Eotaxin has been characterized as a chemokine involved in eosinophil activation; however, mRNA for this C-C chemokine has been shown to be constitutively expressed in thymus. Immunohistochemical analysis showed a punctate distribution pattern, with eotaxin expression localized mainly in the medulla and in Hassle’s corpuscles. Moreover, the receptor for eotaxin, CCR-3, was detected on thymocytes, with the highest level of expression being on the CD8 single-positive population. Equilibrium binding analyses on unfractionated thymocytes demonstrated specific 125I-eotaxin binding profiles comparable with CCR-3 transfectants. Eotaxin induced cell migration and mobilization of intracellular calcium in all thymocytes except the immature CD4−/CD8− population. Eotaxin also induced the secretion of the chemokines interleukin-8, RANTES, and macrophage inflammatory protein-1β from thymocyte cultures in vitro. These results suggest that eotaxin-induced thymocyte activation may have important physiological implications for lymphocyte mobilization within and from this lymphoid organ.
THE CHEMOKINE EOTAXIN was purified and cloned from the bronchoalveolar lavage fluid of allergen-sensitized guinea pigs.1-3 Indeed, expression analysis indicates that eotaxin mRNA is upregulated during allergic inflammation involving significant eosinophil infiltrates.4-10 Additionally, after systemic administration of interleukin-5 (IL-5), increased eosinophil infiltrates are seen at intradermal sites of eotaxin injection.11 Molecular and functional characterizations to identify a specific eotaxin receptor have identified CCR-3 as a specific, high-affinity eotaxin receptor.12-15 In addition to binding eotaxin, CCR-3 also binds the chemokines Eotaxin-2, monocyte chemotactic protein-2 (MCP-2), MCP-3, MCP-4, and RANTES.
Numerous reports have appeared in the literature categorizing eotaxin as being exclusively involved in eosinophil-activation; however, analyses of the tissue distribution of eotaxin have clearly demonstrated constitutive expression of this chemokine in different organs, including lymphoid-specific regions such as the thymus and lymph nodes.9,10 However, with the advent of specific antibodies, the expression of the CCR-3 receptor has also been demonstrated in microglia,16 T lymphocytes (the Th2 subpopulation in clonal T lymphocytes), and basophils.17,18Furthermore, this receptor has been shown to function as a coreceptor for human immunodeficiency virus-1 (HIV-1) on mononuclear cells facilitating viral entry.19 20 Although such findings may account for the activity of RANTES, MCP-3, and MCP-4 in these cell types, little is known about the role of eotaxin in activating cell types other than eosinophils.
Whereas there is a restricted eosinophil presence in the thymus, eotaxin expression in the thymus and its possible biological role(s) have not been assessed in detail. In addition, the lack of overt lymphoid pathology in the thymus of mice with a targetted disruption of the eotaxin gene21 fails to suggest a clear role for thymus-expressed eotaxin. We have undertaken a study to analyze the potential biological effects of eotaxin on thymocytes. Eotaxin exhibits a restricted distribution in the thymus, being associated with cells predominantly in the medulla, as well as Hassle’s corpuscles. CCR-3 mRNA was detected in highly purified populations of thymocytes, with the greatest cell surface expression of receptor on CD8+single-positive thymocytes, as measured using a specific antibody. Thymocytes are induced to migrate in response to eotaxin and receptor ligation-induced intracellular calcium mobilization. Of the biological consequences analyzed, there was no differentiation, proliferation, or cell death induced by eotaxin, but large quantities of the chemokines RANTES, macrophage inflammatory protein-1β (MIP-1β), and IL-8 were secreted from thymocyte cultures in vitro. Although the significance of this biology awaits further experimentation, these data suggest that eotaxin and CCR-3 may play a role in thymic physiology.
MATERIALS AND METHODS
Thymocyte preparation.
Human thymus, removed as a consequence of cardiac surgery (average age, 6 months; age range, 1 week to 15 months; n = 10) was gently teased apart in cold Hanks’ balanced salt solution (HBSS), under sterile conditions. Total thymocytes were separated after Ficoll density gradient centrifugation and multiple HBSS washes at 4°C. Minor contaminating populations of monocytes were removed after Dynal bead separation of antibody-labeled (CD14) cells according to standard protocols. Populations of whole thymocytes were then stored on ice before use or stained for fluorescence-activated cell sorting (FACS) of individual subpopulations.
Immunohistochemistry.
Human thymus was frozen in liquid nitrogen and 7-μm sections were cut and fixed in acetone (5 minutes). Nonspecific antibody binding was prevented by preblocking with 10% human (type AB) and goat serum (Pel-Freez Clinical Systems, Brown Deer, WI) and subsequently with avidin/biotin Blocking Kit (Vector Laboratories, Burlingame, CA). Fixed sections were incubated with mouse monoclonal antibody to human eotaxin (1 hour; 10 μg/mL; R&D Systems, Minneapolis, MN) or an isotype-matched control. Slides were subsequently rinsed in TBS/0.1% bovine serum albumin (BSA) and incubated with biotinylated goat antimouse Ig (2.5 μg/mL; 1 hour). After a second wash, sections were incubated with alkaline phosphatase/avidin-biotin complex for 30 minutes. Sections were incubated with Vector Red alkaline phosphatase substrate for 10 minutes (room temperature), and antibody staining was visualized by microscopy.
FACS.
Thymocytes were labeled with anti-CD4-fluorescein isothiocyanate (FITC) and anti-CD8-phycoerythrin (PE) (Becton Dickinson, Mountain View, CA). After gating on appropriate populations, single-positive (SP; CD4+ or CD8+) populations, CD4+CD8+ double-positive (DP), and CD4−CD8− double-negative (DN) populations were sorted on a FACStar Plus (Becton Dickinson) cell sorter using standard methods. Sorted populations were used immediately for bioassay or frozen at −80°C before RNA preparation.
Chemotaxis assay.
Chemotaxis of thymocytes was performed as previously described22 using the 48-well modified Boyden chamber from NeuroProbe (Cabin John, MD). Cells that had migrated onto the underside of an 8-μm polyvinyl-pyrrolidone free (PVPF) polycarbonate membrane after 1 hour of incubation at 37°C were quantified under high power microscopy (400×), and results are expressed as the mean ± SEM of 5 high power fields.
Calcium mobilization analyses.
Whole thymocyte populations were labeled with Indo-1AM (3 μmol/L final concentration; Molecular Probes, Eugene, OR) for 45 minutes at room temperature. After labeling, cells were resuspended in 1 mL HBSS containing 1% BSA. Measurement of calcium flux was performed according to previously published methods23 using a PTI fluorimeter (South Brunswick, NJ).
Reverse transcriptase-polymerase chain reaction (RT-PCR) analyses of thymocytes.
Unfractionated thymocytes (107 cells) were washed once in phosphate-buffered saline (PBS), and the pellets were used for DNA-free RNA isolation using the SNAP Total RNA Isolation Kit (Invitrogen, San Diego, CA) according to the manufacturer’s protocol. First-strand cDNA synthesis for use in PCR reactions was generated using the cDNA Cycle Kit (Invitrogen). Parallel reactions were performed without AMV Reverse Transcriptase enzyme to control for genomic DNA contamination in the subsequent PCR reactions. PCR analysis was then performed using primers specific for human CCR-3 (sense, 5′-ATGACAACCTCACTAGATACAGTTG; antisense, 5′-CTAAAACACAATAGAGAGTTCCGG)12 for 40 cycles of 94°C for 1 minute, 55°C for 2 minutes, and 72°C for 3 minutes. Similar reactions were run with human CCR-3 cDNA (positive control) and water in place of first-strand cDNA (negative control). Amplified product (1.1 kb) was resolved on a 1% agarose gel containing ethidium bromide. Subsequently, the gel was denatured in Hybridization denaturing solution (1.5 mol/L NaCl, 0.5 mol/L NaOH; 5-Prime–3-Prime, Boulder, CO) for 30 minutes at room temperature. The PCR products were then transferred in 20× SSC (3 mol/L NaCl, 0.3 mol/L Na citrate, pH 7.0; Boehringer Mannheim, Indianapolis, IN) to Nytran nitrocellulose membranes (Schleicher & Schuell, Keene, NH) overnight. The transferred products were cross-linked to the membrane (UV Stratalinker; Stratagene, San Diego, CA), and the membrane was neutralized for 30 minutes in Hybridization neutralization solution (1 mol/L Tris-HCl, pH 7.4, 1.5 mol/L NaCl; 5-Prime–3-Prime). Prehybridization was performed at 42°C in Church buffer (7% sodium dodecyl sulfate [SDS], 0.5 mol/L NaHPO4, 0.5 mmol/L EDTA, pH 8), and the membrane was probed with 106 cpm/mL32P-labeled oligomer corresponding to an internal sequence of the CCR-3 gene: 5′-CGTTATGGCCATCTGCTACACAGG-3′. The blots were washed 3 times in 0.2% SSC/0.1% SDS (42°C) and then exposed on Biomax radiography film (Eastman Kodak, Rochester, NY).
FACS analysis of CCR-3.
Expression of CCR-3 on human thymocytes was analyzed using specific antibody 7B1124 (NIH AIDS Reagent Program, Rockville, MD). Highly purified thymocytes were stained according to standard methods with 7B11 for 30 minutes (1/500 dilution) and then with secondary FITC-coupled antibody (goat antimouse IgG2a; Pharmingen). Cells were washed once and stained with a cocktail of CD4-PE and CD8-PE-Cychrome (Becton Dickinson) for three-color analyses using a Becton Dickinson FACScan.
Radio-ligand binding analyses.
Equilibrium binding of 125I-labeled eotaxin in the presence of various concentrations of unlabeled human chemokines was performed as previously described.25125I-labeled eotaxin was obtained from DuPont NEN (Boston, MA). Recombinant chemokines were obtained from PeproTech (Rocky Hill, NJ) or R&D Systems (Minneapolis, MN). Briefly, 2.5 × 106 highly purified thymocytes were suspended in a 200 μL reaction volume containing 0.1 nmol/L125I-labeled eotaxin and various concentrations of unlabeled chemokines in the following buffer (25 mmol/L HEPES, 1 mmol/L CaCl2, 5 mmol/L MgCl2, and 0.5% BSA, adjusted to pH 7.4) for 90 minutes at 22°C. All assays were performed with 6 identical reactions per condition. The reactions were then filtered onto PEI-treated Whatman GF/C filters using a Packard cell harvester (Meriden, CT). Filters were washed with the following buffer (25 mmol/L HEPES, 1 mmol/L CaCl2, 5 mmol/L MgCl2, and 0.5 mol/L NaCl, adjusted to pH 7.4). Radioactivity retained on the filters was quantified using a Packard Top Count (Downers Grove, IL). The data obtained were analyzed using WaveMetrics IGOR software (Lake Oswego, OR) and/or the LIGAND program to determine the displacement binding characteristics, dissociation constants (kd), and average number of binding sites per cell.
Cytokine and chemokine enzyme-linked immunosorbent assay (ELISA) measurements.
Isolated human thymocytes (108/well of a 6-well Costar culture plate) were placed in culture for 48 hours in the presence of increasing concentrations of recombinant human eotaxin (PeproTech, Rocky Hill, NJ), hrIL-2, hrIL-7 (DNAX Research Institute, CA), or phytohemagglutinin (PHA; Wellcome, Chapel Hill, NC). Cell-free supernatants were then collected and frozen at −80°C before assay. Commercially available ELISA kits for the chemokines IL-8, MIP-1α, MIP-1β, and RANTES and the cytokines IL-4, IL-5, IL-10, and γ-interferon (γ-IFN; R&D Systems) were used according to the manufacturer’s protocols.
RESULTS
Eotaxin shows restricted staining patterns in the thymic medulla.
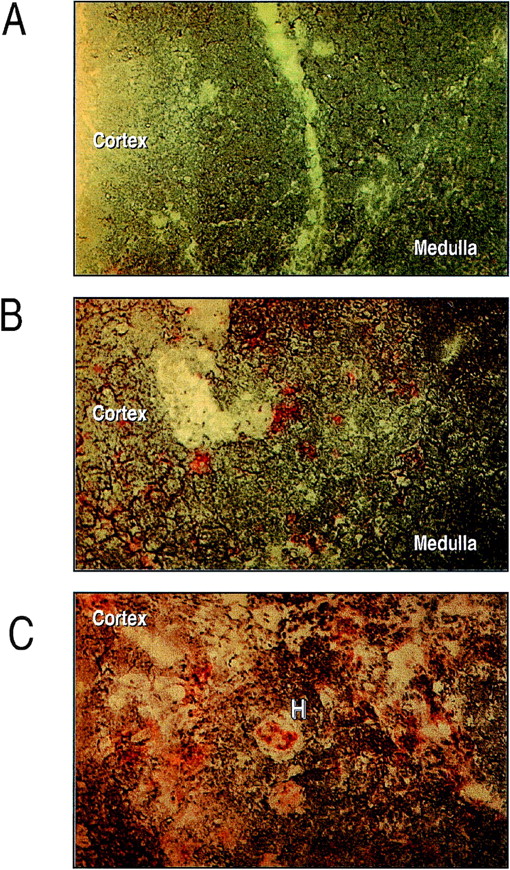
Immunohistochemical analyses of eotaxin, using an antihuman eotaxin antibody showed a distinct pattern of expression in human thymus. Sections showed a punctate staining in large cells with dendritic morphology, primarily in the medulla (Fig1). A significant proportion of eotaxin-positive cells also expressed CD11b in double-staining experiments (not shown). Additionally, there was staining of eotaxin in most Hassle’s corpuscles examined.
Immunohistochemical analysis of eotaxin expression in human neonatal thymus. Human neonatal thymus (1 week) was sectioned and prepared for eotaxin staining as detailed in Materials and Methods. (A) Negative control. (B and C) Staining of eotaxin around cells with dendritic morphology and in Hassle’s corpuscles, respectively. Results are representative of n = 3 separate donors.
Immunohistochemical analysis of eotaxin expression in human neonatal thymus. Human neonatal thymus (1 week) was sectioned and prepared for eotaxin staining as detailed in Materials and Methods. (A) Negative control. (B and C) Staining of eotaxin around cells with dendritic morphology and in Hassle’s corpuscles, respectively. Results are representative of n = 3 separate donors.
125I-eotaxin binds to human thymocytes.
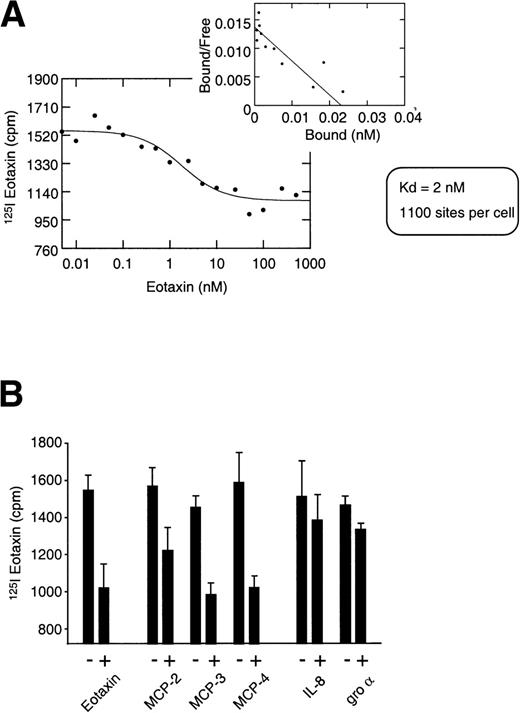
To further establish the significance of eotaxin expression in the thymus, equilibrium binding experiments assessing the interaction of125I-eotaxin with highly purified thymocytes were performed. These binding analyses indicated that a receptor for eotaxin was expressed on thymocytes (Fig 2A). Eotaxin displayed a binding affinity of 2 nmol/L, and Scatchard analysis indicated the expression of an average of 1,100 binding sites per cell over the entire population. Heterologous chemokine competition for 125I-eotaxin binding to thymocytes was consistent with thymocyte expression of CCR-3, the receptor known to bind eotaxin,12-14 because eotaxin tracer was competed from thymocytes by MCP-2, MCP-3, and MCP–4 that are also bound by CCR-3 (Fig 2B). Additionally, IL-8 and Gro-α were used as negative controls and in both cases failed to compete for eotaxin binding. Although this specific, displaceable binding is represented in Fig 2, in both homologous and heterologous competition analyses we noted a considerable level of nonspecific binding of the125I-eotaxin to thymocytes. The nature of such binding is unresolved and under investigation; however, we cannot rule out a role for cell surface glycosaminoglycans and or other cell surface moieties.
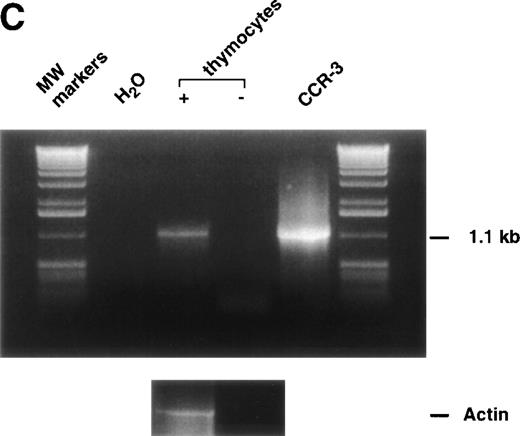
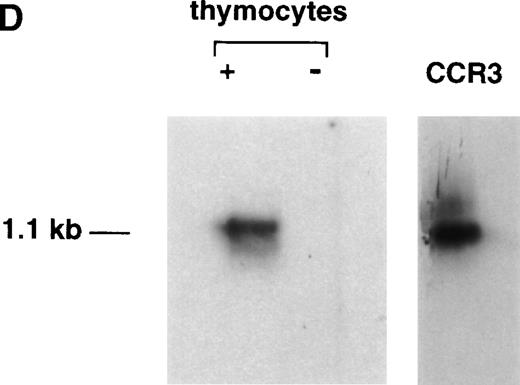
125I-Eotaxin binds human thymocytes with high affinity. (A) Homologous competition using unlabeled eotaxin shows displacement of labeled eotaxin from rigorously purified thymocytes. Scatchard analysis shows a kd of 2 nmol/L and approximately 1,100 eotaxin-binding sites per cell across the entire population. The displacement and Scatchard curves are representative of n = 4 experiments. (B) Heterologous competition of 125I-Eotaxin using unlabeled MCP-2, MCP-3, MCP-4, IL-8, and Gro-. The histograms are the mean ± SE mean displacements from a single experiment in which each point was performed 6 times. The experiment is representative of n = 3 other analyses. (C) RT-PCR analysis of CCR-3 expression on the entire thymocyte population. PCR was performed using cloned full-length CCR-3 cDNA as a control. (D) The PCR product was transferred and probed using an internal 32P-labeled oligomer to human CCR-3, as described in Materials and Methods.
125I-Eotaxin binds human thymocytes with high affinity. (A) Homologous competition using unlabeled eotaxin shows displacement of labeled eotaxin from rigorously purified thymocytes. Scatchard analysis shows a kd of 2 nmol/L and approximately 1,100 eotaxin-binding sites per cell across the entire population. The displacement and Scatchard curves are representative of n = 4 experiments. (B) Heterologous competition of 125I-Eotaxin using unlabeled MCP-2, MCP-3, MCP-4, IL-8, and Gro-. The histograms are the mean ± SE mean displacements from a single experiment in which each point was performed 6 times. The experiment is representative of n = 3 other analyses. (C) RT-PCR analysis of CCR-3 expression on the entire thymocyte population. PCR was performed using cloned full-length CCR-3 cDNA as a control. (D) The PCR product was transferred and probed using an internal 32P-labeled oligomer to human CCR-3, as described in Materials and Methods.
CCR-3 message is expressed in human thymocytes.
As a further test of specificity, we analyzed unfractionated thymocytes for the presence of CCR-3 message by RT-PCR. Figure 2c shows the PCR data, with the presence of a specific 1.1-kb message in the fractions containing reverse transcriptase, but not in the −RT or water controls. To further confirm the specificity of this message, we demonstrated that the 1.1-kb mRNA bound a 32P-labeled oligonucleotide corresponding to an internal sequence in the CCR-3 gene (Fig 2D).
CCR-3 is expressed on the surface of human thymocytes.
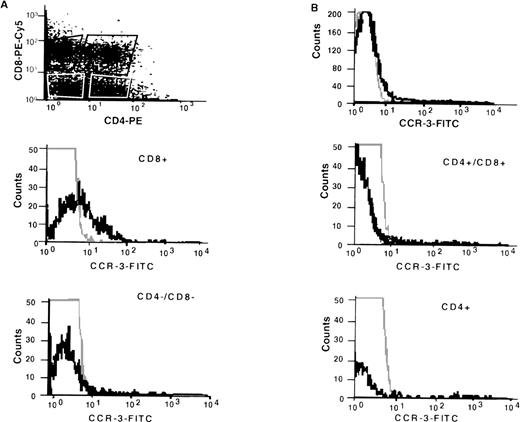
Using a specific anti–CCR-3 antibody (7B11), CCR-3 expression was shown on human thymocytes (Fig 3B) when compared with an isotype-matched negative control. Further analyses, gating on individual subsets of thymocytes, showed differences in expression intensity; CD8+ SP cells stained with greatest intensity with little if any staining on CD4+/CD8+ DP or CD4−/CD8− DN cells. CD4+ SP staining was more ambiguous, with expression observed in 3 of 4 thymocyte samples. However, even in the positive samples, there was a high intensity of staining only on a very small percentage of cells.
FACS analyses of CCR-3 expression on fractionated human thymocytes. Thymocytes were prepared as described in Methods and stained with anti-CCR-3 (7B11; IgG2a) according to standard protocols. After secondary antibody labeling, the cells were stained with anti-CD4-PE and anti-CD8-PE-Cychrome. Three-color analyses were performed according to standard methodology on gated subpopulations as shown by dot-plot. The results are representative of n = 3 separate donors.
FACS analyses of CCR-3 expression on fractionated human thymocytes. Thymocytes were prepared as described in Methods and stained with anti-CCR-3 (7B11; IgG2a) according to standard protocols. After secondary antibody labeling, the cells were stained with anti-CD4-PE and anti-CD8-PE-Cychrome. Three-color analyses were performed according to standard methodology on gated subpopulations as shown by dot-plot. The results are representative of n = 3 separate donors.
Eotaxin induces migration and calcium mobilization in human thymocytes.
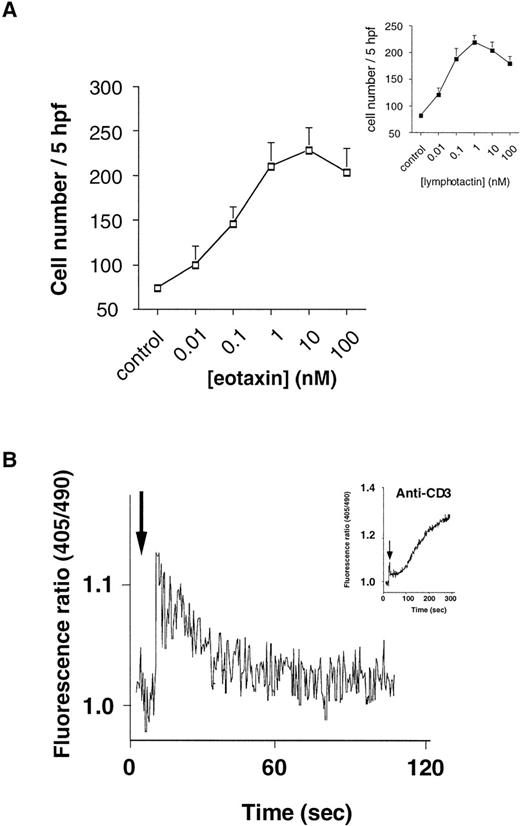
We next investigated the biological activity of eotaxin on human thymocytes, preliminarily analyzing standard readouts for chemokine-induced cell activation, including chemotaxis and calcium mobilization. Up to 10% of the unfractionated thymocytes were found to migrate in response to eotaxin in a dose-dependent manner (Fig 4A). Significant migration was obtained with 0.01 nmol/L eotaxin and maximal eotaxin-induced migration was observed at 10 nmol/L, in contrast to the positive control lymphotactin, which induced maximal migration at 1 nmol/L. Additionally, chemotaxis in response to either chemokine was abolished by preincubation of thymocytes with 100 ng/mL pertussis toxin (PTX; not shown), suggesting coupling of these functional chemokine receptors to a Gαi G-protein. Further analysis of the effects of eotaxin on thymocytes showed that this chemokine was capable of inducing a rapid transient mobilization of Ca2+, typical of chemokine-induced Ca2+ flux profiles in leukocytes (Fig4B), but contrasting that induced by anti-CD3 monoclonal antibody, which was more prolonged (Fig 4B, inset).
Eotaxin-induced migration of human thymocytes. (A) Dose-response curve of eotaxin-induced thymocyte migration. Migration assay was performed as previously described.22 Each point represents mean ± SE mean number of migrated cells in 5 high power fields (×400) from n = 5 experiments performed in duplicate. (Inset) Dose-response curve for human recombinant lymphotactin-induced migration, as a positive control, from the same n = 5 experiments performed in duplicate. (B) Eotaxin-induced calcium mobilization in unfractionated Indo-1AM–loaded thymocytes. Fluorimetry was performed as described in Bacon et al23 using a PTI fluorimeter. The trace is representative of n = 5 separate experiments.
Eotaxin-induced migration of human thymocytes. (A) Dose-response curve of eotaxin-induced thymocyte migration. Migration assay was performed as previously described.22 Each point represents mean ± SE mean number of migrated cells in 5 high power fields (×400) from n = 5 experiments performed in duplicate. (Inset) Dose-response curve for human recombinant lymphotactin-induced migration, as a positive control, from the same n = 5 experiments performed in duplicate. (B) Eotaxin-induced calcium mobilization in unfractionated Indo-1AM–loaded thymocytes. Fluorimetry was performed as described in Bacon et al23 using a PTI fluorimeter. The trace is representative of n = 5 separate experiments.
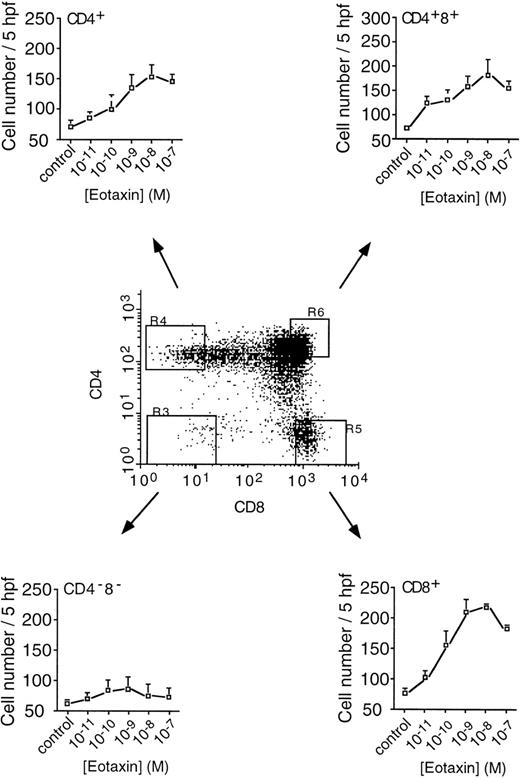
On investigation of fractionated thymocyte subpopulations, it was observed that the DP (CD4+ CD8+) and SP CD4+ and CD8+ populations migrated in response to eotaxin, whereas the DN (CD4−CD8−) cells failed to migrate (Fig 5). In the three responder populations, maximal migration was also observed at 10 nmol/L eotaxin; however, significant numbers of thymocytes had migrated in response to 0.01 nmol/L eotaxin, indicating a potent action of eotaxin in this assay. In addition, it was interesting to note that, in accordance with the higher levels of CCR-3 on CD8+ thymocytes than on CD4+ thymocytes, a considerably greater number of CD8+ SP cells than CD4+ SP cells migrated in response to eotaxin. In a similar manner to unfractionated cells, migration was completely inhibited by PTX pretreatment (not shown).
Migration of fractionated thymocytes. Human thymocytes, labeled with fluorescent antibodies to CD4 and CD8, were sorted into DN (CD4−CD8−), DP (CD4+CD8+), SP (CD4+), and SP (CD8+) populations for analysis of migration. Cells were used immediately after sorting and migration assessed as previously described.22 Each point represents the mean ± SE mean number of migrated cells per 5 high power fields (×400) from n = 3 experiments performed in duplicate. The dot-plot is representative of the gated fields from which the pure populations were obtained. Each population was greater than 99.6% pure (n = 3).
Migration of fractionated thymocytes. Human thymocytes, labeled with fluorescent antibodies to CD4 and CD8, were sorted into DN (CD4−CD8−), DP (CD4+CD8+), SP (CD4+), and SP (CD8+) populations for analysis of migration. Cells were used immediately after sorting and migration assessed as previously described.22 Each point represents the mean ± SE mean number of migrated cells per 5 high power fields (×400) from n = 3 experiments performed in duplicate. The dot-plot is representative of the gated fields from which the pure populations were obtained. Each population was greater than 99.6% pure (n = 3).
Eotaxin induces chemokine release from human thymocytes.
To further assess the biological activities of eotaxin, thymocytes were cultured in the presence of increasing concentrations of eotaxin to determine whether there were any proliferative or phenotypic changes. Over a period of 48 hours, eotaxin induced little if any change in CD25, CD3, CD4, and CD8 expression, as measured by FACS analysis (not shown). In addition, there was no change in total cell numbers or3H-thymidine incorporation, in contrast to IL-7, IL-2, or PHA, which induced thymocyte proliferation as expected (not shown). However, interestingly, eotaxin induced significant increases in chemokine release from thymocytes at doses of 1 to 100 nmol/L (Table 1). Specifically, a mean of 1.4 ng of MIP-1β (range, 1.17 to 1.84 ng) was released from 106thymocytes stimulated with 100 nmol/L eotaxin, whereas a mean of 100 pg (range, 82 to 131 pg) was released from PBS-stimulated thymocytes. Likewise, 205 pg (range, 65 to 371 pg) and 229 pg (range, 106 to 345 pg) of RANTES and IL-8, respectively, were secreted from eotaxin-stimulated thymocytes. In contrast, only 35 pg (range, 16 to 55 pg) and 130 pg (range, 45 to 198 pg) of these two chemokines were detected in PBS-stimulated thymocytes.
DISCUSSION
We have demonstrated that eotaxin protein is expressed in human thymus and that, upon binding its specific receptor CCR-3, it elicits chemotaxis, Ca2+ mobilization, and cytokine release by thymocytes. These results provide information on a potentially novel biological function for eotaxin, beyond the activation of eosinophils and Th2 lymphocytes. The functional correlate of this restricted expression of eotaxin in the medullary region and Hassle’s corpuscles of the thymus remains to be elucidated. Although the eotaxin knock-out mouse does not show compromized T-cell development, a systematic analysis of the migratory patterns of thymic precursors and thymocytes has yet to be undertaken. T-cell development in the thymus requires the regulated migration of selected progenitors from the cortex to the medulla, ultimately resulting in the emigration of selected T cells into the periphery. Interestingly, CD8+ cells expressed the highest levels of CCR-3 receptor, and a greater number of cells in this subpopulation migrated in response to eotaxin, suggesting a preferential activation role for eotaxin in CD8+thymocytes. Although far from unequivocal, the data presented here suggest that eotaxin may be involved in the regulation of cell trafficking in the thymus, because (1) its expression is highly localized to the medulla and (2) thymocytes at different stages of maturation respond differently to eotaxin stimulation. In contrast, Zhang et al26 did not detect any lymphoid cell staining when using the 7B11 antibody directed against CCR-3. The reasons for this discrepancy remain unclear, although it is relevant to note that the antibody is specific for human CCR-3; thus, species differences as well as methodological and preparative differences in obtaining thymocytes may account for any contrasts between the two studies.
Eotaxin induced a greater than 10-fold increase in the chemokines RANTES and MIP-1β, suggesting a prominent role for these two chemokines in thymic physiology. However, the significance of such a robust chemokine release in the thymus is unclear. Of the chemokines measured, only MIP-1β has thus far been shown to have significant effects on thymocytes,27 although both RANTES and MIP-1β can bind to the same receptor (CCR-5). Interestingly though, both RANTES and IL-8 have been shown to play significant migration and activation roles in lymphoid cells.28-30 The induction of these chemokines may therefore represent an autocrine feedback mechanism in the regulation of thymocyte migration within and from the thymic environment. However, the validation of such an hypothesis does await further experimentation. It is relevant to point out that recent studies have demonstrated the induction of IFN-γ and tumor necrosis factor-α release after T-cell activation by various chemokines.31 Although we have not directly measured other cytokines, it is interesting to speculate that this precedent for chemokine-induced chemokine release may be of functional significance in lymphocyte populations.
Although we have not begun to investigate the maturation stages of the lymphocyte populations expressing CCR-3 or their Th1/Th2 polarization, our findings of CD8+ lymphocyte expression of CCR-3 supports the recent findings of CD8+ T-cell clone (Th2) expression.32 Whereas CD4+ cells were less responsive to eotaxin in a chemotaxis assay than CD8+cells, the CD4+ SP population still showed fairly robust migratory potential to this chemokine. Because the levels of detectable CCR-3 on CD4+ SP cells were very low, the reason for this biological effect of eotaxin is as yet unclear. However, it may support the phenomenon of spare receptors or suggest that eotaxin can act through an alternative receptor, such as CXCR-333; alternatively, there may be indirect activation capacities through the release of other chemoattractant factors. The secretion of MIP-1β, RANTES, and IL-8 may indeed fulfill such a role.
Although CCR-3 has been shown to function as a coreceptor for HIV-1,19,20 the role of this receptor in HIV-1 infection in the embryo and neonate remains questionable. There are significantly fewer CCR-3 than CXCR-4 and CCR-5 receptors in the thymus (Res et al, manuscript submitted; and Zhang et al26); however, its presence complements the repertoire of primary HIV coreceptors at this site of T-cell development and thus may provide a mechanism for efficient HIV entry.
These observations detail the expression of both eotaxin and its specific receptor in a tightly regulated T-cell environment. Whereas the biological importance of this action is, as yet, not fully understood, these data suggest that eotaxin may play an important role in thymic physiology.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Kevin B. Bacon, PhD, Neurocrine Biosciences, Inc, 10555 Science Center Dr, San Diego, CA 92121; e-mail: Kbacon@neurocrine.com.