depicted by the triangle is a deletion/insertion polymorphism in theETV6 gene. Restriction enzyme sites are B,BamHI; G, BglII; R, EcoRI; S, SstI; M, MluI; H,HindIII and X, XbaI. The 1.3-kbSstI-XbaI probe used to identify the deletion/insertion polymorphism on Southern blot is indicated .
Map of the ETV6 gene between exon 5 and 6 containing the ETV6 breakpoint cluster region (bcr). Chromosomal orientation from telomere to centromere is as indicated. Exons 5 and 6 are shown as solid boxes. The breakpoints in the different patients are shown as downward arrows with the patient identification numbers marked above. Patients 41 and 44 have two breakpoints each, designated 41a and 41b, and 44a and 44b, respectively. The portion of the gene between these breakpoints is deleted in each patient. Only the derivative 21 was cloned from patients 23 and 55. The 0.3-kb area in the ETV6 gene, shown as a speckled box, represents a pu/py repeat region. The 0.7-kb region depicted by the triangle is a deletion/insertion polymorphism in theETV6 gene. Restriction enzyme sites are B,BamHI; G, BglII; R, EcoRI; S, SstI; M, MluI; H,HindIII and X, XbaI. The 1.3-kbSstI-XbaI probe used to identify the deletion/insertion polymorphism on Southern blot is indicated .
Sequences flanking the breakpoints in four patients withETV6-AML1 rearrangements. RUPN refers to unique patient number. Sequences from the ETV6 germline (GL ETV6), AML1germline (GL AML1), and the two derivatives, derivative 12 (DER 12) and derivative 21 (DER 21), are shown. Nontemplated nucleotides are shown in lower case. Nucleotide sequences with at least a 5/7 match to the consensus V(D)J heptamer recognition sequence (CAC A/T GTG) are shown with asterisks.
Sequences flanking the breakpoints in four patients withETV6-AML1 rearrangements. RUPN refers to unique patient number. Sequences from the ETV6 germline (GL ETV6), AML1germline (GL AML1), and the two derivatives, derivative 12 (DER 12) and derivative 21 (DER 21), are shown. Nontemplated nucleotides are shown in lower case. Nucleotide sequences with at least a 5/7 match to the consensus V(D)J heptamer recognition sequence (CAC A/T GTG) are shown with asterisks.
One of the breakpoints found in lymphoblasts from patient 41 occurred within this pu/py tract, whereas the other breakpoint from this patient’s lymphoblasts was 316 nucleotides 5′ of this tract. Similarly, the ETV6 breakpoints in patient 44 were 726 nucleotides (44a of Fig 5) and 67 nucleotides (44b of Fig 5) 5′ of the pu/py tract. The ETV6 sequence between the corresponding breakpoints was found to be deleted in each patient (540 nucleotides deletion in patient 41, 659 nucleotides deletion in patient 44). Of interest, a deletion/insertion polymorphism resulting in a deletion of 738 nucleotides with respect to the published ETV6 sequence (nucleotide 31601 to 32338 of GenBank accession no. U61375 ) was found in the ETV6 gene 172 nucleotides telomeric to the pu/py tract (Fig 5). This seems to be a stable polymorphism; of the six germline and rearranged ETV6 alleles that we characterized, three had this sequence and three did not.
Analyses of the nucleotide sequences from the germline ETV6 andAML1 alleles in the vicinity of the breakpoints showed several additional features. There were 5/7 matches for “cryptic” V(D)J heptamer signal sequences in the four patients studied as indicated in Fig 6. No nonamer-like sequences were found. Additionally, there were nontemplated nucleotides added at the translocation fusion sites in these three patients reminiscent of “N-region” nucleotide addition that is known to occur in physiological V(D)J recombination. Lastly, in addition to the pu/py tract in the ETV6 gene, an alternating “TGGA” repeat region of 225 bp was found at one of the AML1 gene breakpoints from patient 41.
DISCUSSION
Characterization of chromosomal translocation breakpoints at a molecular level has resulted not only in identifying known and potential proto-oncogenes, but has also provided valuable information regarding the possible mechanisms that cause these chromosomal rearrangements. Whereas the genomic organization of a large part of the ETV6 gene, including a t(12;21) breakpoint cluster region, has been described,35 clustering of the AML1 breakpoints has not been reported. In fact, until now, the precise size of AML1intron 1 was not known.17 With the data presented in this paper we report mapping of the AML1 gene between exons 1 and 3, localization of the t(12;21) breakpoints in this region, and characterization of the nucleotide sequence flanking breakpoints in both the ETV6 and AML1 genes.
We identified and characterized overlapping PAC clones spanning theAML1 gene from exons 1 through 3. AML1 intron 1 is large and spans approximately 165 kb. Most of the t(12;21) breakpoints have been thought to occur in intron 1, based on the observation thatETV6-AML1 fusion cDNAs fuse ETV6 exon 5 to AML1exon 2; a minority of ETV6-AML1 fusion cDNAs fuse ETV6exon 5 to AML1 exon 3,28 leading to speculation that these might arise from patients who have AML1 breakpoints within intron 2. All of the 4 patients who we characterized had breakpoints within intron 1; 3 out of 4 were in a 40-kb region of intron 1 immediately 5′ of exon 2. When we screened our panel of 13 patients for breakpoint clustering, we were unable to detect tight clusters near any of the breakpoints we identified.
To gain insight into the mechanism of ETV6-AML1 translocations, we cloned and sequenced breakpoints from individual patients. Illegitimate V(D)J recombination is a well-known cause for chromosomal translocations resulting in juxtaposition of an immunoglobulin (Ig) or T-cell receptor (TCR) gene to a proto-oncogene; in many translocations involving Ig or TCR genes, “cryptic” V(D)J heptamer and nonamer consensus sequences in the immediate vicinity of the breakpoint can be found.20,21 Although we found features that suggested that illegitimate V(D)J recombination might occur in this region, the evidence for illegitimate V(D)J recombinase activity was not as convincing as that observed in more compelling cases.20 21Significant similarity to the V(D)J heptamer consensus sequence (CACA/TGTG) was found near breakpoints of all of our patients, but only on the ETV6 side of the breakpoints; no heptamer sequences were found near the AML1 breakpoint regions. Nontemplated nucleotides were added at the breakpoint junction in the derivative chromosomes; however, this feature, although regarded as a hallmark of normal V(D)J recombination, is not an exclusive feature of V(D)J recombinase activity.
One of the AML1 breakpoints from patient 41 occurred within a 225-bp (TGGA)n repeat region. Tetranucleotide repeats such as (AGGC)n, (TCTG)n, and (TGGA)n have previously been identified at recombination hotspots within the HLA class II region, and it has been speculated that this (TGGA)n repeat may predispose DNA to recombination events.36
We also identified a pu/py tract in the vicinity of several breakpoints in the ETV6 gene. Additionally, the single genomicETV6-AML1 genomic fusion previously reported is also within this pu/py tract.27 Boehm et al24 have described the presence of pu/py tracts in proximity to some translocation breakpoints and have speculated that DNA at these sites may exist in a Z-DNA conformation which then leads to increased recombination frequency. This contention was supported by the existence of DNAse I hypersensitive sites adjacent to pu/py tracts24 in that study. Interestingly, we found a 738-bp deletion with respect to the published ETV6 gene sequence in the ETV6 gene immediately 5′ to the pu/py tract; this has recently been reported as a stable polymorphism in the ETV6gene.35 Therefore it would seem that this pu/py tract within the ETV6 gene is a hotspot for recombination events, including insertions/deletions as well as chromosomal translocations. Additionally, (CA)n repeat regions are well known to undergo expansion and contraction both in vivo and in vitro presumably because of “stuttering” of DNA polymerases.37,38Intriguingly, triplet repeat regions on plasmid templates have recently been shown to be site of replication fork pausing in Escherichia coli,39 raising the possibility that these nucleotide repeat regions may be more accessible to recombination events caused by replication fork pausing.
Recently, several investigators have noted that patients with B-cell precursor ALL and an ETV6-AML1 fusion have an improved prognosis, and have recommended that these patients be treated with regimens that emphasize antimetabolite chemotherapy.5 6 If patients are to be stratified for therapeutic purposes based onETV6 gene rearrangements, it is important to emphasize the insertional polymorphism that occurs within the ETV6 breakpoint cluster region, to prevent patients from being incorrectly genotyped.
We have shown that the AML1 breakpoints in the t(12;21) occur in the large intron 1 of this gene; the ones we have characterized are dispersed throughout intron 1. The breakpoints in the ETV6 gene most commonly occur in a region of intron 5 close to a pu/py tract. Although characteristics of DNA sequence in the region of the breakpoints such as cryptic heptamer signal sequences and nontemplated nucleotide addition suggest that recombination may be mediated in part by aberrant V(D)J recombinase activity, the lack of precise clustering argues against this possibility. Intriguingly, several breakpoints as well as a stable insertion polymorphism mapped near an extended polymorphic pu/py repeat region, suggesting that this region may be predisposed to DNA breakage-religation events, including chromosomal translocations.
ACKNOWLEDGMENT
We thank the staff of the RPCI Biopolymer facility, Thomas Isac for oligonucleotide synthesis, and Michelle Detwiler for sequencing. We thank Elena Greco for illustrations. We also thank Dr Gary Gilliland for providing the ETV6 cDNA and Dr Misao Ohki for the AML1cDNA.
Supported in part by grants from the Roswell Park Alliance Foundation, the National Institutes of Health (CA 73773), and a 1997 Developmental Fund award from the Roswell Park Cancer Center Support Grant (CA16056) to P.D.A. and HG01165 to P.J.D. P.D.A. is a scholar of the Leukemia Society of America.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Peter D. Aplan, MD, Department of Pediatrics, Roswell Park Cancer Institute, Buffalo, NY 14263; e-mail:paplan@sc3101.med.buffalo.edu.