Abstract
High incidences of graft failure and graft-versus-host disease in the recipients of bone marrow transplantations (BMT) from unrelated donors (URD) may reflect the existence of allelic disparities between the patient and the URD despite apparent HLA identity at HLA-A, HLA-B, and HLA-DRB1 loci. To identify the extent and pattern of allelic disparities at HLA-A and HLA-B loci, 128 patients and 484 potential URD were evaluated by DNA typing. DNA typing for HLA-A, HLA-B, and HLA-DRB1 was performed at Memorial Sloan Kettering Cancer Center. HLA-A and HLA-B serotyping on URD was provided by the registries. By original typing (serology for HLA-A and HLA-B; DNA typing for DRB1) 187, 164, and 133 URD were 6/6, 5/6, and 4/6 matches, respectively. Following DNA typing, however, only 52.9% of the originally 6/6 matched URD remained 6/6, while 38.5%, 7.5%, and 1.1% were found to be 5/6, 4/6, and 3/6 matches. The level of disparity was higher in the originally 5/6 (P< .01) and 4/6 (P < .01) matched URD. A higher level of disparity was seen for HLA-B as compared to HLA-A. In addition, a serotype related variation was also noticed. For example, 24.1% of HLA-A2 and 60.1% of HLA-B35 seromatched URD were genotypically disparate, but no disparities were seen for HLA-A1 and HLA-B8. A higher percentage of HLA-A (67.4%) compared with HLA-B (35.4%) serologic homozygous URD remained genotypically homozygous (P = .01). The level of allelic disparity was lower (P < .01 for 6/6; P = .02 for 5/6) if the patient had one of the 15 most common haplotypes (A1B8DR3, A2B7DR15, A3B7DR15, etc) in comparison to the rest of the group. Outcome studies will answer the question whether these disparities are associated with a higher rate of immunological complications seen with URD-BMT.
A SUITABLE HLA-MATCHED donor may not be available within the immediate or extended family for as many as 70% of patients who could benefit from a bone marrow transplantation (BMT).1,2 For these patients an unrelated donor (URD) offers an alternate source of stem cells (marrow, cord blood, or peripheral blood stem cells). A steady increase in the number of URD-BMT in the last few years3,4-7 would not have been possible without the tremendous expansion of many national and international bone marrow donor registries, like the National Marrow Donor Program (NMDP), that identify suitable URD.3,4,8-10Unfortunately, serious immunological complications including graft-versus-host disease (GvHD), graft rejection, and delayed immune recovery are far more severe and frequent following URD-BMT.3,10-13 These complications may be the result of genetic disparities between patients and their URD despite histocompatibility matching by conventional typing. Most transplantation centers use serology to type for HLA-A and HLA-B and DNA typing for class II HLA loci.14,15 Siblings and other close family members that match by serology are very likely to be genotypic matches because the whole major histocompatibility complex (MHC) segregates as a single haplotype. In contrast, unrelated individuals with the same serotypes may have different alleles. Serology cannot resolve all alleles, so 83 HLA-A and 186 HLA-B alleles resolve into only 28 HLA-A and 59 HLA-B serotypes.16Furthermore, some serotypes are exceptionally polymorphic. For example, A2 and B35 represents 18 and 20 different alleles, respectively.16 Following the development of DNA typing for HLA class II loci a few years ago, analysis of URD-patient pairs revealed a significant frequency of disparities between patients and serologically HLA-DR matched URD.17 Furthermore, it has been shown that allelic matching for class II loci offers outcome advantages compared to phenotypic matching.18-20 We have recently developed and successfully shown the feasibility of polymerase chain reaction-sequence specific oligonucleotide probe (PCR-SSOP) typing for class I alleles.21
Genotypic identity for HLA-A and HLA-B (as is the case for HLA-DRB1) between the patient and URD can be achieved if typing is performed by this DNA-based molecular method rather than the traditionally used serology. In view of the marked polymorphism of HLA-A and HLA-B loci, it can be expected that URD-patient pairs matched by serology may often be mismatched genotypically. We would also expect that HLA matching of donor-recipient pairs using molecular typing for HLA-A and HLA-B would potentially improve the outcome of URD-BMT. Therefore, to assess the usefulness of HLA-A and HLA-B DNA typing for URD-BMT, it is imperative that precise determination of the existence and the nature of genetic disparities between serologically matched URD-patient pairs be made. Allelic disparities have been detected following oligotyping for HLA-A2, HLA-A3, and HLA-B44 between patients and their seromatched URD.22 Direct DNA sequencing in another study identified 3 allelic mismatches in 9 HLA-A and HLA-B seromatched URD.23In the present study, we have performed DNA typing by PCR-SSOP for HLA-A and HLA-B alleles on a large number of patients and their URD identified by marrow donor registries. These cohorts include individuals expressing a wide range of HLA serotypes. Our aim in this study was to (1) identify HLA-A and HLA-B allelic disparities in patient-URD pairs matched by serology for HLA-A and HLA-B and by DNA typing for HLA-DRB1; (2) analyze the pattern of allelic disparities for different HLA-A and HLA-B serotypes; (3) analyze any locus-specific differences; (4) determine the effect of DNA typing on serologic “blanks”; (5) determine the influence of the racial/ethnic background of the patient on HLA-A and HLA-B allelic disparity; and (6) determine if the subgroup of patient-URD pairs containing one of the common haplotypes show a different degree of allelic disparity compared with the rest of the group.
MATERIALS AND METHODS
Samples.
The study was conducted between January 1996 and July 1997 at Memorial Sloan Kettering Cancer Center (MSKCC). Of all the potential URD identified for 128 patients, 484 were found to be either a 4/6, 5/6, or 6/6 match by HLA-A and HLA-B serology and HLA-DRB1. The sources of these URD were the NMDP (n = 462), other national and international registries (n = 19), and the New York Blood Center Cord Blood Registry (n = 3). Eighty-two of 128 patients (Pts) were self-reported as Caucasians. A total of 298 URD (ptCAUC) were selected for these 82 patients. The remaining 186 URD (ptNonCAUC) were selected for 46 non-Caucasian (Hispanic, 18; African American, 12, Asian, 11; others, 5) patients.
Typing technique.
The DNA typing for HLA-A, HLA-B, and HLA-DRB1 for the patients and URD was done at MSKCC. The serological typing on the URD was provided by the registries. Our pilot studies to evaluate the need for reserotyping all samples from URD indicated that this expensive and time-consuming testing was unnecessary. The level of discrepancy was very low. Those URD who matched for “split” antigens as defined by the World Health Organization (WHO) Nomenclature Committee24 were considered serological matches. The DNA typing for HLA-A and HLA-B was performed by DNA-based PCR-SSOP technique as previously described,21,25 and HLA-DRB1 typing was performed according to the well established protocol.26 Alleles were assigned as per the DNA sequence published by the “Nomenclature for the Factors of HLA system” committee.24 The nucleated cells from the blood samples were collected, and genomic DNA was isolated. Locus-specific PCR amplification of the full length of exons 2 and 3 of the genomic DNA was performed using primers derived from introns 1 and 3 flanking the polymorphic exons 2 and 3. The PCR product was applied to a positively charged nylon membrane (Boehringer Mannheim, Mannheim, Germany), and the DNA was cross-linked by exposing the membrane to ultraviolet light in a Stratalinker 2400 (Stratagene, La Jolla, CA). The membranes were hybridized with digoxigenin-ddUTP–labeled SSOP. The membranes were washed and treated with anti–digoxigenin-Fab antibody conjugated to alkaline phosphatase (Boehringer Mannheim). Subsequently, they were treated with Lumiphos 480 (Boehringer Mannheim) and exposed to Kodak XAR (Eastman Kodak, Rochester, NY) photographic film. A panel of 44 and 52 probes designed from the sequences of exons 2 and 3 for the HLA-A and HLA-B loci were used, respectively. A locally developed computer program analyzed the hybridization pattern for each sample and assigned alleles. The results were checked and confirmed manually. Heterozygotes were identified by the presence of two different hybridization score patterns.
Data analysis and statistics.
URD were stratified according to the degree of HLA-A, HLA-B serology and HLA-DRB1 matching into 6/6, 5/6, and 4/6 subgroups. Allelic disparity was graded according to the total number of new mismatches revealed following DNA typing. In a small percentage of cases (7.5%), SSOP typing could not resolve the allelic identity down to a single allele. For most of these samples the identity was restricted to two possible alleles, eg, A*0201/0206. If the URD typed as above and the patient was A*0202, the two were considered to be allelic mismatches. All tests of no association between categorical variables including the frequency of genotypic disparity for various serotypes and ptCAUC versus ptNonCAUC URD were computed using Fisher’s exact test. The relationship between the frequency of disparities within each serotype and the number of alleles in that serotype was analyzed for HLA-A and HLA-B locus using the rank correlation coefficient, Kendall’s tau.27
RESULTS
URD and patient characteristics: Original typing.
A total of 484 URD for 128 patients were identified. Nineteen HLA-A and 34 HLA-B serotypes were observed in the patient population. HLA-A2 and HLA-B35 were the most common serotypes. Phenotype frequencies of HLA-A and HLA-B serogroups in the patient population and the gene frequency of alleles within each serogroup are presented in Tables 1 and 2. By original typing (serology for HLA-A and HLA-B; DNA typing for HLA-DRB1), 187 URD were found to be 6/6 match; 164 were 5/6 match; and 133 were 4/6 match. The URD in the 4/6 group were selected as 5/6 or 6/6 matches by HLA-A, HLA-B, and HLA-DR serology and were found to be 4/6 match following HLA-DRB1 DNA typing. The details of the matching status are presented in Table 3.
URD matching status following DNA typing.
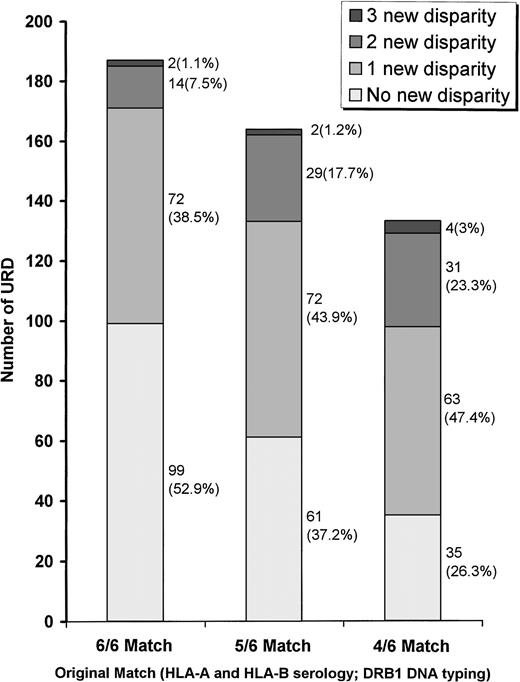
Following PCR-SSOP, only 99 of 187 (52.9%) originally 6/6 matched URD remained 6/6 identical (Fig 1). One disparity was identified in 72 of 187 6/6 URD (38.5%), and therefore their matching status was downgraded to a 5/6. Fourteen (7.5%) 6/6 URD were downgraded to a 4/6, and 2 (1.1%) to a 3/6 match status. In comparison to the 6/6 group, the degree of disparity identified by PCR-SSOP in the originally 5/6 and 4/6 matched pairs was more pronounced (P < .01 and P < .01, respectively). No difference was found between the 5/6 and the 4/6 groups. In the originally 5/6 match group, only 61 (37.2%) URD remained 5/6 matches. One new disparity in 72 (43.9%), two new disparities in 29 (17.7%), and three new disparities in 2 (1.1%) URD were identified resulting in a matching status downgrade to a 4/6, 3/6, and 2/6, respectively. From the original 4/6 group only 35 (26.3%) remained 4/6 matches.
Level of HLA-A and HLA-B genotypic mismatching identified following DNA typing for the URD in the 6/6, 5/6, and 4/6 original match subgroups. In comparison to the original 6/6 group, degree of genotypic mismatching was significantly higher for the 5/6 (P< .005) and the 4/6 (P < .001) group. There was no statistical difference between 4/6 and 5/6 original subgroups. The number of URD in each category is shown next to the column with percentage values in the parenthesis.
Level of HLA-A and HLA-B genotypic mismatching identified following DNA typing for the URD in the 6/6, 5/6, and 4/6 original match subgroups. In comparison to the original 6/6 group, degree of genotypic mismatching was significantly higher for the 5/6 (P< .005) and the 4/6 (P < .001) group. There was no statistical difference between 4/6 and 5/6 original subgroups. The number of URD in each category is shown next to the column with percentage values in the parenthesis.
Locus- and serotype-specific variations.
The degree of serological matching at HLA-A was compared with that at HLA-B loci and was not found to be different (Table 3) in either the subgroups of URD (6/6, no HLA-A or HLA-B disparity; 5/6, P = .76; and 4/6, P = .90) or the whole URD group (P > .90). However, following DNA typing a higher number of disparities were detected for HLA-B in comparison to that for HLA-A locus (P = .01) in the whole group of 484 URD (Table 4). A similar difference was seen in the original 6/6 match subgroup (P = .01). However, the difference in the level of HLA-A and HLA-B genotypic disparities in the originally 4/6 (P = .06) was less pronounced. No difference was noted in the 5/6 subgroup.
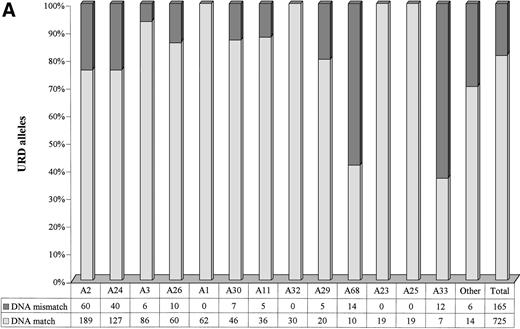
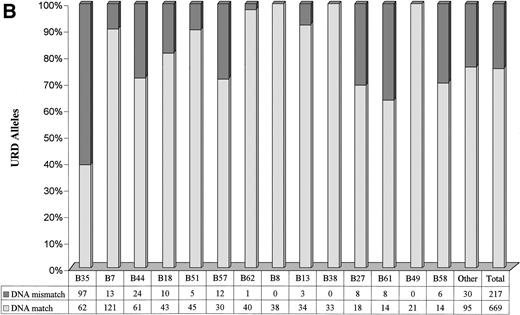
A marked variation in the level of genotypic disparity was identified for specific HLA-A and HLA-B serotypes (Fig 2A and B). For example, 24.1% of donors seromatched for HLA-A2 and 60.1% of donors seromatched for HLA-B35 (the most common serotypes in the study population) were found to be genotypic mismatches. Genotypic disparities within A11, A24, B58, A29, B44, B57, B27, and B61 serotypes were found to be between 21% to 37%. A much higher proportion of A68 (58.33%) and A33 (63.16%) seromatches were genotypic mismatches. No genotypic disparities were identified by PCR-SSOP for A1, A23, A25, A32, B8, B38, and B49 serotypes. The alleles observed within each HLA-A and HLA-B serotypes in the URD population are presented in Tables 5 and 6. It is of note that only 1.86% of the URD had a typing result outside of the serotype either at HLA-A or HLA-B locus following DNA typing. For example, only 3 out of 274 HLA-A2 serotypes typed as alleles (Table5) that did not belong to A2 serotypes (A*3201, A*6802, and A*7401). Similarly, only 2 out of 138 HLA-B7 serotypes typed as alleles (Table6) that did not belong to B7 serotypes (B*1801 and B*8101).
Level of genotypic disparities identified within each HLA-A (A) and HLA-B (B) serotype. Each column represents the number of times a particular serotypes was matched between patients and their URD. The dark segment represents the genotypic or DNA mismatch within each serotype and the light segment represents genotypic matches. All the columns have been scaled to 100% and the data table along the x-axis shows the actual occurrences. The columns have been positioned from the left to the right in a decreasing order of prevalence of the serotypes in the URD group. Marked variation in the level of genotypic disparities between patient and sero-matched URD for different serotypes is apparent.
Level of genotypic disparities identified within each HLA-A (A) and HLA-B (B) serotype. Each column represents the number of times a particular serotypes was matched between patients and their URD. The dark segment represents the genotypic or DNA mismatch within each serotype and the light segment represents genotypic matches. All the columns have been scaled to 100% and the data table along the x-axis shows the actual occurrences. The columns have been positioned from the left to the right in a decreasing order of prevalence of the serotypes in the URD group. Marked variation in the level of genotypic disparities between patient and sero-matched URD for different serotypes is apparent.
The effect of DNA typing on serologically “blank” patients and URD.
By serological typing, one “blank” allele at the HLA-A and HLA-B loci was found in 18 (14.06%) and 7 (5.47%) patients, respectively (Table 7). These patients were confirmed to be homozygous for the expressing allele by family studies. Following PCR-SSOP typing, 10 of 18 HLA-A and 3 of 7 HLA-B serologic homozygous patients remained genotypically homozygous. Similarly, 44 of 68 (64.7%) HLA-A and 17 of 48 (35.4%) HLA-B serologic homozygous URD remained genotypically homozygous. Compared to HLA-A, a higher proportion of the HLA-B serologic homozygotes (P = .01) were detected to be genotypically heterozygotes by PCR-SSOP. There was wide variation in the percentage of genotypic homozygosity among those serologically homozygous for specific HLA-A and HLA-B serotypes. Although 66% of serological blanks within HLA-A2 and 100% within HLA-B18 were genotypically homozygous, only 18.7% of HLA-B35 were genotypically homozygous.
The influence of common haplotypes.
Haplotypes could be assigned for 109 of 128 patients on the basis of family studies. Out of 484 URD, 407 had been selected for these 109 patients. Thirty-five patients had one of the 15 most common haplotypes28 (A1B8DR3, A24B35DR11, A29B44DR7, A2B18DR11, A24B7DR15, A30B13DR7, A2B7DR11, A2B7DR15, A3B7DR15, A26B35DR11, A26B38DR4, A2B15DR4, A2B44DR4, A2B57DR7, and A3B35DR1). None of the patients had two of the above haplotypes. URD (n = 161) who were selected for these 35 patients comprised the “CH (common haplotype)” subgroup, and the URD (n = 246) identified for the rest of the patients (n = 74) comprised the “NoCH” group. One or more new allelic disparities were seen in a lower percentage of donor-recipient pairs in the “CH” group in comparison to the “NoCH” group within the 6/6 (33.8% v 56.5%; P <.01) and the 5/6 (53.6% v 65.9%; P = .02) subgroups. The numbers of 4/6 “CH” donors were too small to analyze.
Effect of patient ethnicity/race.
Of 298 “ptCAUC” URD, 146, 92, and 60 were 6/6, 5/6, and 4/6 matches, respectively, by original typing. Following DNA typing within the “ptCAUC” group, one or more allelic disparities were found in 62 (43.8%) of 6/6 matches, 54 (57.7%) of 5/6 matches, and 41 (68.3%) of 4/6 URD. Of 186 “ptNonCAUC” URD, 41, 72, and 73 were 6/6, 5/6, and 4/6 matches, respectively, by original typing. Within the “ptNonCAUC” group, one or more allelic disparities were found in 24 (58.5%) of 6/6 matches, 49 (68.1%) of 5/6 matches, and 57 (78.1%) of 4/6 URD. Overall, a higher percentage of “ptNonCAUC” compared to “ptCAUC” URD (69.9% v 53.4%; P < .01) were found to have one or more allelic disparity following DNA typing. The differences between “ptCAUC” and “ptNonCAUC” URD were not found to be significant when the 6/6 (P = .11), 5/6 (P= .26), and 4/6 (P = .24) matching categories were analyzed separately. The effect of race/ethnicity of patient on the frequency and identity of mismatched alleles within each serotype was examined. The frequency of allelic disparity was higher in the “ptNonCAUC” compared to the “ptCAUC” URD within HLA-A2 (P = .01), A24 (P < .01), A26 (P < .01), A68 (P = .05), B51 (P = .01), and B52 (P = .01) serotypes (data not shown). For all other HLA-A and HLA-B serotypes no difference was found.
DISCUSSION
The outcome of bone marrow transplants, particularly those involving URD, is dependant on optimizing the histocompatibility matching. It is well known, however, that antigen-based typing (serology and Isoelectric focusing) cannot detect all alleles. Studies conducted following the development of DNA typing techniques for HLA class II loci have revealed that many URD matched by HLA-DR serology are in fact genotypic mismatches.17-20 Furthermore, HLA-DRB1 disparities detected by DNA techniques have been shown to correlate with clinical outcome following URD-BMT.18 Similar analyses for class I loci have yet to be performed due to the delay in the development of DNA typing methods. The class I region contains numerous nonclassical genes and pseudogenes as well as complex nucleotide substitutions and polymorphisms of nucleotide sequences across 2 exons.21,29 We have developed and tested SSOP typing for class I alleles recently.21 The present study was undertaken to investigate the degree and the pattern of HLA-A and HLA-B genotypic disparity between URD and patient that had been matched by HLA-A and HLA-B serology and HLA-DRB1. Our results clearly show that HLA-A and HLA-B genotypic disparities exist within URD-patient pairs who have been matched for HLA-A and HLA-B by serology and that these disparities can be easily detected by the use of DNA typing for URD selection. In view of the presence of allelic disparities in 47.1% of 6/6 and in an even higher proportion of 5/6 and 4/6 URD, it is apparent that serotyping for HLA-A and HLA-B may not be optimal for identifying histocompatible URD. Such a finding, while significant, is not surprising in view of the marked genotypic polymorphism for both HLA-A and HLA-B and the limited capacity of serology to distinguish different alleles. As many as 186 HLA-B and 83 HLA-A alleles are represented by 59 HLA-B and 28 HLA-A serotypes, a ratio of 3:1. When the originally 6/6 and 5/6 matched groups were analyzed together (n = 351), only 28.21% (n = 99) and 37.89% (n = 133) remained 6/6 and 5/6 genotypic matches, respectively. Additionally, approximately 7%, 21%, and 48% of the originally 6/6, 5/6, and 4/6 URD, respectively, were found to be disparate at HLA-A as well as HLA-B loci. It is widely believed that registry-selected URD for Caucasian patients are more likely to be histocompatible at the allelic level. Our analysis of the patient’s ethnic/racial background on the level of disparity revealed two important points. The first observation that the level of allelic disparities was higher if patients were non-Caucasian rather than Caucasian is largely expected. However, a significant level of allelic disparity existed even for Caucasian patients. As many as 43.8% of 6/6 URD for Caucasian patients showed one or more allelic disparity. It is possible that genetic disparity within the “ptCAUC” URD is a reflection of the wide ethnogeographic origins of the Caucasian population of the United States. However, there may be bias arising from voluntary and self-reported racial designations. Furthermore, HLA assignment by serology qualified by ethnogeographic background would likely be of limited additional value in the context of URD selection from large computerized databases like the NMDP.
Our results are in accordance with the preliminary reports of HLA class I genotypic disparities between seromatched patients and their URD, but our estimates of the level of mismatching exceed what has been described so far. In a recent study that used limited HLA-B molecular typing, disparities were observed in about a quarter of 61 seromatched URD.30 However, some of these URD may also have been mismatched at the DRB1 locus, because the initial selection criteria included serotyping for HLA-A, HLA-B, and HLA-DR. Allelic disparities were also detected in a small study between patients and their seromatched URD following oligotyping for HLA-A2, A3, and B44.22 Direct DNA sequencing in another study identified 3 allelic mismatches in 9 HLA-A and HLA-B seromatched URD.23A lower degree of HLA-A and HLA-B allelic disparity was identified in patients and HLA-A, HLA-B, and HLA-DR seromatched kidney donors.31 In our study a significantly higher frequency of genetic disparity was identified for HLA-B (38.5%) compared with the HLA-A (18.2%) alleles in the originally 6/6 matched URD, a difference likely to reflect the higher degree of polymorphism for HLA-B alleles. Furthermore, many patients and URD that were perceived to be homozygous by serology were in fact found to be allelic heterozygotes. This has implications for the calculation of vectors for GvHD or graft rejection. The vector for GvHD will increase if a homozygous patient is found to be heterozygous. Conversely the vector for rejection will increase if the homozygous URD is a heterozygote. The above observations are particularly important for common serotypes like HLA-B35 (81.3% serohomozygotes were allelic heterozygotes) and HLA-A2 (34% serohomozygotes were allelic heterozygotes). As observed by our group with IEF typing, a higher percentage of HLA-B homozygotes compared with HLA-A homozygotes were found to be genotypically disparate.32 Our results are comparable to a previous report of 25% incidence of allelic heterozygosity in 40 individuals who were homozygous for HLA-B by serology.33
A marked variation in the frequency of allelic disparities was observed for different serotypes. For example, 60.1% of seromatches for HLA-B35 (the most polymorphic HLA-B serotype with 20 known alleles) were allelic disparates. A high frequency of allelic disparity (28%) was also detected for B44, another common HLA-B serotype. Conversely, no allelic disparities were identified within B8, B38, and B49. In the case of B49, lack of disparity is expected because it contains only one allele. Analogous to HLA-B, a marked variation in genetic disparity was also observed for HLA-A serotypes. Disparity was identified for a quarter of URD seromatched for HLA-A2 and HLA-A24, the two most common HLA-A serotypes. In addition, as many as 58.3% of A68 and 63.2% of A33 seromatches were genetically disparate. Fortunately, A68 and A33 are uncommon. A similar variation in the degree of discrepancy between serologic and allelic assignment for HLA-A was recently been described.34 Lack of a monotone relationship (ie, as the number of alleles within a serotype increases, the level of allelic disparity within that serotype also increases) between the number of alleles and the degree of allelic disparity for each HLA-B serotype (P = .4) in this study may be due to the fact that many WHO-defined alleles are rare, are seen only in isolated populations, and are not represented in our study subjects. However, for HLA-A locus that has a significantly lower number of alleles, there is some evidence (P = .07) of this relationship. The level of disparity within 6/6, 5/6, and 4/6 groups as well as the frequency of disparity within each serotype remained similar if the analysis was limited to only one randomly selected URD per patient.
In a small proportion of cases, HLA-A (1.86%) as well as HLA-B (1.86%) alleles of a different serotype were identified following DNA typing. These observations support the results of our pilot studies to evaluate the need for reserotyping all samples from URD. Those studies had indicated that this expensive and time-consuming testing was unnecessary. Most of the discrepancies in the present study were seen between closely related “split” serotypes. For example, one B52 typed as a B*5301, a B53 typed as a B*5201, and an A30 typed as A*3101. A slightly higher incidence of alleles of a different serotype was seen in a study of 69 URD.35 In 11 of 60 samples in another study, HLA-B alleles different from the serotype were found. According to the authors of the preceding study, their sample had a higher representation of serotypes that are common in ethnic minorities and are known for serotyping difficulties.36
The patients with common haplotypes are more likely to find URD from the registries. Our analysis show that patients with 1 of the 15 most common haplotypes in the US population28 have another advantage. The URD who have been selected for these patients by HLA-A and HLA-B serology are less likely to have allelic disparities. The A1B8DR3 haplotype is additionally favorable because no allelic disparities were identified for either A1 or B8. Therefore, an URD identified by HLA-A and -B serology for a patient carrying A1B8DR3 is very likely to be matched for HLA-A and HLA-B alleles. Furthermore, in view of a very tight linkage association between B*0801 and Cw*0701,37 they are also likely to match for HLA-C. Unfortunately, despite being the commonest, A1B8DR3 has a haplotype frequency of only 6.6% in the United States.28
It is conceivable that a high incidence of genetic disparities at HLA-A and HLA-B loci in seromatched URD-patient pairs as shown in this study may contribute to the high incidence of immunological complications including GvHD and graft rejection in recipients of URD marrow. It has previously been reported that even a single amino acid difference in HLA-B between patient and URD can have serious implications for the graft survival38 as well as GvHD.39 Class I mismatching by serology has previously been shown to result in a poorer outcome.40 Recently, mismatching by Isoelectric focusing was also shown to increase the risk of GvHD and transplantation-related mortality.41 Based on the pivotal role played by HLA-A and HLA-B in the generation of alloreaction and immune responses, it can be hypothesized that these genetic disparities will be clinically significant in the context of URD-BMT. The class I gene products are expressed on the surface of almost all nucleated cells and hold foreign peptides in a groove formed by their α1 and α2 domains to present them to the T-cell receptors (TCR) on the CD8+ cytotoxic T-lymphocytes, the cells that are critical mediators of GvHD and graft rejection.39 Novel in vitro studies to identify specific allelic mismatches with a high alloreactive potential may enhance our ability to improve the outcome for the recipients of URD marrow grafts. In addition, future prospective studies as well as retrospective outcome analyses will determine if the use of molecular typing to define HLA-A and HLA-B identity offers improved survival and decreased complication rate following URD-BMT. Similar analyses to understand the role played by HLA loci other than HLA-A, HLA-B, and HLA-DRB1 as well as by minor histocompatibility loci will also be rewarding.
ACKNOWLEDGMENT
The authors thank Carol Landrey for immense help with sample and serology data collection on the URD. We thank the staff members of the histocompatibility laboratory and deeply appreciate their technical support. The authors thank Frank Lewis of Information Service at MSKCC for his help with ethnicity/race data collection on the patients.
Supported by Grant No. P01 CA23766 from the National Cancer Institute, National Institutes of Health (N.A.K, G.H., R.J.O. and S.Y.Y.), Butler Foundation Inc (N.A.K.), Vincent Astor Chair in Clinical Research (R.J.O.), and Navy Medical Research Grant N00014-97-1-1044 (S.Y.Y.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Vinod K. Prasad, MBBS, MD, MRCP (London), Box 379, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10021; e-mail: Prasadv@mskcc.org.