The t(11;19)(q23;p13.1) translocation is frequently found in adult myeloid leukemia. In the MLL/MEN fusion protein generated by this translocation, most of the coding region of the MEN protein, an RNA polymerase II elongation factor, is fused to the N-terminal third of the MLL protein, a possible transcriptional regulator. However, the molecular mechanism of leukemogenesis by the fusion protein remains unclear. We investigated the effects of the fusion protein on p53 function using luciferase assays. Overexpression of the fusion protein suppressed the transactivation ability of p53. This negative effect of the fusion protein on p53 function was dependent on the region derived from MEN. Moreover, p53 coimmunoprecipitated with MLL/MEN as well as MEN, suggesting that the fusion protein binds to p53 through the MEN region. We found that MEN binding to p53 was mediated by its N-terminal region and repression of p53 transcriptional activity was mediated by its C-terminal region. We also found that these two functional regions were essential for the transformation of Rat1 cells mediated by MEN. Although we could not demonstrate a functional difference between MLL/MEN and MEN in this study, these data suggest that the MLL/MEN chimeric transcriptional regulator may exert its oncogenic activity by inhibiting the function of the p53 tumor-suppressor protein by binding to it. Our findings provide a novel insight into the leukemogenic mechanism exerted by the t(11;19)(q23;p13.1) translocation.
THE 11q23 TRANSLOCATIONS, in which the breakpoints of chromosome 11 are commonly mapped to band q23, are the most frequently observed chromosomal abnormalities in human leukemia.1,2 The MLL gene is located at the breakpoint of 11q23 and is rearranged by the translocations. The MLL gene product, although not yet fully clarified, contains AT-hooks in its N-terminal portion and two zinc finger domains in its middle3,4 and appears to encode a putative transcriptional regulator. Upon chromosomal translocation, the MLL gene is disrupted between the AT-hooks and the zinc finger domains and fused with various partner genes. These partner genes encode various kinds of proteins with known structures,2 including serine/proline rich sequences,5 zinc finger motifs, leucine zipper motifs6,7 found in transcriptional factors, and a Src homology 3 module8 found in signaling molecules. The resulting fusion proteins appear to differ functionally. Among the 16 genes identified as partners for MLL in the 11q23 translocations, the functions of only six are known. These six genes are MEN/ELL (RNA polymerase II elongation factor) at 19p13.1,9-11 CBP at 16p13.3,12 p300 at 22q13,13 AF6 (Ras-binding protein) at 6q27,14,15 hCDCrel (cell division cycle protein) at 22q11.2,16 and ABI-1 (Abl binding protein) at 10p11.2.17 However, the molecular mechanisms underlying leukemogenesis by these chimeric genes remain unclear.
We have cloned the MLL/MEN chimeric cDNAs generated by the t(11;19)(q23;p13.1),9 which is specifically found in adult myeloid leukemia. This fusion protein consists of N-terminal 1406 amino acids (aa) of MLL, followed by the entire coding region of MEN except for its N-terminal 45 amino acids. MEN is an RNA polymerase II elongation factor that enhances elongation by suppressing transient pausing by polymerase at many sites along DNA.11 MEN, as well as ELL2, which shares significant homology with MEN, exhibit the elongation enhancing activity conferred by their N-terminal regions.18,19 MEN also contains an RNA polymerase II interaction domain in its most N-terminal area. This region is capable of negatively regulating the polymerase activity in promoter-specific transcription initiation.10,20 The MLL/MEN fusion protein, in which this most N-terminal domain of MEN has been lost, may lead to overactivation of elongation by failing to inhibit initiation.18 Therefore, loss of the region for RNA polymerase II interaction could be a cause for the leukemogenesis induced by the MLL/MEN fusion protein. In addition, overexpression of the MEN protein in Rat1 cells leads to increased colony formation.21 MEN also stimulates AP-1 activity by increasing Fos expression, and these two functions are lost when MEN lacks the lysine-rich region in its C-terminal portion. Therefore, MEN may possess an oncogenic function by enhancing AP-1 activity. The MLL/MEN fusion protein contains most of the coding region of MEN and therefore may induce strong AP-1 activity. If the AP-1 activity induced by the fusion protein is stronger than that induced by the MEN protein, it could lead to malignant transformation of myeloid cells.
The p53 tumor-suppressor gene is one of the key genes involved in human malignancies.22 Mutations and/or allelic losses in the gene have been found at high frequencies in acute leukemia,23-25chronic myelocytic leukemia in blastic crisis,26,27 and myelodysplastic syndrome.28,29 p53 acts not only as a transcriptional activator of genes containing a p53 binding site in their regulatory regions,30 but also as a repressor of genes containing a TATA box but lacking a p53-binding site.31 p53 regulates cell cycle progression by modulating the expression of genes such as p21 (WAF-1). p53 is also involved in apoptotic induction after DNA damage, possibly by transactivating Bax,32 a proapoptotic member of the bcl-2 family proteins.
To analyze the effects of the MLL/MEN fusion protein on cell cycle regulation, we investigated the effect of this chimeric protein on p53 function. We found an inhibitory effect of MLL/MEN on the transactivating ability of p53. This inhibitory effect appears to be mediated by direct interaction between MLL/MEN and p53. We found that MEN binds to p53 through its N-terminal p53 binding domain.
In addition, we found that MEN alone represses p53-mediated transcription almost to the same level as MLL/MEN. This function is mediated through both its N-terminal p53 binding domain and its C-terminal repression domain. These two functional domains of MEN are necessary for the increased colony formation of Rat1 cells by the induced MEN protein. Our results provide another interesting model for leukemic transformation of hematopoietic cells by the abnormal expression of a transcriptional elongation factor.
MATERIALS AND METHODS
Plasmid construction.
We previously constructed the pME18S-MEN(HA), the pME18S-MLL/MEN(HA), and the pME18S-tMLL(HA) plasmids, which contain the entire MEN and MLL/MEN chimeric cDNAs, and the N-terminal MLL part of MLL/MEN chimeric cDNA (truncated MLL [tMLL]), respectively, followed by the sequence coding for nine amino acids of the influenza hemagglutinin (HA) epitope (TACCCATACGACGTCCCAGACTACGCT) in their C-terminals.20 In this study, the pME18S-MEN(FLAG) and pME18S-MLL/MEN(FLAG) plasmids were generated by replacing the coding sequence of the HA epitope with that of the FLAG epitope (GACTACAAGGACGACGATGACAAG). To construct the pME18S-GAL4/VP16, a DNA fragment containing the coding region of GAL4/VP16, in which the yeast GAL4 DNA binding domain (1-147 aa) was followed by a part of the herpesvirus transcriptional activator VP16 (413-490 aa), generated by polymerase chain reaction (PCR) and inserted into theEcoRI-Spe I sites of the pME18S expression vector. A series of MEN deletion mutants named pME18S-dMEN1-7(HA) were constructed by digesting the pME18S-MEN(HA) plasmid with appropriate restriction enzymes, blunting with either T4 DNA polymerase or Klenow fragment, and religating. The EcoRI fragments from these constructs were cloned in the sense orientation into the EcoRI site downstream of the 5′ long terminal repeat of the retroviral vector pSRMSVtkneo and named pSRMSVtkneo-MEN and pSRMSVtkneo-dMEN1-7, respectively. The empty pSRMSVtkneo vector (pSRMSVtkneo-Mock) was used as a negative control.
A reporter plasmid, 2x RGC-Luc, was generated by introducing double-stranded oligonucleotides containing the sequence of two ribosomal gene cluster (RGC) p53 sites (5′-GTTGCCTGGACTTGCCTGGCCTTGCCTTTTC-3′)33upstream of the tk promoter of the tk-Luc plasmid. pXP2-HH0.34 was generated by inserting genomic cyclin G promoter sequence, HH0.34,34 upstream of luciferase coding sequence of the pXP2 plasmid. HH0.34 was a gift from Dr Koji Okamoto (Columbia University, New York, NY). GAL4-Luc was generated by inserting two tandemly repeated GAL4 binding sites into theBamHI site upstream of the tk promoter of the tk-Luc plasmid.
pRC/CMV-p53 (CMVp53) was generated by introducing the entire coding region of the human p53 cDNA into the expression vector, pRC/CMV (Stratagene, La Jolla, CA).
Cell culture.
COS1, COS7, and Rat1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), and HeLaS3 cells were cultured in Ham’s F12 medium. The media were all supplemented with antibiotics and 10% fetal calf serum (FCS). HeLaS3 cells, originally developed by Drs T.T. Puck, P.I. Marcus, and S.J. Cieciura, were given by the Health Science Research Resources Bank (Osaka, Japan).
Transfection and luciferase assay.
Cells were plated at a density of 3.0 × 105 cells per 6-cm dish 12 hours before transfection. Reporter and expression plasmids together with 250 ng of pCMV-βgal (Stratagene) were transfected into the cells by the calcium phosphate precipitation method as described previously.35 To keep the transfection efficiency as constant as possible among the samples to be compared, the total amount of DNA in terms of weight was adjusted to be equal by adding pME18S plasmid DNA. After incubation for 36 hours, cells were harvested and subjected to the luciferase assay using a Luciferase Assay System (Promega, Madison, WI) and Lumat LB-9501 (BSI Instruments Inc, Aliquoppa, PA). The data were corrected by the β-galactosidase activity of each cell lysate assayed using Galacto-Light Plus (Tropix, Bedford, MA). All transfection experiments were performed three times, and the representative data are presented.
Immunoprecipitations and Western blotting.
COS1 cells were transfected using the diethyl aminoethyl (DEAE)-dextran method36 and harvested 48 hours later in phosphate-buffered saline (PBS). After centrifugation, they were sonicated by the Sonicator 250 (Branson Ultrasonics, Danbury, CT) in lysis buffer (25 mmol/L HEPES-KOH [pH 8.0], 150 mmol/L KCl, 2 mmol/L EDTA-2Na, 1 mmol/L dithiothreitol [DTT], 1 mmol/L phenylmethyl sulfonyl fluoride [PMSF], 0.1% NP-40) and centrifuged at 15,000 rpm for 5 minutes. The supernatants were incubated with Protein A-sepharose conjugates (Sigma, St Louis, MO) for 6 hours at 4°C that were pretreated with anti-FLAG antibody (M2; Eastman Kodak Co, New Haven, CT) or anti-p53 antibody (DO-1; Santa Cruz Biotechnology, Inc, Santa Cruz, CA). After they were washed 3 times in lysis buffer, the immunoprecipitates were electrophorased in 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nylon membranes (Millipore, Bedford, MA). The membranes were blocked with 10% skim milk treated with anti-HA (HA.11; BAbCO, Richmond, CA) or anti-p53 antibody, washed, and reacted with mouse or rabbit anti-IgG antibody coupled to alkaline phosphatase (Promega, Madison, WI). The blots were visualized by incubation with nitroblue tetrazolium-bromochloroindolyl phosphate (Promega).
Viral infection.
To prepare the retrovirus stocks, 10 μg of pSRMSVtkneo-Mock, pSRMSVtkneo-MEN, pSRMSVtkneo-dMEN4, and pSRMSVtkneo-dMEN7 constructs were transfected with 40 μg of ψ packaging plasmid into 1 × 106 COS7 cells by the DEAE-dextran method. The culture medium containing viruses was harvested 96 hours after transfection. Viral titers were determined and normalized. Viral infections were performed by exposing 5 × 104 Rat1 cells to 1 mL of virus stocks for 8 hours. G418-resistant populations were selected in medium containing 800 μg/mL G418 after an additional incubation for 48 hours in medium without G418. The following experiments were performed with uncloned cell populations.
Assays for transformation.
For the soft agar assay, cells of each transfected derivative were trypsinized, suspended in DMEM containing 0.3% agar and 20% FCS, and plated onto a bottom layer containing 0.6% agar. Cells were plated at a density of 2 × 104 cells/3.5 cm dish in tetraplicate, and colonies greater than 0.125 mm in diameter were enumerated after 14 days. The numbers of colonies were presented as a mean value.
RESULTS
MLL/MEN chimeric protein suppresses the transcriptional activity of p53.
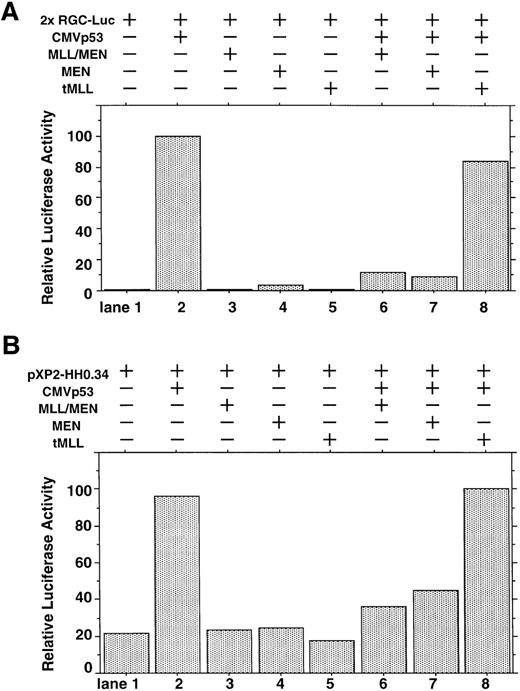
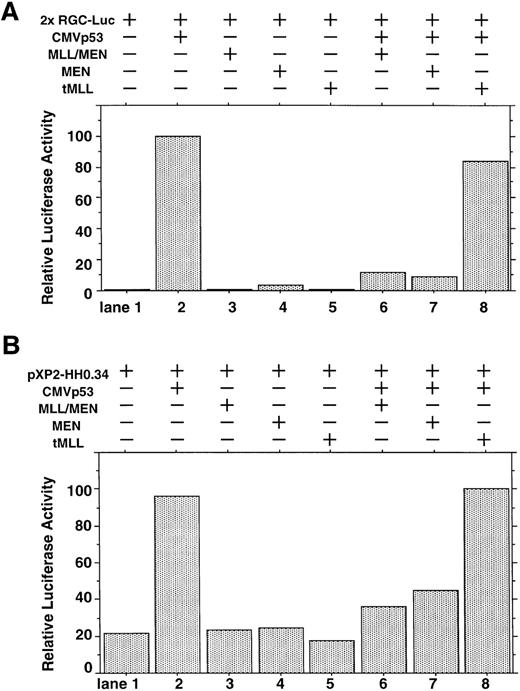
We investigated whether the MLL/MEN chimeric protein has an effect on p53-dependent transcription. Luciferase assays were performed using a reporter plasmid containing two p53 binding sites upstream of tk basal promoter region (2x RGC-Luc), which has widely been used in assays of p53-mediated transcriptional activity.37,38 We used HeLaS3 cells for our assays because both alleles of the p53 gene are deleted in them.39 The p53 expression vector (CMVp53), alone or in combination with the MLL/MEN expression vector [pME18S-MLL/MEN(HA)], was transfected into HeLaS3 cells together with 2x RGC-Luc. Transfection of CMVp53 led to a marked luciferase activity as expected37 (Fig 1A, lane 2). Expression of the MLL/MEN chimeric protein alone in HeLaS3 cells had no effects on the reporter plasmid (lane 3). Surprisingly, the luciferase activity mediated by CMVp53 significantly decreased when coexpressed with the MLL/MEN chimeric protein (lane 6). Therefore, the MLL/MEN chimeric protein repressed the transcriptional activity of p53.
Effect of the MLL/MEN chimeric protein on p53-dependent transcription. (A) HeLaS3 cells were transfected with 2 μg of 2x RGC-Luc, 2 μg of CMVp53, and 2 μg of pME18S-MEN(HA), pME18S-MLL/MEN(HA), or pME18S-tMLL(HA) by using a calcium phosphate method. The total amounts of the transfected DNAs were equalized in all samples with the backbone vector, pME18S. Equal amounts of the cell lysates were assayed for luciferase activities 24 hours after transfection. The highest value was arbitrarily set as 100 and all others were consequently normalized. (B) HeLaS3 cells were transfected with 2 μg of pXP2-HH0.34, 2 μg of CMVp53, and 2 μg of pME18S-MEN(HA), pME18S-MLL/MEN(HA), or pME18S-tMLL(HA) by using a calcium phosphate method and were assayed in the same way as (A).
Effect of the MLL/MEN chimeric protein on p53-dependent transcription. (A) HeLaS3 cells were transfected with 2 μg of 2x RGC-Luc, 2 μg of CMVp53, and 2 μg of pME18S-MEN(HA), pME18S-MLL/MEN(HA), or pME18S-tMLL(HA) by using a calcium phosphate method. The total amounts of the transfected DNAs were equalized in all samples with the backbone vector, pME18S. Equal amounts of the cell lysates were assayed for luciferase activities 24 hours after transfection. The highest value was arbitrarily set as 100 and all others were consequently normalized. (B) HeLaS3 cells were transfected with 2 μg of pXP2-HH0.34, 2 μg of CMVp53, and 2 μg of pME18S-MEN(HA), pME18S-MLL/MEN(HA), or pME18S-tMLL(HA) by using a calcium phosphate method and were assayed in the same way as (A).
Next, we determined which portion of the MLL/MEN chimeric protein is responsible for repression of p53-induced transcriptional activity. Luciferase assays were performed in the same way using the MEN expression vector [pME18S-MEN(HA)] and the tMLL expression vector [pME18S-tMLL(HA)], instead of the pME18S-MLL/MEN(HA). As did the MLL/MEN chimeric protein, expression of either the MEN protein or the tMLL protein alone in HeLaS3 cells had no effects on the level of reporter activation (lanes 4 and 5). In the cotransfection experiments, the tMLL protein, which does not include the MEN part of the chimeric protein, could not repress p53-dependent transcription at all (lane 8). However, the MEN protein was able to repress the activity of p53 to a the similar level as the MLL/MEN chimeric protein (lane 7). Therefore, the MEN portion of the MLL/MEN chimeric protein is responsible for repression of p53-mediated transcriptional activity. This finding was confirmed by similar results obtained with luciferase assays using a reporter plasmid containing cyclin G native promoter sequence (Fig 1B). These repressive effects do not seem to be mediated through the direct suppression of the promoter, because neither MLL/MEN nor MEN inhibited the luciferase activity of the reporter (Fig 1A and B, lanes 3 and 4).
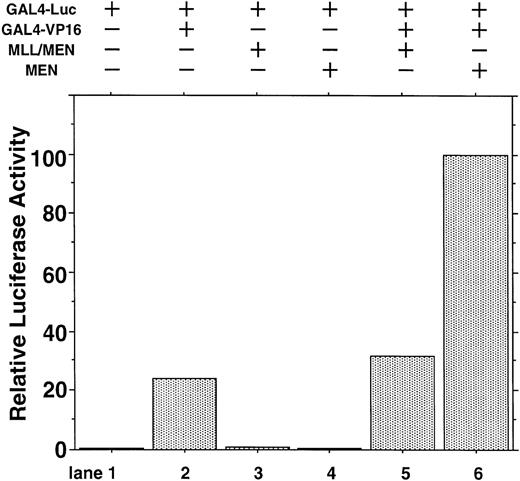
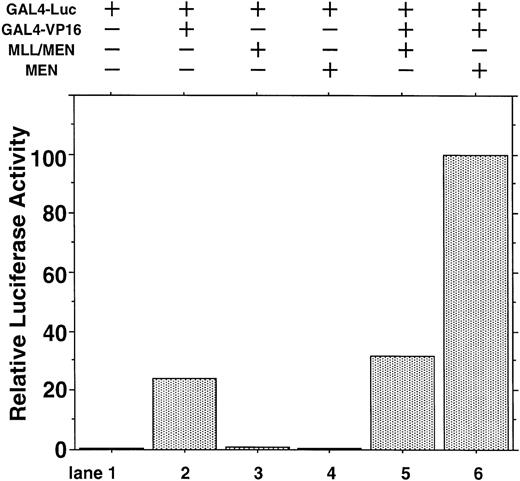
To exclude the possibility that MEN or MLL/MEN could be a global suppressor of transcription, we performed luciferase assays using a luciferase reporter plasmid with two tandemly repeated GAL4 binding sites (GAL4-Luc) and pME18S-GAL4/VP16, which is an expression vector GAL4 DNA containing the binding domain fused to the herpesvirus transcriptional activator VP16 (GAL4-VP16). In this experiment, neither MEN nor MLL/MEN repressed luciferase activities exhibited by GAL4-VP16 (Fig 2). Therefore, both MEN and MLL/MEN seem to be specific suppressors of p53-mediated transcription, but not global suppressors of transcription. As shown in lane 6, the MEN protein coexpressed with GAL4-VP16 produced a higher luciferase activity than GAL4-VP16 alone (lane 2). We speculate that MEN, being an RNA polymerase II elongation factor, might work as a potential activator of transcription.
Effect of the MLL/MEN chimeric protein on p53-independent transcription. HeLaS3 cells were transfected with 2 μg of GAL4-Luc, 2 μg of pME18S-GAL4/VP16, and 2 μg of pME18S-MEN(HA) or pME18S-MLL/MEN(HA) by using a calcium phosphate method and were assayed in the same way as in Fig 1.
Effect of the MLL/MEN chimeric protein on p53-independent transcription. HeLaS3 cells were transfected with 2 μg of GAL4-Luc, 2 μg of pME18S-GAL4/VP16, and 2 μg of pME18S-MEN(HA) or pME18S-MLL/MEN(HA) by using a calcium phosphate method and were assayed in the same way as in Fig 1.
MLL/MEN chimeric protein binds to p53 in vivo.
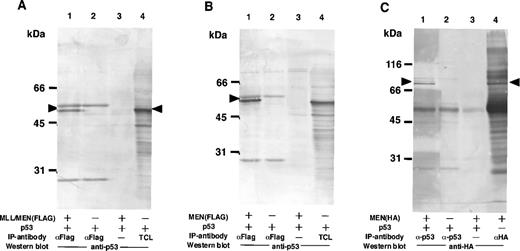
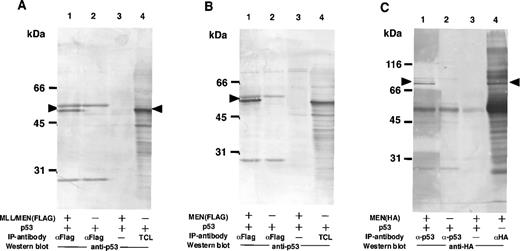
Based on the results shown above, we suspected that the suppressive effect of the chimeric protein on the activity of p53 was mediated by through interaction between these two proteins. To examine the possibility that the MLL/MEN chimeric protein binds to p53, immunoprecipitations followed by Western blotting were performed (Fig 3A). COS1 cells were transfected with the p53 expression vector (CMVp53) alone or in combination with the MLL/MEN expression vector [pME18S-MLL/MEN(FLAG)]. Forty-eight hours later, cell extracts were subjected to immunoprecipitation with the monoclonal anti-FLAG antibody. Western analysis using the anti-p53 antibody showed that p53 was coimmunoprecipitated with the anti-FLAG antibody in the presence of the chimeric protein (lane 1), but not in its absence (lane 2) or without the anti-FLAG antibody (lane 3). Lane 4 shows the total cell lysate from COS1 cells transfected with CMVp53 as a control. These results indicate that p53 is associated with the MLL/MEN chimeric protein in vivo. The MLL/MEN chimeric protein could hardly be detected in the immunoprecipitates with the anti-p53 antibody, possibly due to the relative difficulty in detecting a large molecule such as the MLL/MEN protein (data not shown).
The MLL/MEN chimeric protein binds to p53. (A) Cell extracts derived from COS1 cells transfected with CMVp53, alone or in combination with pME18S-MLL/MEN(FLAG), were immunoprecipitated with anti-FLAG monoclonal antibody (lanes 1 and 2) or with the protein A-Sepharose beads only (lane 3). Western blot was performed using anti-p53 monoclonal antibody (DO-1). Lane 4 contains the total cell lysate (TCL) from COS1 cells transfected with CMVp53 as a control. (B) Cell extracts derived from COS1 cells transfected with CMVp53, alone or in combination with pME18S-MEN(FLAG), were immunoprecipitated with anti-FLAG monoclonal antibody (lanes 1 and 2) or with the protein A-Sepharose beads only (lane 3). Western blot was performed using anti-p53 monoclonal antibody (DO-1). Lane 4 contains the TCL from COS1 cells transfected with CMVp53 as a control. (C) Cell extracts derived from COS1 cells transfected with CMVp53, alone or in combination with pME18S-MEN(HA), were immunoprecipitated with anti-p53 monoclonal antibody (lanes 1 and 2) or with the protein A-Sepharose beads only (lane 3). Western blot was performed using anti-HA polyclonal antibody (HA.11). Cell extracts derived from COS1 cells transfected with pME18S-MEN(HA) were immunoprecipitated with anti-HA polyclonal antibody and subjected to Western blot using anti-HA polyclonal antibody (lane 4) as a control.
The MLL/MEN chimeric protein binds to p53. (A) Cell extracts derived from COS1 cells transfected with CMVp53, alone or in combination with pME18S-MLL/MEN(FLAG), were immunoprecipitated with anti-FLAG monoclonal antibody (lanes 1 and 2) or with the protein A-Sepharose beads only (lane 3). Western blot was performed using anti-p53 monoclonal antibody (DO-1). Lane 4 contains the total cell lysate (TCL) from COS1 cells transfected with CMVp53 as a control. (B) Cell extracts derived from COS1 cells transfected with CMVp53, alone or in combination with pME18S-MEN(FLAG), were immunoprecipitated with anti-FLAG monoclonal antibody (lanes 1 and 2) or with the protein A-Sepharose beads only (lane 3). Western blot was performed using anti-p53 monoclonal antibody (DO-1). Lane 4 contains the TCL from COS1 cells transfected with CMVp53 as a control. (C) Cell extracts derived from COS1 cells transfected with CMVp53, alone or in combination with pME18S-MEN(HA), were immunoprecipitated with anti-p53 monoclonal antibody (lanes 1 and 2) or with the protein A-Sepharose beads only (lane 3). Western blot was performed using anti-HA polyclonal antibody (HA.11). Cell extracts derived from COS1 cells transfected with pME18S-MEN(HA) were immunoprecipitated with anti-HA polyclonal antibody and subjected to Western blot using anti-HA polyclonal antibody (lane 4) as a control.
Given that the MEN portion of the MLL/MEN chimeric protein is sufficient for repression of p53-mediated transcriptional activity, we investigated whether this region is responsible for binding of the MLL/MEN chimeric protein to p53. COS1 cells were transfected with the p53 expression vector (CMVp53), alone or in combination with the MEN expression plasmid, pME18S-MEN(FLAG), or pME18S-MEN(HA), and immunoprecipitations followed by Western analysis were performed (Fig3B and C). Western analysis of the immunoprecipitates showed that p53 was coimmunoprecipitated with the anti-FLAG antibody in the presence of the MEN protein (Fig 3B, lane 1), but not detected in its absence (Fig3B, lane 2) or without the anti-FLAG antibody (Fig 3B, lane 3). Lane 4 shows the total cell lysate from COS1 cells transfected with CMVp53 as a control. Conversely, Western blot analysis of the immunoprecipitates with the anti-p53 antibody showed the presence of the MEN protein of 80 kD associated with p53 (Fig 3C, lane 1). Lane 4 shows the MEN protein detected in the immunoprecipitates with the anti-HA antibody. These results showed that the MLL/MEN chimeric protein binds to p53 in vivo, and that the MEN portion of the chimeric protein is sufficient for binding to p53.
The regions of MEN responsible for repression of p53-mediated transactivation.
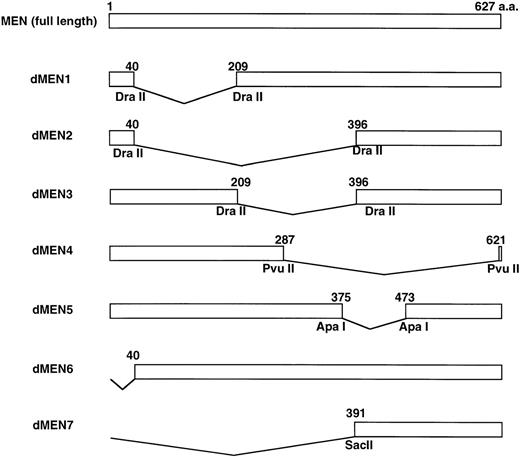
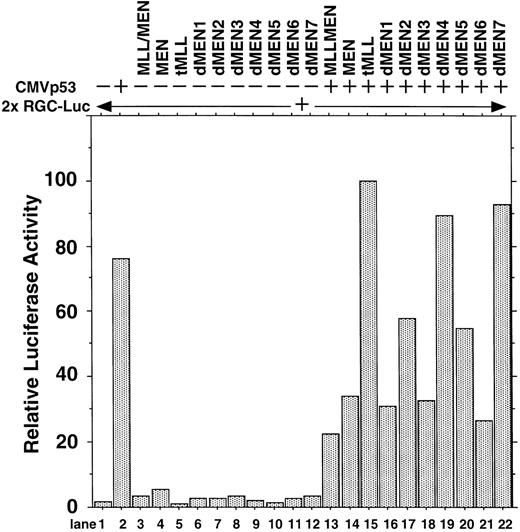
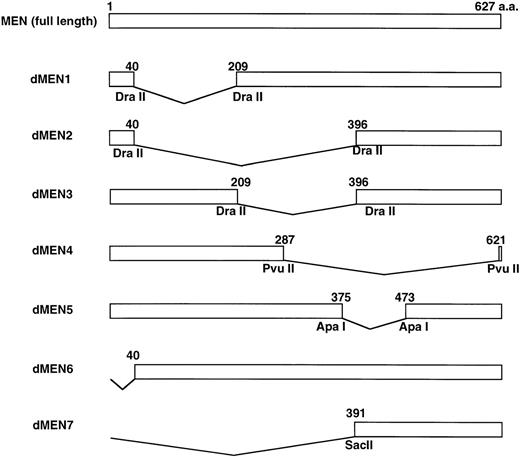
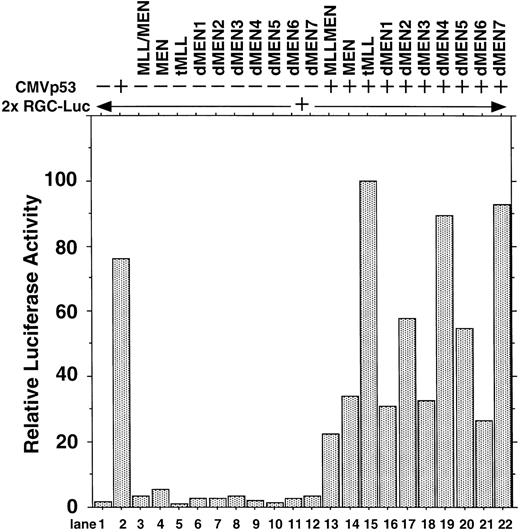
To determine the regions of the MEN protein responsible for repression of p53-mediated transactivation, we constructed a series of deletion mutants of MEN, named dMEN1-7 (Fig 4). Luciferase assays were performed as described above using 2x RGC-Luc as a reporter plasmid. As expected, tMLL could not inhibit p53-mediated transcription of the 2x RGC promoter. Among the deletion mutants, only dMEN4 and dMEN7, which lack roughly the C-terminal half and the N-terminal half of the MEN protein, respectively, were unable to suppress the luciferase activity mediated by p53 (Fig 5, lanes 19 and 22). Because some mutants such as dMEN2 and dMEN5 could partially suppress p53-mediated transactivation, the extensive deletions in dMEN4 or dMEN7 appear to be necessary for complete disruption of the repressive effect on p53-mediated transactivation.
Deletion mutants of the MEN protein. Structures of full-length MEN and seven deletion mutants are shown. Restriction enzyme sites used to construct the mutants are indicated. aa, amino acids.
Deletion mutants of the MEN protein. Structures of full-length MEN and seven deletion mutants are shown. Restriction enzyme sites used to construct the mutants are indicated. aa, amino acids.
Deletion mutants of MEN that disrupt inhibitory functions on p53-mediated transactivation. HeLaS3 cells were transfected with 2 μg of 2x RGC-Luc, 2 μg of CMVp53, and 2 μg of pME18S-MEN(HA), pME18S-MLL/MEN(HA), pME18S-tMLL(HA), or one of the expression vectors of MEN mutants by using a calcium phosphate method and were assayed in the same way as in Fig 1.
Deletion mutants of MEN that disrupt inhibitory functions on p53-mediated transactivation. HeLaS3 cells were transfected with 2 μg of 2x RGC-Luc, 2 μg of CMVp53, and 2 μg of pME18S-MEN(HA), pME18S-MLL/MEN(HA), pME18S-tMLL(HA), or one of the expression vectors of MEN mutants by using a calcium phosphate method and were assayed in the same way as in Fig 1.
The regions of MEN responsible for binding to p53.
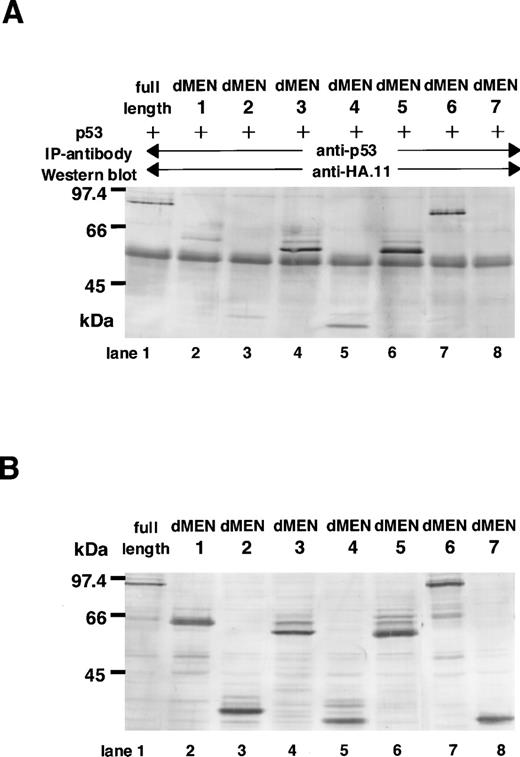
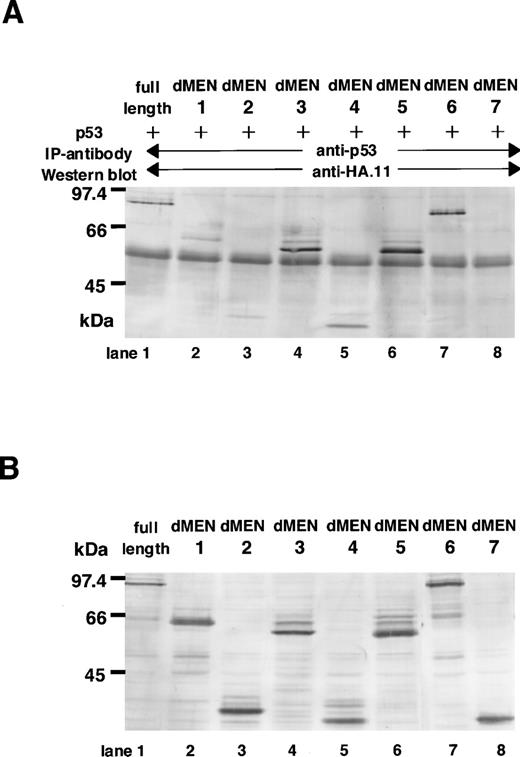
Next, to determine the region(s) of MEN required for binding to p53, immunoprecipitations followed by Western blotting were performed. COS7 cells were transfected with CMVp53, in combination with one of the expression vectors of MEN [pME18S-MEN(HA)] or its deletion mutants [pME18SdMEN1-7(HA)]. Forty-eight hours later, cell extracts were subjected to immunoprecipitation with the monoclonal anti-p53 antibody. The amounts of these proteins used for the immunoprecipitation were comparable, as shown in Fig 6B. Western analysis using the anti-HA polyclonal antibody showed that only dMEN7, which could not suppress the transcriptional activity of p53 in luciferase assays, was not coimmunoprecipitated with the anti-p53 antibody (Fig 6A, lane 8). Although all other mutants, including dMEN4, were coimmunoprecipitated with anti-p53 antibody (Fig 6A, lanes 2 through 7), the binding between p53 and dMEN1 or dMEN2 seems to be relatively weak. This indicates that the binding region for p53 is partially deleted in these mutant proteins. Because the only significant difference between dMEN2 and dMEN7 is the presence of the N-terminal 40 amino acids, these amino acids must be important for p53 binding. All these results strongly suggest that the region of the MEN protein responsible for binding p53 is located in the N-terminal half of the protein.
Deletion mutants of MEN that disrupt the binding with p53. (A) Cell extracts derived from COS1 cells transfected with CMVp53 and pME18S-MEN(HA) or pME18S-dMEN1-7(HA) were immunoprecipitated with anti-p53 monoclonal antibody. Western blot was performed using anti-HA polyclonal antibody (HA.11). (B) Lanes 1 through 8 contain the equal amount of the TCLs from the same samples used for the immunoprecipitation assays in (A). Western blot was performed using anti-HA polyclonal antibody (HA.11).
Deletion mutants of MEN that disrupt the binding with p53. (A) Cell extracts derived from COS1 cells transfected with CMVp53 and pME18S-MEN(HA) or pME18S-dMEN1-7(HA) were immunoprecipitated with anti-p53 monoclonal antibody. Western blot was performed using anti-HA polyclonal antibody (HA.11). (B) Lanes 1 through 8 contain the equal amount of the TCLs from the same samples used for the immunoprecipitation assays in (A). Western blot was performed using anti-HA polyclonal antibody (HA.11).
Although dMEN4 was unable to repress p53-mediated transactivation in the luciferase assays, it coimmunoprecipitated with anti-p53 antibody. It is possible that the C-terminal half of MEN, which is deleted in dMEN4, is responsible for repression of p53-mediated transcription, but not for binding p53. As is the case with the binding region, it seems that the repression domain broadly spans the C-terminal half of the MEN protein. In support of this, dMEN5, which lacks a smaller part of the C-terminal region than does dMEN4, was able to repress p53-mediated transactivation in the luciferase assays, although to a lesser extent than MEN (Fig 5, lane 20).
These results suggest that MEN contains two functional domains, a binding domain and a repression domain, which can be assigned to the N-terminal half and the C-terminal half of the protein, respectively.
Correlation of p53 repressive activity and Rat1 transformation.
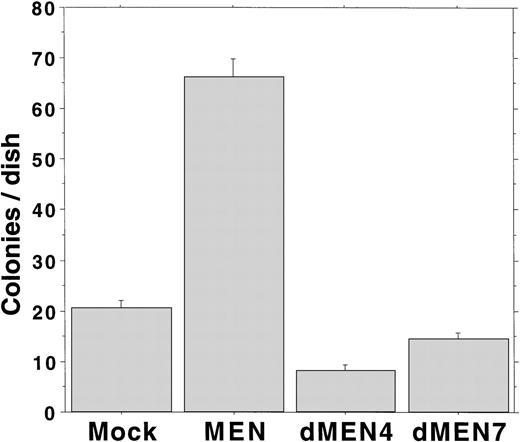
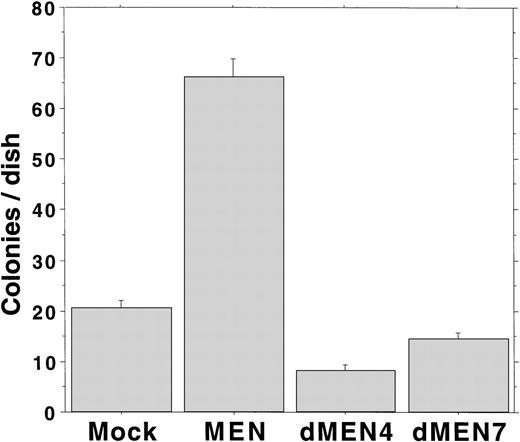
We have previously shown that overexpression of the MEN protein leads to Rat1 cell transformation.18 To analyze the transforming ability of dMEN4 and dMEN7, which could not suppress the transcriptional activity of p53 due to loss of the repression domain and the binding domain, respectively, we evaluated the capacity for anchorage-independent growth by the Rat1 transformation assay. Rat1 derivatives expressing the corresponding mutants were seeded in DMEM containing 0.3% agar and 20% FCS, and colony formation was estimated as an anchorage-independent growth ability. Mock transfectants barely made colonies larger than 0.125 mm in diameter. However, as expected MEN-expressing cells formed many colonies within 14 days. Surprisingly, like mock transfectants, both dMEN4 and dMEN7 transfectants barely made colonies (Fig 7). These results show that both the N-terminal half and the C-terminal half regions of the MEN protein are essential for transformation of Rat1 cells.
Soft agar assays examining Rat1 transformation ability of MEN mutants. Soft agar colony counts of Rat1 derivatives expressing each mutant were shown. Colonies greater than 0.125 mm in diameter were counted on day 14. MEN-expressing cells formed significantly increased numbers of macroscopic colonies compared with mock, dMEN4, and dMEN7 transfectants.
Soft agar assays examining Rat1 transformation ability of MEN mutants. Soft agar colony counts of Rat1 derivatives expressing each mutant were shown. Colonies greater than 0.125 mm in diameter were counted on day 14. MEN-expressing cells formed significantly increased numbers of macroscopic colonies compared with mock, dMEN4, and dMEN7 transfectants.
Therefore, p53-repressive activity correlates with the transforming ability of Rat1 cells, and the oncogenic ability of MEN seems to be exerted, at least to a certain extent, through repression of p53 activity.
DISCUSSION
In this study, we have shown that overexpression of the MLL/MEN chimeric protein suppresses the transactivation ability of p53, using reporter plasmids containing either the cyclin G native promoter sequence or a minimal promoter with two p53 binding sites. This inhibitory effect of the fusion protein on p53 function is shown to be dependent on the MEN portion, as the MEN protein alone can repress p53-mediated transcription to almost the same level as MLL/MEN. We have also demonstrated that p53 physically associates with both MLL/MEN and MEN, as p53 was coimmunoprecipitated with these two Flag-tagged proteins by anti-Flag antibody. In addition, the MEN protein itself was coimmunoprecipitated by anti-p53 antibody. Therefore, the fusion protein may bind to p53 through its MEN domain. The suppressive effect of the fusion protein appears to be by this functional interaction with p53, rather than by a direct effect on the reporter plasmid, as it did not have a suppressive effect on the reporter in the absence of p53. This is the first report to describe an interaction between the MLL/MEN chimeric protein and the tumor-suppressor protein, p53. The inhibitory effect of the chimeric protein on the antioncogene p53, mediated by this protein-protein interaction, may be critical to the leukemogenesis associated with the t(11;19)(q23;p13.1) translocation.
Through analysis of the deletion mutants of MEN, we have shown that the MEN protein may contain two functional regions: a binding domain for p53 located in the N-terminal half, and a repression domain of p53-mediated transcription located in the C-terminal half. The binding domain is sufficient for association with p53, but not for inhibition of its function. Blockade of p53-mediated transactivation also requires the repression domain. While it is reasonable to consider that this interaction with p53 is the first step by which the MLL/MEN or MEN protein exert an inhibitory effect on p53, the exact molecular mechanism that is involved remains unclarified. It is possible that the repression domain recruits some kind of histone deacetylase to the promoters activated by p53. Alternatively, this domain may block the binding of p53 to promoters or cofactors that are needed for p53 transactivation. It is not clear whether the association between p53 and MLL/MEN or MEN is direct, as we have shown the interaction by coimmunoprecipitation assays. The protein-protein interactions demonstrated by this method do not necessarily prove direct association. Thus, the association of p53 with the MLL/MEN chimeric protein and the MEN protein may be indirect and mediated by other intervening molecules that work as a bridge or an adaptor between these two proteins.
We have also demonstrated that deletion mutants of MEN lacking either of the N-terminal or C-terminal domains could not transform Rat1 cells. As we have shown previously, these cells are transformed by the MEN protein of full length.21 These data suggest that the binding domain and the repression domain demonstrated by the molecular experiments are also essential for the transformation ability of MEN shown in the biological experiments using Rat1 cells.
Whether there are significant functional differences between the MLL/MEN and the MEN proteins remains to be determined. Although the MLL/MEN protein possesses an additional MLL portion in its N-terminal region that does not exist in the MEN protein, these two proteins suppressed the effect of p53 to almost the same level. In the coimmunoprecipitation experiments, both proteins were coimmunoprecipitated with p53. Therefore, the functions of these two proteins in terms of binding to and suppressing p53 seem to be identical. Furthermore, we have already examined the subcellular localization of the MLL/MEN fusion and the MEN proteins in COS7 cells and showed the same nuclear localization pattern for both proteins.20 Thus, there appears to be no evidence that the chimeric protein translocates with p53 from the region where the MEN protein originally residues to another location, thereby sequestering p53 away from its normal site. These findings raise the question of why the generation of the novel fusion protein resulting from chromosomal translocation leads to leukemic transformation, even in a situation in which endogenous MEN protein already exists.9 There are several potential answers to this question. First, the chimeric protein might be more stable than the MEN protein. The fusion protein has acquired the N-terminal sequence derived from MLL. This may increase stability allowing the fusion protein to exert its function for a longer period than the MEN molecule. Second, the chimeric protein may be expressed more strongly or steadily than the MEN protein in hematopoietic stem cells. As a result of the translocation, the expression of the MLL/MEN fusion protein is driven by the MLL promoter. The higher expression of the fusion protein could be achieved by two possible mechanisms. One possibility is that the MLL promoter is stronger than the MEN promoter. The other possibility is that the MEN promoter is precisely regulated by other transcription factors. For example, these factors might be dependent upon the differentiation stage or the cell cycle, or negatively regulated by excessive expression of the MEN protein itself. The MLL/MEN chimeric protein driven by the MLL promoter may escape from the precise control exerted by the MEN promoter, leading to greater expression of the MLL/MEN protein relative to the MEN protein. Consequently, the overall inhibitory influence on p53 function could be increased or dysregulated.
MEN is a multifunctional molecule and acts not only as an RNA polymerase II elongation factor, but also as a direct suppressor of p53. We have already shown that the MEN protein stimulates AP-1 activity through its elongation activity and increases the colony formation of Rat1 cells.19 This function is dependent on the lysine-rich region in the MEN molecule. The oncogenic activity of MEN may be observed in various kinds of cells that express Jun and Fos proteins. It is possible that the MLL/MEN protein exerts a greater oncogenic effect than the MEN protein itself through stimulation of AP-1 activity. In this study, we spotlighted the MLL/MEN and the MEN proteins as inhibitors of p53. This function appears to be unrelated to the elongation activity. Wild-type p53 functions in normal hematopoietic cell differentiation in multilineage cells40-44 and may play pivotal roles in hematopoietic cell maturation, possibly by inhibiting proliferation during terminal differentiation. It has been reported that the p53 genes are rearranged or mutated in a variety of leukemic cells.23-29 As is the case with the lack of p53 expression in homozygous deletion, inhibition of the tumor-suppressive effect of p53 by the MLL/MEN protein could cause leukemias. Therefore, repression of p53 tumor-suppressive transcriptional activity by the MLL/MEN chimeric protein may play an important role in leukemogenesis by the t(11;19)(q23;p13.1) translocation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hisamaru Hirai, MD, Department of Hematology and Oncology, Faculty of Medicine, University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-8655, Japan.