Abstract
Alloreactive T lymphocytes that respond directly to foreign major histocompatibility complex (MHC) molecules and bound peptide are known to be central mediators of graft-versus-host disease (GVHD) and allograft rejection. We have recently identified a peptide from the human protein, cytochrome P450 (isotypes IIC9, 10, or 18), that is recognized in association with human leukocyte antigen (HLA) B*3501 by alloreactive cytotoxic T lymphocytes (CTLs). These CTLs with this specificity were isolated from several unrelated individuals and were found to express a common T-cell receptor (TCR). Synthetic analogs of the cytochrome P450 peptide were generated by introducing single amino acid substitutions at putative TCR contact positions. Four altered peptide ligands were powerful competitive antagonists of these CTL clones, reducing lysis levels of target cells expressing the alloantigen HLA B*3501 by over 80%. This first demonstration that it is possible to suppress CTL alloreactivity with structural variants of allodeterminants raises the prospect that such TCR antagonists could be exploited within the clinical arena to specifically modulate GVHD and allograft rejection.
T CELLS CAN RECOGNIZE foreign major histocompatibility complex (MHC) antigens by two distinct routes. The direct pathway involves the recognition of foreign MHC antigens as intact molecules on the surface of allogeneic stimulator cells. In most cases, endogenous peptides, which constitutively bind to MHC antigens, are thought to be an integral part of the epitopes recognized via this route.1 The indirect pathway of allorecognition requires the presentation of peptides, derived from foreign MHC antigens, on an MHC molecule shared with the alloreactive T cell.2 Although this self-restricted, indirect presentation of allopeptides may play an important role in chronic allograft rejection, the direct pathway of allorecognition is the principal contributor to cytotoxic T lymphocyte (CTL) responses mediating graft-versus-host disease (GVHD) and early allograft rejection episodes.3
Although the repertoire of T cells available for use in an alloresponse against a single allo-MHC molecule is diverse,1 the actual repertoire used may be highly selected. Studies of the clonal heterogeneity of alloreactive T-cell populations infiltrating human allografts undergoing rejection have shown highly selected T-cell receptor (TCR) usage, with a single clone predominating in some cases.4-6 In vivo clonal expansions of T cells with selected TCR usage have also been identified during acute GVHD and persistence of these T cells for up to 1 year has been reported.7,8 Although the basis for this limited diversity is unclear, it is possible that preexisting expansions of alloreactive T cells, which have recently been demonstrated in healthy individuals, could be involved. For example, a CTL clonotype with specificity for an alloantigen was shown to be expanded in the periphery of a healthy individual who had never been exposed to that alloantigen.9This was thought to represent a cross-reactive memory T-cell population that had been raised against a self-MHC–restricted foreign antigen, which could not be identified.
Studies from our laboratory have also characterized alloreactive T-cell expansions in healthy individuals. These were shown to be driven by cross-reactive stimulation with the persistent herpes virus Epstein-Barr virus (EBV).10,11 These EBV/allo-cross–reactive CTL expansions were so large that such T cells dominated conventional mixed lymphocyte cultures from some individuals. This graphically illustrates how a prior history of infection with an immunogenic virus such as EBV can influence an individual’s level of responsiveness to an alloantigen; such mechanisms may underlie the observed clinical association between herpes virus exposure and GVHD.12
The limited use of the TCR repertoire in GVHD and allograft rejection may provide the opportunity to therapeutically disrupt the alloresponse by targeting a selected T-cell population for inactivation, as has been achieved in experimental animal models.13 In the present report, we describe the inactivation of a CTL clonotype that displays direct alloreactivity for HLA B*3501. We have found previously that this clonotype recurs in unrelated healthy individuals and is preexpanded due to cross-reactive antigenic stimulation with a self-MHC–presented epitope from Epstein-Barr nuclear antigen 3A.11 A peptide corresponding to regions of the human isoenzymes cytochrome P450 IIC 9, 10, and 18 is recognized by this clonotype in association with the alloantigen HLA B*3501. We now show that it is possible to use analogs of this cellular peptide to specifically suppress the direct alloreactivity of these CTL clones, via altered peptide ligand antagonism.
MATERIALS AND METHODS
Establishment and maintenance of cell lines.
Lymphoblastoid cell lines (LCLs) were established by exogenous transformation of peripheral B cells with EBV derived from the B95.8 cell line.14 Phytohemagglutinin (PHA) blasts were generated by stimulating peripheral blood mononuclear cells (PBMCs) with PHA (CSL, Melbourne, Australia), and after 3 days, growth medium containing supernatant from the MLA-144 cell line and recombinant interleukin-2 (rIL-2) was added. PHA blasts were propagated with biweekly replacement of rIL-2 and MLA-144 supernatant (PHA free) for up to 8 weeks. The CTL clones used in this study have been described previously.11 All cell lines were regularly screened for mycoplasma contamination.
Cytotoxicity assay.
CTL clones were tested in duplicate for cytotoxicity in the standard 5-hour chromium release assay, using an effector:target ratio (E:T) of 2:1. Where synthetic peptide was involved, it was added directly to51Cr-labeled targets and incubated for 1 hour before CTL addition and remained present throughout the assay. Peptides were synthesized by Chiron Mimotopes (Chiron Corp, Emeryville, CA) on a 1-mg scale using Pin-Technology.15 Toxicity testing of all peptides was performed before use by adding peptide to51Cr-labeled PHA blasts in the absence of CTL effectors. A beta scintillation counter (Topcount Microplate; Packard Instrument Co, Meriden, CT) was used to measure 51Cr levels in assay supernatant samples. The mean spontaneous lysis for target cells in culture medium was always less than 20%, and the variation about the mean specific lysis was less than 10%.
RESULTS AND DISCUSSION
Specific inhibition of T cells participating in alloimmune responses is the ultimate goal of research in transplantation because currently available immunosuppressive therapy is nonspecific, impairing the entire immune system. Previous studies of self-MHC–restricted T cells have shown that analogs of immunogenic peptides, so-called altered peptide ligands (APLs), may profoundly reduce the magnitude of the response to the wild-type epitope.16-22 To evaluate the potential of APLs as specific modulators of alloreactivity, a CTL clonotype, previously characterized by our group, was examined further. These CTL clones that use identical TCRs display direct alloreactivity for HLA B*3501 and were isolated from two unrelated individuals.11 Although a variety of cell types that express this alloantigen are lysed by these clones (including LCLs, Burkitt’s lymphoma cell lines, and a bladder cancer cell line), HLA B*3501+ PHA-stimulated T-cell blasts are not recognized. The peptide KPIVVLHGY, derived from human cytochrome P450 IIC 9, 10, or 18, was found to be recognized by these cross-reactive clones when bound to the alloantigen HLA B*3501 on PHA blasts.11
Fifty-seven monosubstituted peptide analogs of KPIVVLHGY were synthesized in which the potential TCR contact residues at positions 6, 7, and 8 were sequentially replaced with all other genetically coded amino acids. Because only analogs with significantly less activity than the parent allodeterminant are potential antagonists of this CTL clonotype, each analog was first screened for activity as an agonist by testing for the capacity to sensitize HLA B*3501+ PHA blasts to lysis. CTL clone JL12, a representative clone expressing this alloreactive TCR, was used as an effector in a standard51Cr-release assay against PHA blast target cells from donor LP (HLA A2, A32, B*3501, B62) after pretreatment with three different concentrations of each peptide. As shown in Fig 1, 51 of 57 peptides were either inactive or required peptide concentrations over 100-fold higher than the parent peptide for comparable lysis levels by CTL clone JL12. The only amino acid replacements tolerated well by the clone were phenylalanine, histidine, methionine, or tyrosine instead of leucine at position 6, and phenylalanine or tyrosine instead of histidine at position 7. All amino acid substitutions at position 8 of KPIVVLHGY resulted in either loss of, or a large reduction in, allospecific lysis of the PHA blast target cells.
Recognition by CTL clone JL12 of HLA B*3501+ PHA blasts with the addition of monosubstituted peptide analogs of KPIVVLHGY. Every one of the 20 genetically coded amino acids was tested in each of positions 6, 7, and 8 within the parent sequence KPIVVLHGY. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [ ], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
Recognition by CTL clone JL12 of HLA B*3501+ PHA blasts with the addition of monosubstituted peptide analogs of KPIVVLHGY. Every one of the 20 genetically coded amino acids was tested in each of positions 6, 7, and 8 within the parent sequence KPIVVLHGY. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [ ], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
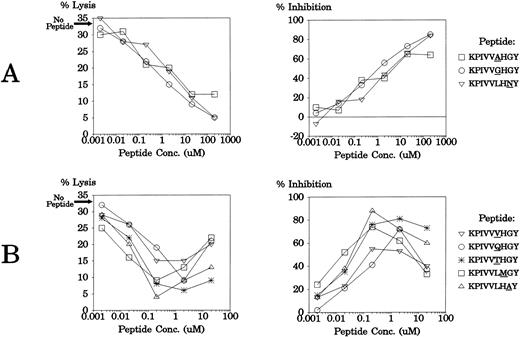
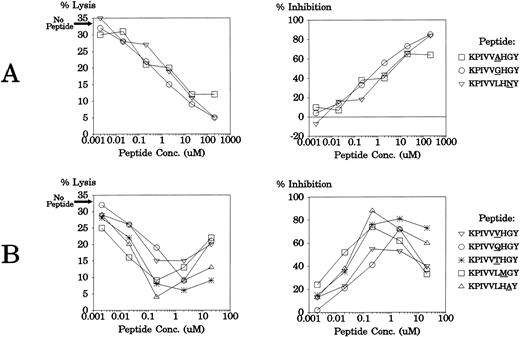
The 51 monosubstituted peptide analogs of KPIVVLHGY that had significantly less agonist activity than the parent peptide were then tested for their ability to inhibit alloreactivity by this CTL clonotype. LCLs from donor AF (HLA A1, A11, B*3501, B51) were treated with 200 μmol/L, 2 μmol/L, or 0.02 μmol/L of each peptide before being tested for lysis by CTL clone JL12. These LCLs were also used as targets for the clone without peptide pretreatment, and the level of allospecific lysis was 33.1%. Data are presented in Fig 2 as percent inhibition of lysis, relative to this lysis level observed without exogenous peptide addition. As shown in Fig 2, many of the peptide analogs showed the capacity to significantly reduce allospecific lysis below 33.1%. Eight APLs were powerful competitive antagonists of these CTL clones, reducing lysis levels of the HLA B*3501+ LCL target cells by over 50%. In some cases, these antagonist peptides inhibited CTL lysis most efficiently at 200 μmol/L, the highest concentration (KPIVVAHGY, KPIVVGHGY, and KPIVVLHNY). Other APLs (KPIVVQHGY, KPIVVTHGY, KPIVVVHGY, KPIVVLMGY, KPIVVLHAY), showed differential effects on the CTL clone, inhibiting lysis most effectively at 2 μmol/L, but increasing lysis levels at 200 μmol/L (negative inhibition values are shown as 0% in Fig 2).
Inhibition of anti-HLA B*3501 allospecific lysis by CTL clone JL12 with peptide analogs of KPIVVLHGY. An HLA B*3501+ LCL was tested for lysis by CTL clone JL12 after pretreatment with selected monosubstituted peptide analogs of KPIVVLHGY. The level of lysis of these LCLs without peptide pretreatment was 33.1%. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue; the vertical axis displays the percent inhibition of lysis relative to the level of lysis of the LCL without exogenous peptide addition. Where peptide increased lysis of the B*3501+ LCL, a value of 0% inhibition is shown. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [ ], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
Inhibition of anti-HLA B*3501 allospecific lysis by CTL clone JL12 with peptide analogs of KPIVVLHGY. An HLA B*3501+ LCL was tested for lysis by CTL clone JL12 after pretreatment with selected monosubstituted peptide analogs of KPIVVLHGY. The level of lysis of these LCLs without peptide pretreatment was 33.1%. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue; the vertical axis displays the percent inhibition of lysis relative to the level of lysis of the LCL without exogenous peptide addition. Where peptide increased lysis of the B*3501+ LCL, a value of 0% inhibition is shown. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [ ], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
These eight APLs were then tested at six different peptide concentrations for antagonistic activity using a different CTL clone (JL20), isolated from the same individual, that shares an identical TCR with JL12.11 The target cells used were LCLs from donor CS (HLA A3, A23, B*3501, B44) and these were lysed at 32.7% without peptide addition. As shown in Fig3, all peptides again showed strong antagonistic effects on the TCR. As in the earlier experiment, some of the APLs displayed optimal inhibitory effects at the highest concentration (Fig 3A), while others blocked lysis most efficiently at 0.2 μmol/L or 2 μmol/L (Fig 3B; data not shown for the 200-μmol/L peptide level where these APLs showed agonistic activity).
Antagonism of CTL clone JL20 with eight different APLs. An LCL from donor CS (HLA A3, A23, B*3501, B44) was tested for lysis by CTL clone JL12 after pretreatment with a range of concentrations of synthetic peptide APLs. The data are presented as percent lysis and percent inhibition of lysis relative to the level of lysis of the LCL without exogenous peptide addition (32.7%). The E:T ratio was 2:1.
Antagonism of CTL clone JL20 with eight different APLs. An LCL from donor CS (HLA A3, A23, B*3501, B44) was tested for lysis by CTL clone JL12 after pretreatment with a range of concentrations of synthetic peptide APLs. The data are presented as percent lysis and percent inhibition of lysis relative to the level of lysis of the LCL without exogenous peptide addition (32.7%). The E:T ratio was 2:1.
The discovery that TCR antagonist peptides can inhibit the function of T cells specific for conventional self-MHC–restricted antigens has raised hopes for their clinical application in modulating harmful immune responses.16-22 Indeed, such novel reagents based on the structure of autoantigenic epitopes have enjoyed some measure of success in the treatment of experimental models of autoimmunity.19-22 The present report reinforces such hopes in relation to the potential use of APLs as specific therapeutics for GVHD and allograft rejection in humans by showing, for the first time, that CTL alloreactivity can be inhibited with analogs of an allo-MHC–bound peptide ligand. Also supporting this notion is a recent demonstration that indirect alloreactivity by T helper cells can be suppressed by TCR antagonism.23 A report from Paul Allen’s laboratory also demonstrated the effectiveness of APLs in inhibiting the proliferation and activation of alloreactive T helper cells.24 The relevance of this latter model to human transplantation is less clear, however, because the TCR antagonist peptides were presented by a self-MHC molecule that was coexpressed on the stimulator cells with the allo-MHC molecule. Nonetheless, this murine study corroborates our basic observation that direct alloreactivity can be just as susceptible to APL antagonism as self-MHC–restricted T-cell reactivity. This result is surprising given that recent studies have shown that TCR affinities for allo-MHC–restricted ligands tend to be higher than for self-MHC–restricted ligands.25 26
The present report describes the antagonism of a particularly interesting class of alloreactive T cells, ie, clonotypes that are preexpanded in healthy individuals due to cross-reactivity with common environmental stimuli. Although the importance of such T cells in transplantation is not yet clear, it seems possible that the APLs defined in this study could ultimately be used clinically to block these preactivated T-cell populations in appropriate transplant recipients to modulate either the graft-versus-host or anti-allograft response.
Conclusion.
To achieve long-lasting, antigen-specific unresponsiveness is the ultimate goal in transplant biology. As the underlying mechanisms that contribute to allorecognition become more clearly defined, the prospect of finding effective methods to specifically prevent the clinical complications associated with alloreactivity is enhanced. The biggest obstacle to such approaches is likely to be the potential diversity of the TCR repertoire in the response to alloantigens. However, because the alloreactive T cells infiltrating human allografts undergoing rejection often use a highly selected TCR repertoire, it is certainly possible that specific immunomodulating techniques such as APL antagonism will find application in human transplantation.
Supported by grants from the National Health and Medical Research Council.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Scott R. Burrows, PhD, Queensland Institute of Medical Research, The Bancroft Centre, 300 Herston Rd, Brisbane 4029, Australia; e-mail: scottB@qimr.edu.au.

![Fig. 1. Recognition by CTL clone JL12 of HLA B*3501+ PHA blasts with the addition of monosubstituted peptide analogs of KPIVVLHGY. Every one of the 20 genetically coded amino acids was tested in each of positions 6, 7, and 8 within the parent sequence KPIVVLHGY. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/3/10.1182_blood.v93.3.1020/5/m_blod40306001x.jpeg?Expires=1766375053&Signature=NQtVElgAN3NRKeMRiudgQQOV4ASauUrJ8BJOqQ8RjeHLBMLTmR8x2kBLeqQQMlqk0YvDCOJt3IqdlRkDc2BJiywUqohtmvq~Gy5Mt1RVHgVtMZ2LBHZAlbb5yXAWk8Nv2K0CO3Hf23LiM7fPreDcVsaPtj4-t7zGWqgLNrnrIABIYHZC2rmVAmWQkTGOX7nPlzcu~c54Bkuks7-7mLybKmsxEp0wLibSIGIc5ojEXUo9U3pHgW6BeRNlyCt2J~JRsM97b0i2~PmZH21Gmsmv52C5FU3pCHHKwYHsZXRddot5uuU6CE3qnc-uzh2LcDJrlR3Tq6GwAtniFBOwPJzbvg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Inhibition of anti-HLA B*3501 allospecific lysis by CTL clone JL12 with peptide analogs of KPIVVLHGY. An HLA B*3501+ LCL was tested for lysis by CTL clone JL12 after pretreatment with selected monosubstituted peptide analogs of KPIVVLHGY. The level of lysis of these LCLs without peptide pretreatment was 33.1%. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue; the vertical axis displays the percent inhibition of lysis relative to the level of lysis of the LCL without exogenous peptide addition. Where peptide increased lysis of the B*3501+ LCL, a value of 0% inhibition is shown. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/3/10.1182_blood.v93.3.1020/5/m_blod40306002x.jpeg?Expires=1766375053&Signature=nUEXM1zCJA4ffWtwQKzKgPFWLjAtXhkZesetg4ylhIG~MjJMOaLM05LIq14w6~qgA1w11BdFU8Bm~Lu1Rb21R~xjhWWKczq2sD0kl40Q7fmZ-yuWZGMyr0qtGHTOOTYfUNt9MAkau387UR7B8aF18FUOvJbMp8Cmd5Eb8bNP7h7ukeGqZMLcQtUvKZuJ4Z~3bS7sRv~fB~gUMUfLa1N5hgi6l4K4ThX7kW7bJQFPhoXnPDvcX6fgR2dUxeuJ54VSZWkXpkdTyEjCDU7RXLkdVIgLCcdZ3qiIjyDbr8Po3VFpBgYCCrPJI84aWbJTnAmRth3cq2F2R1GByr941coOFw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 1. Recognition by CTL clone JL12 of HLA B*3501+ PHA blasts with the addition of monosubstituted peptide analogs of KPIVVLHGY. Every one of the 20 genetically coded amino acids was tested in each of positions 6, 7, and 8 within the parent sequence KPIVVLHGY. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/3/10.1182_blood.v93.3.1020/5/m_blod40306001x.jpeg?Expires=1766375054&Signature=B7G-mv4zM7-TXZt5Fwr3TmkgQ6bsevBAnwQIIg7aF5QDCjd~trzPwaGfefAyf8rRpFEu3xgvPDmPLxInooxACtp~GIR7oL3-Jt0fOMnatld4g7EN4i3sMCBCKhxh~WsRt7fCYiMhXRJZqMrSxzfSIVcJpGbavAB~d0cP-~sDCk80occJE4iaRJQKRxWAKJv3oPzV7zOMox9EVTdQwDI3BkcjczK2gx3tC7myKcXSKdZDvwMvhL5FXusA6Ja7UuLUDL6wTUtZwrdCQ1c-5B-YM7cNEEIpuuWxVv05O5t73r6hXtdPthEACnCA4NZcoo540jMHtSTHTcIWAKmUvAywTQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
 ], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.
], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.![Fig. 2. Inhibition of anti-HLA B*3501 allospecific lysis by CTL clone JL12 with peptide analogs of KPIVVLHGY. An HLA B*3501+ LCL was tested for lysis by CTL clone JL12 after pretreatment with selected monosubstituted peptide analogs of KPIVVLHGY. The level of lysis of these LCLs without peptide pretreatment was 33.1%. The letter within each graph represents the parent residue being replaced; the horizontal axis lists the residue replacing the parent residue; the vertical axis displays the percent inhibition of lysis relative to the level of lysis of the LCL without exogenous peptide addition. Where peptide increased lysis of the B*3501+ LCL, a value of 0% inhibition is shown. Three different peptide concentrations were used (200 μmol/L [▪], 2 μmol/L [], and 0.02 μmol/L [□]) and the E:T ratio was 2:1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/3/10.1182_blood.v93.3.1020/5/m_blod40306002x.jpeg?Expires=1766375054&Signature=osK-K54ka6W9hQPm-mTbKZJizjuX7MLo4vbNc7exL7qwhuJgwzO3TPu2SYTqR2Zgi62rvHqDdlZz6Acl~NuHnsSq9JCx229RQz9jnqZvh1ZD4UxnB5Zx6MRQHVs86KWdPAYjtHHrrZvqAuMQK1cAV36e5bJST39nihMf9-UwrAcMWitQE-BrHzjZVqTjYqHCRszWnw2QR6MiKTYltkIm-Lpj00cGentEJ92CVPQh~ZpXvq1U~8Wn9aiik6d90cgs5SKISGNI4Lr7Qyz4~vOv11lSOzs4y~3C576~BdqQFsksNoTjxGFksDSKEQGe4fN4T47B8KuOfP049K29NGf~QQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)