Abstract
The majority of the human population harbors latent cytomegalovirus. Although CD14+ peripheral blood mononuclear cells have been implicated as sites of latency, the conformation of the latent viral genome in these cells is unknown. In this study, the conformation of viral genomic DNA was assessed in CD14+ cells from healthy virus seropositive carriers using an electrophoretic separation on native agarose gels in combination with polymerase chain reaction detection. Here we show that the viral genome migrates as a circular plasmid with a mobility equivalent to a circular 230-kb Shigella flexneri megaplasmid marker. Neither linear nor complex or integrated forms of the viral genome were detected. This report provides further evidence that the CD14+ cell population is an important site of viral latency in the naturally infected human host. Detection of the viral genome as a circular plasmid during latency suggests that this virus maintains its genome in a manner analogous to other herpesviruses where latent viral genome conformation has been studied.
HUMAN CYTOMEGALOVIRUS (CMV), a large 230- to 240-kb dsDNA member of the betaherpesvirus subfamily, is harbored latently in 50% to 100% of healthy adults.1,2 The virus can infect the human host directly through mucous membrane contact, or by infected cells present in blood transfusions or tissue transplants. Infections caused by CMV are a leading infectious disease cause of morbidity in allograft transplant recipients as well as other immunocompromised hosts (eg, human immunodeficiency virus [HIV] infected).3 They are also the predominant cause of congenital infection in the Western world.4 CMV infection is a significant risk after blood transfusion, because circulating mononuclear cells of the peripheral blood (PB) provide a reservoir for the latent virus.5-7 Reactivation of latent virus occurs in the face of diminished immune surveillance and contributes more to the incidence of serious disease in immunocompromised hosts than primary infection. Despite the important role latency and reactivation play in the pathogenesis of CMV disease, knowledge of the underlying mechanisms controlling these processes remains limited.
Although viral DNA has been detected in circulating PB mononuclear cells (PBMCs) from healthy seropositive individuals,8,9 the conformation of this DNA has not been investigated. Identification of the CMV genome form in blood cells can provide insight into understanding mechanisms of viral persistence and dissemination in the human host. For herpesviruses where this has been studied, latent infections are characteristically associated with a circular plasmid form of the viral genome.10-14 In contrast, productive herpesviral infection is associated predominantly with linear viral genome forms.12,15 It has long been known that the latent Epstein-Barr virus (EBV) is a circular extrachromasomal plasmid in cultured B-lymphoblastoid cells.10,11 The latent herpes simplex virus-1 genome exists in an endless state based on DNA blot analysis16 and an endless state has also been predicted for the latent varicella-zoster virus genome.17 While these latter findings are consistent with a circular genome conformation analogous to EBV, tandemly integrated genomes or concatemers remain a possibility. To analyze genome conformation specifically, a native agarose gel system has been used to characterize large bacterial plasmids18 and circular or linear forms of herpesvirus saimiri, herpesvirus ateles, EBV, and human herpesvirus 8 genomes.12-14 Rare cells in the PB CD14+population, which can only be detected after polymerase chain reaction (PCR) amplification, are known to carry CMV without evidence of productive infection8,9,19 and can reactivate CMV after allogeneic stimulation.7 The focus of our study was to apply native agarose gel electrophoresis, combined with PCR amplification,13 to determine the CMV genome conformation in PB mononuclear CD14+ cells from healthy seropositive carriers. The results of this study show that the CMV genome migrates as a circular plasmid form, suggesting that this virus maintains its genome in the PB CD14+ cell population in a manner analogous to other herpesviruses where latent viral genome conformation has been studied.
MATERIALS AND METHODS
Isolation of CD14+ cells.
Individuals were identified as CMV seropositive using a CMV IgG ELISA (Sigma, St Louis, MO). Healthy adult CMV-seropositive volunteers donated according to institutional review board guidelines. Fifty to 100 mL of blood was obtained by percutaneous venepuncture and anticoagulated with EDTA (final concentrations, 5 mmol/L). A volume of 250 to 300 mL of pooled blood was prepared for each sample set. PBMCs were isolated on Lymphoprep gradients (Nycomed, Oslo, Norway). CD14+ cells were selected using magnetic cell sorting with microbeads (Miltenyi Biotec, Auburn, CA) according to manufacturer’s instructions, using two successive column purifications for each preparation. Purity analysis was performed using fluorescein isothiocyanate (FITC) anti-CD14 clone MφP9 (Becton Dickinson, San Jose, CA) followed by flow cytometry.
Cells and viruses used in control samples.
Primary human fibroblasts were infected at a multiplicity of infection (MOI) of 5 plaque forming units (PFU) using CMV strain Toledo. Cells were obtained 7 days after infection, for preparation of positive controls. The bacteria Shigella flexneri (gift from Stanley Falkow, Stanford University) contains a circular megaplasmid of approximately 230 kb, and served as a circular plasmid size marker. Bacteria were cultured in Luria broth, using appropriate biohazard precautions.
Preparation of agarose blocks for gel analysis.
Cell samples were prepared in agarose blocks using previously described methods,20 and the CD14+ cells were prepared at 2.5 × 106 cells per 100 μL volume in each block. Two agarose blocks (stacked front to back) per lane were cast directly into gels.
Electrophoretic gel analysis.
Horizontal agarose gels were prepared according to methods previously described.12 Low melting point (LMP) agarose (0.75%) in 0.5× Tris-Borate-EDTA buffer was used to cast 25 × 20-cm gels. After the gel had solidified, a section of the gel at the origin (3 × 15 cm) was excised, and the comb placed in the cutout section. The section was filled with 0.8% LMP agarose containing 2% sodium dodecyl sulfate (SDS) and 1 mg/mL of Proteinase K added after gel temperature had cooled to 50°C. Bacteria samples were loaded live onto gels using 108 cells per lane with previously described methods.12 Gels were run at 4°C for 3 hours at 0.8 V/cm, followed by 24 hours at 4.5 V/cm.
After electrophoresis, portions of the gel containing the controls were stained in ethidium bromide solution (1 μg/mL) and photographed. For PCR amplification, sample lanes were scored into sequential cores starting from the origin, with the location of each recorded on a gel template. Each agarose core was melted at 65°C for 15 minutes, then digested with beta-agarase (FMC Bioproducts, Rockland, ME) according to the manufacturer’s instructions. DNA was precipitated with glycogen (10 μg per sample), ammonium acetate (Sigma) added to a final concentration of 2.5 mol/L, and 2.5 vol of 100% ethanol added. The resulting pellets were resuspended in 10 μL of TE buffer (10 mmol/L TRIS-HCl pH 8, 1 mmol/L EDTA), and the entire volume was added to each PCR reaction.
PCR detection assays.
To reduce the risk of losing PCR amplification due to viral sequence heterogeneity among different isolates, primer pairs directed to the major early beta 2.7 gene were selected. This primer set recognizes a conserved region of the CMV genome, and has been applied successfully to detection of CMV isolates from eight different individuals and one laboratory strain (AD 169) as previously reported.21Forward primer, reverse primer, and probe oligonucleotides are as previously reported.21 PCR amplifications (50-μL vol) used reaction mix as reported with the following exceptions: Tris-HCl pH 8.3, 3 mmol/L MgCl2, 1.25 U of Amplitaq Gold (Perkin Elmer, Norwalk, CT), and 1.25 U of Taq Extender (Stratagene, La Jolla, CA). The primary PCR amplification product was 315 bp. PCR reactions were performed in a Perkin Elmer 2400 thermocycler under the following conditions: 10 minutes at 94°C was used to activate the Amplitaq polymerase, followed by 50 cycles of 20 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 70°C. After the cycles were completed, a final extension at 70°C for 7 minutes was performed. For nested PCR conditions, the forward primer 5′ CCG GTC GGC TTC TGT TTT AT 3′ and reverse primer 5′ TCT CTT GTT GGG AAT CGT CG 3′ were used. PCR amplifications (30 cycles, 20-μL vol) used reaction mix as reported above with the following exceptions: Tris-HCl pH 8.7, 0.1% Triton X-100, and 0.1 U of native Taq polymerase (Promega, Madison, WI). One microliter of the primary PCR reaction was added as template. PCR conditions were 5 minutes at 94°C, followed by 30 cycles of 20 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 72°C. A final extension at 72°C for 7 minutes was then performed. Nested PCR yielded a 202-bp product. The internal oligonucleotide probe used was the same as above. CD14+cell samples were confirmed to contain CMV DNA before gel analysis, using total cell DNA prepared and PCR amplified as described above. When using limiting dilutions of positive control viral DNA as template, PCR reactions were sensitive enough to detect two template copies, without requiring a nested amplification.
The detection limits of the combined gel resolution and PCR approach were determined, using limiting dilutions of CMV infected cells in a background of negative cells. Viral DNA was detected down to a dilution of ≈3 infected cells in a background of 106 uninfected PBMCs collected from a seronegative donor (see Fig 2E). This amount of viral DNA is expected to be in the range of 3 × 103 to 3 × 104 copies, well below the detection limits for direct blot hybridization.22 To further establish the limit of detection by PCR after recovery of DNA after gel electrophoresis and beta agarase digestion, known copy numbers of CMV cosmid clones were used as template. It was determined that the level of sensitivity was 50 copies (data not shown). Detection was lost below this level.
No inhibition of the PCR reactions was observed when low copy numbers of CMV DNA were tested in the presence of melted agarose gels collected from linear regions of uninfected cell DNA. Cores collected adjacent to low copy positive sample lanes were also routinely negative. No inhibition of the PCR reactions was observed, when tested using low copy positive controls in the presence of beta agarase digested agarose cores (data not shown).
DNA blot analysis of PCR products.
Ten percent of each PCR reaction volume was analyzed on 6% polyacrylamide gels, with a 1-kb ladder marker (GIBCO-BRL, Gaithersburg, MD). DNA was transferred to Zetaprobe membrane (Boehringer Mannheim, Indianapolis, IN), by capillary transfer in 0.4 N sodium hydroxide. The oligonucleotide probe (above) was prepared using previously described methods.9
RESULTS
Resolution of viral genome forms using native agarose gels.
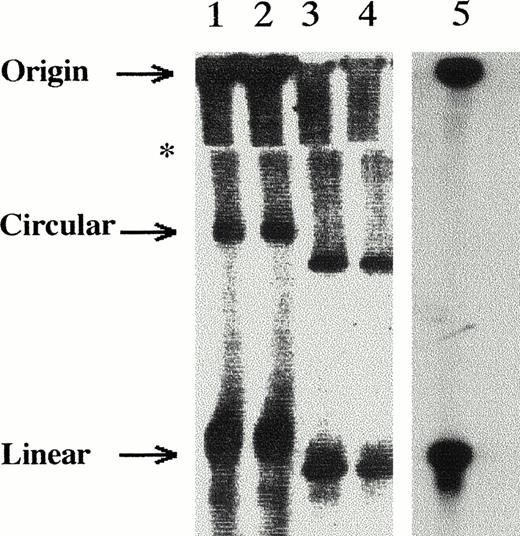
The Southern blot in Fig 1 shows that circular forms of DNA are clearly resolved from those of unit-length linear structures. The results obtained from a megaplasmid of approximately 230 kb (similar in size to the CMV genome) carried inS flexneri bacteria are shown in duplicate lanes 1 and 2. Probe-positive linear and circular forms of the megaplasmid are observed. The linearized form seen is caused by the harsh lysozyme treatment and shearing of some of the megaplasmid DNA.18Duplicate lanes 3 and 4 show the distinct separation of probe-positive linear and circular forms for EBV, a herpesvirus that is closely related in size to CMV (≈170 kb). For EBV analyses, the B95-8 cell line was used. In these cells, both circular and linear forms of the EBV viral genome are present, because 5% of the cells are known to be producing virus with the remaining number carrying latent circular viral genome.13 23 Finally, in lane 5, CMV productively infected cells (collected 7 days postinfection) show only a distinct probe-positive unit-length linear 230-kb band and no circular form, as would be predicted in a productive infection. In addition, some viral DNA is retained at the well origin for all lanes. The linearized megaplasmid form (lanes 1 and 2) migrates with a similar mobility as the unit length linear form of the CMV genome and closely with the linear EBV form. Overall, the clear resolution of circular versus linearized genome forms is shown using the native agarose gel system.
Southern blot showing resolution of DNA forms. Lanes loaded as the following: lanes 1 and 2, duplicate samples of 108 cells S flexneri bacteria carrying ≈230-kb circular megaplasmid, loaded live in each lane; lanes 3 and 4, duplicate samples of EBV cell line B95-8 with 106 cells loaded per lane; lane 5, 106 CMV lytically infected cells. Southern blot was sectioned, and each section hybridized with probe specific for those particular samples. The blots were then realigned for figure. The (∗) symbol marks the lower edge of the proteinase K/SDS/agarose trough.
Southern blot showing resolution of DNA forms. Lanes loaded as the following: lanes 1 and 2, duplicate samples of 108 cells S flexneri bacteria carrying ≈230-kb circular megaplasmid, loaded live in each lane; lanes 3 and 4, duplicate samples of EBV cell line B95-8 with 106 cells loaded per lane; lane 5, 106 CMV lytically infected cells. Southern blot was sectioned, and each section hybridized with probe specific for those particular samples. The blots were then realigned for figure. The (∗) symbol marks the lower edge of the proteinase K/SDS/agarose trough.
Purity of isolated CD14+ cell populations and detection of CMV DNA before native agarose gel analysis.
CD14+ cells were isolated from PBMCs using positive selection as described in Materials and Methods. The purity of the CD14+ cells was routinely 95% or higher, based on flow cytometry analysis (data not shown). Typical yields ranged from 10% to 20% of the original PBMC numbers. The cells displayed a monocyte type morphology, and were able to adhere to glass or plastic.
Demonstration of free episomal form of CMV DNA from CD14+ cell populations in healthy carriers.
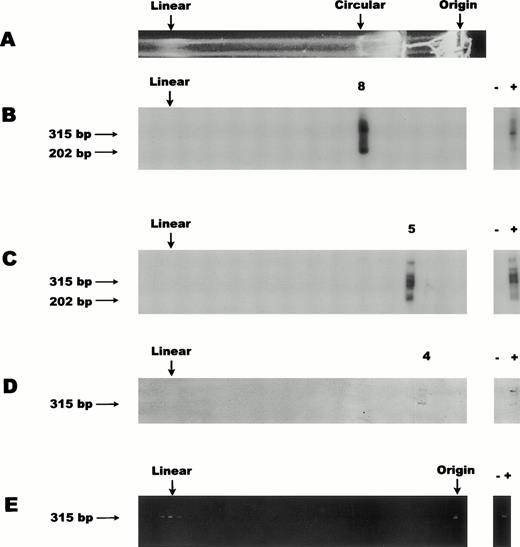
To characterize the conformation of the CMV genome in PB mononuclear CD14+ cells, we analyzed three independent sets of pooled cells from healthy seropositive individuals. Figure 2 shows the results obtained after selection of CD14+ cells, electrophoretic separation, and PCR amplification of DNA recovered from individual gel cores followed by blot hybridization. A clear CMV probe-positive signal was observed in each of three analyses, sets 1 through 3 (panels B through D), from respective single cores with similar mobility to that of the circular 230-kb megaplasmid marker control (panel A). In all three sets, a CMV DNA positive signal was detected in only one discrete agarose core.
CMV genome conformation in PB CD14+ cells. For all panels the top (Origin) of the gel is represented on the right and the bottom of the gel is depicted on the left. Symbols + and − indicate positive and negative controls for PCR. Numbers 8, 5, and 4 label the probe-positive cores for panels (B), (C), and (D), respectively. Size markers (315 bp and 202 bp) for the primary and nested PCR products, respectively, are represented. (A) Ethidium bromide–stained gel showing a representative lane containing 108 cells of S flexneri bacteria carrying the ≈230-kb circular megaplasmid marker. Ethidium bromide staining of bacterial chromosomal DNA is associated with the region near the well origin (Origin). The lower edge of the proteinase K/SDS/agarose trough is seen as a brightly stained region midway between the circular marker and the well origin. The circular megaplasmid control migrated at core 8 for all gels shown in panels (B), (C), (D), and (E). The apparent linear megaplasmid band (Linear) invariably comigrated with the CMV linear band as exemplified in the positive cores after PCR amplification of low copy CMV-infected cells (E). (B) DNA blot analysis of nested PCR products, from an inclusive series of sequential agarose cores collected from the gel lane prepared from set 1 of pooled CD14+ cells from five donors. (C) DNA blot analysis similar to (B), from set 2 (3 donors). (D) DNA blot analysis of primary PCR products, from set 3 (5 donors) and using only 10% of the proteinase K normally used in block preparation. (E) Ethidium bromide–stained gel of PCR amplification of low copy CMV-infected cells.
CMV genome conformation in PB CD14+ cells. For all panels the top (Origin) of the gel is represented on the right and the bottom of the gel is depicted on the left. Symbols + and − indicate positive and negative controls for PCR. Numbers 8, 5, and 4 label the probe-positive cores for panels (B), (C), and (D), respectively. Size markers (315 bp and 202 bp) for the primary and nested PCR products, respectively, are represented. (A) Ethidium bromide–stained gel showing a representative lane containing 108 cells of S flexneri bacteria carrying the ≈230-kb circular megaplasmid marker. Ethidium bromide staining of bacterial chromosomal DNA is associated with the region near the well origin (Origin). The lower edge of the proteinase K/SDS/agarose trough is seen as a brightly stained region midway between the circular marker and the well origin. The circular megaplasmid control migrated at core 8 for all gels shown in panels (B), (C), (D), and (E). The apparent linear megaplasmid band (Linear) invariably comigrated with the CMV linear band as exemplified in the positive cores after PCR amplification of low copy CMV-infected cells (E). (B) DNA blot analysis of nested PCR products, from an inclusive series of sequential agarose cores collected from the gel lane prepared from set 1 of pooled CD14+ cells from five donors. (C) DNA blot analysis similar to (B), from set 2 (3 donors). (D) DNA blot analysis of primary PCR products, from set 3 (5 donors) and using only 10% of the proteinase K normally used in block preparation. (E) Ethidium bromide–stained gel of PCR amplification of low copy CMV-infected cells.
A clear CMV probe-positive signal was observed in sample set 1 (panel B; core 8), at a region comigrating with the circular 230-kb megaplasmid marker (panel A). In addition, DNA blot analyses typically showed both the primary PCR amplification product (315 bp) as well as the nested amplification product (202 bp). Neither the region where linear DNA would migrate nor the well origin showed positive signal. For comparison, the relative mobility of the linear CMV genome on this type of gel system is represented in Fig 2E, starting with ≈3 CMV-infected fibroblasts in a background of 106CMV-negative PB cells.
Set 2 (panel C) exhibited a slightly slower migrating CMV probe-positive core than that of the circular megaplasmid marker or the migration of the CMV DNA probe positive core in sample set 1. For set 2, core 5 (panel C) migrated approximately 1 cm slower than core 8 (panel B), which was the core migrating with similar mobility to the circular megaplasmid. Once again, there was no evidence of a PCR-positive signal in the region of the gel representing linear 230-kb CMV genome and none seen at the well origin, even after nested PCR amplification and DNA blot analysis. For both sets 1 and 2 (panels B and C) the primary PCR amplification showed CMV DNA migrating at a mobility similar to the circular megaplasmid marker DNA and nested PCR was not necessary to see this signal (see panel D where only primary PCR was undertaken). Nested PCR was used to increase the sensitivity of the assay for viral DNA in other regions of the gel. Our results suggest that viral DNA was not present in the linear region of the gel or near the well origin.
One explanation for the mobility differences between set 1 and set 2 might have been varying efficiency of proteinase K digestion due to the fact that the blocks were prepared from such large numbers of cells.12 To address this possibility, an additional pooled set of CD14+ cells was prepared from seropositive individuals similar in average age, gender, and serostatus as those of the first pooled set. The cells were prepared in agarose blocks at the same density (2.5 × 106 cells each) as in the first two sets, but only 10% of the proteinase K was used in the digestion step. As in previous gels, two agarose cell blocks (total of 5 × 106 cells) were loaded per lane. The results (panel D) show the probe-positive signal was detected at the slowest observed mobility, core 4 as compared with that of the circular megaplasmid (core 8) and both previous CMV sample sets, but still in the region of the gel where a circular form would be expected to migrate. Only primary amplification was undertaken in this analysis (panel D), because it had previously been established that a single round of PCR was sufficient to amplify viral DNA in these samples. These results suggest that incomplete proteolysis may contribute to reduced mobility of circular CMV DNA, although these differences in migration may also have other explanations, such as variation in the electrophoretic conditions. Taken together, all three pooled sets of CD14+cells showed probe-positive signal from cores in the region where a circular form would migrate. Given the fact that these gels separate primarily on the basis of structure,12 18 the slight mobility differences do not detract from the overall conclusion.
DISCUSSION
Using a native agarose gel electrophoresis system in conjunction with PCR amplification, we were able to show that CMV genome in CD14+ cells of healthy seropositive adult carriers is detected only as a circular form without evidence of linear or integrated CMV genome. The detection of circular CMV in the CD14+ cell population from PB of healthy hosts indicates that this is the form in which the latent viral genome persists in this population of cells. Because the predicted viral DNA copy number present in healthy subjects lies below the detection limits of direct blot hybridization analysis,8 PCR amplification was used as a means to extend the sensitivity of the detection limits. In our results, the circular form of CMV DNA was consistently detected after only one round of initial PCR amplification. Viral DNA was not detected in the linear region of the gels or in the well origins in any of these analyses, despite the fact that nested PCR conditions and DNA blot hybridization analyses were used in a majority of analyses.
Viral DNA from productive or persistent infections is associated with both large complex structures, such as concatemers or branched forms, as well as linear forms of the viral genome, the products of concatemers cleaved into unit-length linear viral DNA during the packaging and assembly process. In the type of gel analysis used here, branched, highly structured or very large DNA forms are retained at the well origin of the gel, while linear forms of ≈50 to 700 kb migrate with a mobility similar to linear CMV DNA.12,24 25 Both large complex and/or concatemeric viral DNA and unit-length linear forms were detected at the well origin and in the linear region of the gel, respectively, for high copy and low copy CMV productively infected cell controls (Fig 1 and Fig 2, respectively). These linear, large concatemeric or integrated viral DNA do not migrate in the same region of the gel with circular forms. If other forms exist in the viral DNA reservoir during latency, the copy number was below our detection limits.
Studies in healthy carriers have shown the presence of CMV DNA throughout the myeloid lineage.8,26,27 Viral DNA has been detected in CD34+ cell populations that include early bone marrow hematopoietic progenitors27 and CMV DNA and RNA from bone marrow myeloid-committed progenitors coexpressing CD33/15.26 In addition, CD14+ cells from PB, but generally not CD14− cell types in PB, are the predominant site of CMV viral genome.9,21 CMV virus can also be reactivated from CD14+ PB cells from healthy subjects after allogeneic stimulation and prolonged in vitro culture.7 Our results have extended these observations and indicate that latent viral DNA is maintained in CD14+ PB cells as an extrachromosomal circular plasmid.
Finally, a circular plasmid conformation for the CMV genome is consistent with that of other latent herpesviral genomes, and supports studies suggesting the CMV genome is harbored in a latent state in immature and more differentiated myeloid cells.26-29 Future studies should be undertaken to extend the analyses of the CMV genome conformation to include these progressively earlier stages of the myeloid cell lineage.
Taken together, these results are consistent with the proposed model that the latent CMV genome resides in early CD34+ and CD33+ progenitor cells in the bone marrow, and is partitioned to the monocyte or dendritic lineage, where it can be detected in CD14+ cells of the PB.30 In support of this, we have reported the first successful characterization of the CMV genome conformation in CD14+ cells from normal CMV seropositive individuals and demonstrated that the genome persists as a circular plasmid comigrating with a 230-kb circular megaplasmid marker.
ACKNOWLEDGMENT
We thank Mike McVoy, Lenore Pereira, Lucy Rasmussen, and Steve St. Jeor for their critical review of this manuscript.
Supported by National Institutes of Health grants awarded to J.A.W. (K08 AI01193) and E.S.M. (RO1 AI33852). J.A.W. is a recipient of UC Davis Hibbard E. Williams Research Funds and UC Davis Medical Center Children’s Miracle Network Telethon Research Funds.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jean A. Wiedeman, Pediatric Infectious Diseases, Neurosciences Bldg, 1515 Newton Ct, Room 600, Davis, CA, 95616; e-mail: jataylorwiedeman@ucdavis.edu.