Abstract
Oncostatin M (OSM) is a member of the interleukin-6 (IL-6) family of cytokines that share the gp130 receptor subunit. Of these family members, leukemia inhibitory factor (LIF) is most closely related to OSM, and various overlapping biologic activities have been described between human LIF and OSM (hLIF and hOSM). Two types of functional hOSM receptors are known: the type I OSM receptor is identical to the LIF receptor that consists of gp130 and the LIF receptor β subunit (LIFRβ), and the type II OSM receptor consists of gp130 and the OSM receptor β subunit (OSMRβ). It is thus conceivable that common biologic activities between hLIF and hOSM are mediated by the shared type I receptor and OSM-specific activities are mediated by the type II receptor. However, in contrast to the human receptors, recent studies have demonstrated that mouse OSM (mOSM) does not activate the type I receptor and exhibits unique biologic activity. To elucidate the molecular structure of the functional mOSM receptor, we cloned a cDNA encoding mOSMRβ, which is 55.5% identical to the hOSMRβ at the amino acid level. mOSM-responsive cell lines express high-affinity mOSM receptors, as well as mOSMRβ, whereas embryonic stem cells, which are responsive to LIF but not to mOSM, do not express mOSMRβ. mOSMRβ alone binds mOSM with low affinity (kd = 13.0 nmol/L) and forms a high-affinity receptor (kd = 606 pmol/L) with gp130. Ba/F3 transfectants expressing both mOSMRβ and gp130 proliferated in response to mOSM, but failed to respond to LIF and human OSM. Thus, the cloned mOSMRβ constitutes an essential and species-specific receptor component of the functional mOSM receptor. Reminiscent of the colocalization of the mOSM and mLIF genes, the mOSMRβ gene was found to be located in the vicinity of the LIFRβ locus in the proximal end of chromosome 15.
THE INTERLEUKIN-6 (IL-6) family of cytokines includes IL-6, IL-11, leukemia inhibitory factor (LIF), oncostatin M (OSM), ciliary neurotrophic factor (CNTF), and cardiotrophin (CT).1-5 A unique feature of this cytokine family is that the receptors share the gp130 receptor subunit as a common signal transducer.6 Of the family members, LIF and OSM are most closely related structurally7 and their genes are colocalized.8,9 Human OSM (hOSM) and LIF also exhibit various common biologic activities such as growth stimulation of the DA1.a lymphoid subline,10 induction of M1 monocytic leukemia cell differentiation,11 inhibition of embryonic stem (ES) cell differentiation,12 and induction of acute-phase protein expression in hepatocytes.13,14 In addition, hOSM exhibits unique activities including growth inhibition of A375 human melanoma,15,16 growth stimulation of Kaposi’s sarcoma,17 and induction of tissue inhibitor of metalloproteinase-1 (TIMP-1) in fibroblasts.18 Molecular cloning of the hOSM receptor subunit (hOSMRβ) and reconstitution of the functional hOSM receptor indicated two types of functional hOSM receptor. The type I OSM receptor is identical to the high-affinity LIF receptor that consists of gp130 and the LIF binding protein, LIFRβ.19 The type II OSM receptor consists of gp130 and the OSM-specific receptor component, OSMRβ.20,21 gp130 binds hOSM with low affinity19,22,23 and forms a high-affinity hOSM receptor with OSMRβ. The existence of two types of functional hOSM receptors provides a molecular basis for the common biologic activities between LIF and OSM, as well as OSM-specific activities.24
The mouse OSM (mOSM) gene had long been unidentified, but it was recently cloned as a cytokine-inducible gene.25 Biologic characterization of mOSM revealed that its activities are significantly different from those of hOSM. Recent studies have shed light on the unique biologic activities of OSM during mouse development. First, mOSM plays an important role in the expansion of multipotential hematopoietic progenitor cells in the aorta-gonad-mesonephros (AGM) region of the mouse embryo at 11.5 days postcoitum (dpc),26where definitive hematopoiesis is believed to be initiated in the mouse embryo.27 Second, Sertoli cells in neonatal testes express mOSM, and their proliferation is strongly stimulated by mOSM.28 Third, maturation of hepatic cells is induced by mOSM (Kamiya et al, submitted). Interestingly, neither LIF nor hOSM exhibit such activities.
Previously, we demonstrated that although both hOSM and LIF stimulate proliferation of DA1.a, induce differentiation of M1 cells, and inhibit differentiation of ES cells, none of these activities are observed with mOSM.29 Conversely, while mOSM inhibits proliferation of a subline of NIH3T3, neither hOSM nor LIF exhibit such activity. Moreover, Ba/F3 transfectants expressing gp130 and LIFRβ proliferate in response to either LIF or hOSM, but fail to respond to mOSM. These results suggested that the mLIF receptor is activated equally by LIF and hOSM, but mOSM is unable to transduce signals through the mLIF receptor. Consistent with this hypothesis, while we could detect high-affinity LIF receptors on LIF-responsive cell lines, no high-affinity mOSM receptors were present on such cells. Similarly, we found high-affinity mOSM receptors in mOSM-responsive NIH3T3 cells that do not exhibit any LIF binding. Based on these findings, we previously proposed that unlike hOSM, mOSM does not use the type I receptor equivalent to the LIF receptor; instead, mOSM elicits biologic activity only through its specific type II receptor, which is presumably composed of gp130 and a putative mOSMRβ.29
In this report, we describe the molecular cloning of mOSMRβ cDNA and present evidence to prove our model by reconstituting the functional high-affinity mOSM receptor using this molecule.
MATERIALS AND METHODS
Cells and cytokine.
Ba/F3 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), gentamicin (50 μg/mL), and IL-3 (10 ng/mL). M1 cells were cultured in RPMI 1640 containing 10% FBS and gentamicin. NIH3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% calf serum and gentamicin. CCE (embryonic stem [ES] cell line) cells were maintained in DMEM supplemented with 15% FBS, gentamicin, 50 μmol/L 2-mercaptoethanol, and mLIF (10 ng/mL). LO cells were cultured in DMEM supplemented with 15% FBS, gentamicin, 2 mmol/L l-glutamine, 1 mmol/L sodium pyruvate, and mOSM (10 ng/mL). mOSM was produced in COS7 cells and purified.29Recombinant mouse IL-3 was produced in silkworm.30 hOSM and mLIF were purchased from R&D Systems (Minneapolis, MN).
Reverse transcriptase–polymerase chain reaction cloning using degenerated primers.
Poly A+ RNA was prepared from an OSM-dependent cell line, LO (T. Hara, A. Miyajima, unpublished data, October 1997) using the FastTrack kit (Invitrogen, San Diego, CA) according to the manufacturer’s instructions. Two degenerated primers were designed on the basis of homology between hOSMR and LIF (A, 5′-CA(A/G)GAAA(C/T)(A/C)(C/T)A(C/T)AA(C/T)TT(C/T)AC-3′; B, 5′-G(A/G)A(A/C)AG(A/G/T)AT(C/T)TT(A/C/G/T)CC(A/G)TT(A/G/T)GC-3′). The polymerase chain reaction (PCR) was performed using a set of these primers under the following conditions: denaturation for 3 minutes at 94°C before thermal cycling, denaturation for 1 minute at 94°C, annealing for 2 minutes at 46°C, extension for 3 minutes at 72°C for 35 cycles, and a final extension for 7 minutes at 72°C. This program was repeated using 1 μL of the first PCR solution as the template for the second PCR. PCR products were separated on 2% agarose gel. DNA fragments of approximately 500 bp were isolated by the QIAquick Gel Extraction kit (Qiagen, Santa Clarita, CA) and subcloned into the pCRII vector (Invitrogen). The sequence of cloned DNA fragments was determined by an automated DNA sequencer (Applied Biosystems, Foster City, CA).
Screening of cDNA library.
cDNA libraries were constructed using polyA+ RNA from LO cells with oligo dT primers or random primers as described previously.31 The probe for Southern and colony hybridization was prepared by PCRs with a NTP mixture containing digoxigenin-UTP (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer’s protocol. As previously described,31 pools (1,500 independent clones per pool) of the cDNA library were prepared in 96-well plates. DNA mixtures of each row were subjected to Southern blot analysis to identify a positive pool that contains a longer cDNA insert. Overlapping clones that cover the entire open reading frame encoding mOSMRβ were isolated by sibselection. 5′RACE analysis was performed using the 5′-rapid amplification of cDNA end (RACE) System for Rapid Amplification of cDNA Ends, Version 2.0 (GIBCO-BRL, Rockville, MD) according to the manufacturer’s protocol. Each primer (5′-GGAGTCAATGGTAAAGGCTC-3′ and 5′-CTCCAAGACTTCGCTTCGG-3′) was used for synthesis of first-strand cDNA and PCR, respectively.
Northern blot analysis.
PolyA+ RNA was prepared from LO, NIH3T3, M1, Ba/F3, and CCE cells using the FastTrack kit (Invitrogen) according to the manufacturer’s instructions. Standard Northern blots of 1 μg polyA+ RNA were performed. The single-strand antisense probe for Northern blot analysis was made as described before in the absence of a sense primer.
Generation of Ba/F3 transfectants.
mOSMRβ cDNA was placed under the SRα promoter of the expression vector pME18S32 carrying the puromycin-resistant gene. As previously described,31 linearized plasmids (30 μg) were transfected into 5 × 106 Ba/F3 cells by electroporation, and transfectants were selected with puromycin (1 μg/mL) for 10 days. Expression of mOSMRβ was confirmed by flow cytometry using anti-mOSMRβ monoclonal antibody (M. Tanaka, T. Hara, A. Miyajima, unpublished data, November 1997). A double transfectant of Ba/F3 was established by introducing a plasmid DNA carrying gp130 cDNA and the neomycin-resistant gene into the mOSMRβ transfectant.
Radioiodination and binding assay.
The purified recombinant mOSM derived from COS7 cells was radioiodinated with Iodogen (Pierce, Rockford, IL) as previously described.33 The specific radioactivity was determined to be 4.2 × 106 cpm/pmol by self-displacement analysis. Scatchard analyses were performed as previously described.34 Data for the binding assays were analyzed by the LIGAND program.35
Chemical cross-linking experiment.
A chemical cross-linking experiment was performed as described previously.34 In brief, LO cells (4 × 106) were incubated in 250 μL DME-BSA (DMEM containing 1 mg/mL bovine serum albumin [BSA] and 20 mmol/L HEPES, pH 7.4) with 5 nmol/L 125I-mOSM in the presence or absence of a 1,000-fold excess of nonlabeled ligands at 4°C for 4 hours. Cells collected by centrifugation at 5,000 rpm for 30 seconds were washed with 500 μL ice-cold phosphate-buffered saline (PBS) twice, and unbound labeled ligands were removed. Cell-bound labeled ligands were cross-linked with 1 mmol/L disuccinimidyl suberate in 200 μL 0.1 mol/L borate buffer, pH 8.0, at 4°C for 15 minutes. The reaction was quenched by washing twice with 500 μL stopping buffer (10 mmol/L Tris hydrochloride, pH 7.4, 0.14 mol/L NaCl, and 1 mmol/L EDTA). Cross-linked cells were lysed and subjected to 6.5% polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS) followed by autoradiography.
Proliferation assay.
The proliferative response of Ba/F3 transfectants was examined by colorimetric assays using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide ([MTT] Sigma, St Louis, MO) as described previously.36 In brief, 104 cells were incubated in 100 μL medium with various concentrations of mOSM in 96-well plates. After 3 days in culture, 10 μL MTT (5 mg/mL in PBS) was added to each well and further incubated for 4 hours at 37°C. Then, 150 μL 0.04N HCl in isopropanol was added to lyse the cells, and the optical density at 570 nm was measured.
Interspecific mouse backcross mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J × M. spretus) F1 females and C57BL/6J males.37 A total of 205 F2 mice were used to map theOsmr locus (see text for details). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as previously described.38 All blots were prepared with Hybond-N+ nylon membrane (Amersham, Arlington Heights, IL). The probe, about 3.1 kb of XhoI/NotI fragments of mouse cDNA, was labeled with [α32P]dCTP using a random primed labeling kit (Stratagene, LaJolla, CA); washing was performed to a final stringency of 0.8X SSCP and 0.1% SDS at 65°C. Fragments of 16.5, 11.5, 5.9, and 4.7 kb were detected in BglI-digested C57BL/6J DNA, and fragments of 7.5, 7.1, 5.9, 4.6, and 3.6 kb were detected inBglI-digested M. spretus DNA. The presence or absence of the 7.5-, 7.1-, and 3.6-kb BglI M. spretus–specific fragments, which cosegregated, was evaluated in backcross mice. A description of the probes and RFLPs for the loci linked to Osmrincluding Prlr, Lifr, Myo10, and Hspg1 has been reported.8 39 Recombination distances were calculated using Map Manager, version 2.6.5 (K. Manly and R. Cudmore, Roswell Park Cancer Institute, Buffalo, NY). Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
RESULTS
Characterization of mOSM receptor in LO cells.
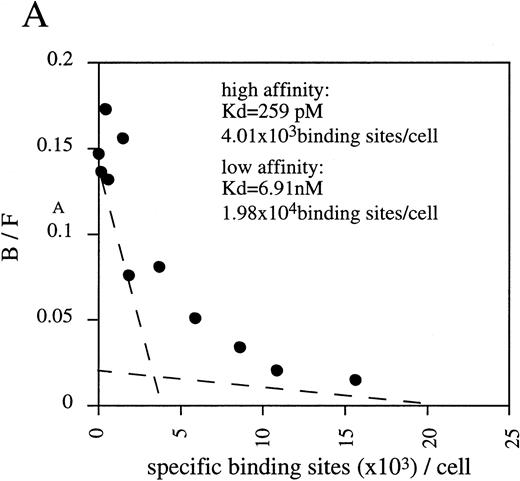
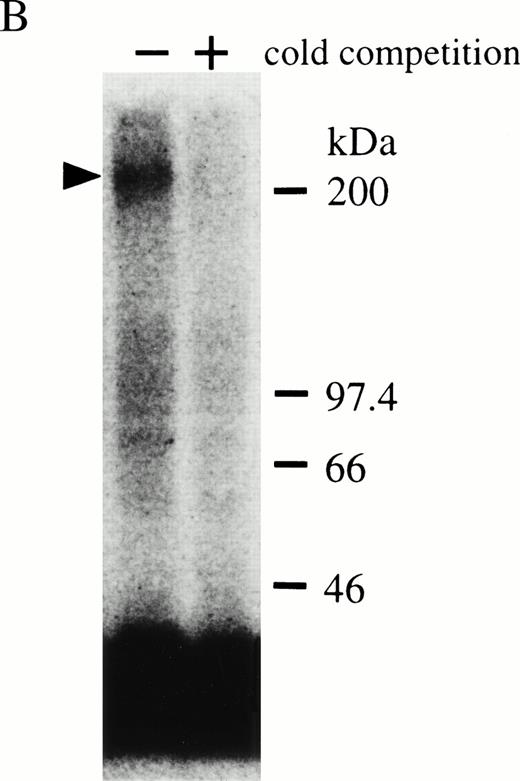
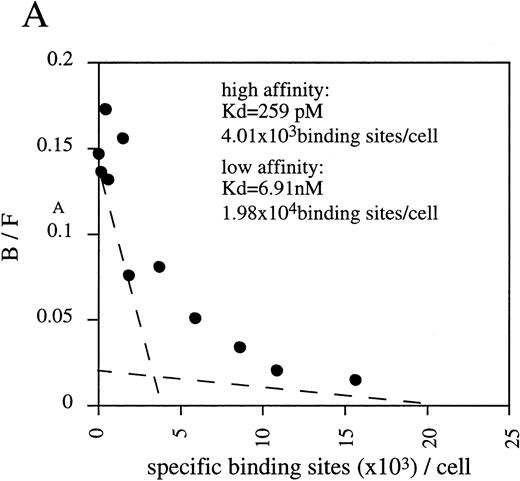
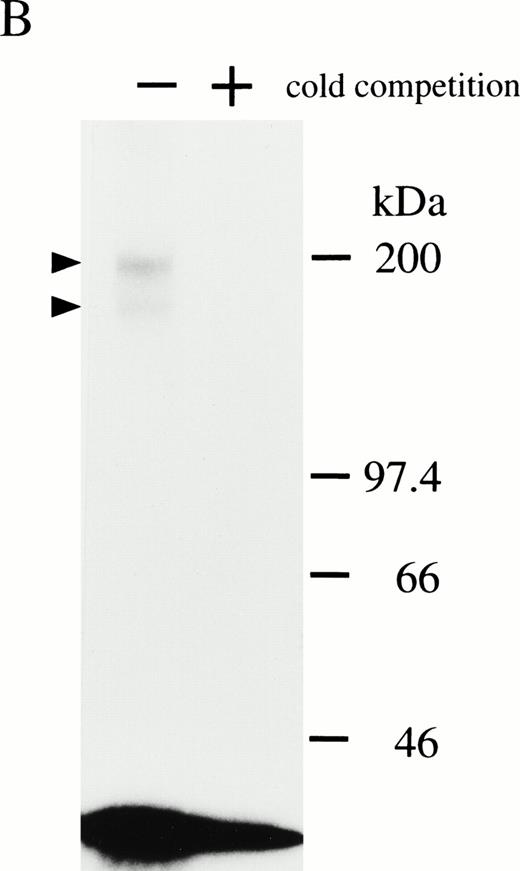
By Scatchard analysis of mOSM binding sites on mOSM-responsive and -nonresponsive cell lines, we previously demonstrated that mOSM did not bind to the mLIF receptor.29 However, the molecular nature of the mOSM receptor remained unknown. To reveal its molecular structure, we performed chemical cross-linking experiments using a newly established cell line, LO. The LO cell line was established from 11.5 dpc mouse embryo as a mOSM-dependent cell line (T. Hara, A. Miyajima, unpublished data, October 1997). Scatchard analysis showed two binding sites with distinct affinities, high-affinity (kd = 259 pmol/L) and low-affinity (kd = 6.91 nmol/L; Fig 1A). The kd value and receptor number of the high-affinity binding site of mOSM were similar in range to those previously reported in mOSM-sensitive NIH3T3 cells.29 Chemical cross-linking experiments using125I-mOSM exhibited two bands of approximately 200 and 180 kD that specifically competed with nonradioactive mOSM (Fig 1B). By subtracting the molecular mass of COS-derived mOSM (36 kD),29 the size of the two cross-linked proteins is estimated to be approximately 160 and 140 kd. As the molecular mass of the hOSMRβ was reported to be 180 kD,21 the 200- and 180-kD bands appear to represent mOSMRβ and gp130, respectively.
(A) Scatchard plot analyses of mOSM binding to LO cells. LO cells were incubated with various concentrations of125I-labeled mOSM in the presence or absence of a 1,000-fold excess of unlabeled mOSM. After 3 hours of incubation at 4°C, free mOSM was washed out through a Whatman GF/C glass filter (Maidstone, UK), and the bound radioactivity was measured by a gamma counter. Specific binding was obtained by subtracting nonspecific binding from total binding. Data are plotted according to the Scatchard transformation using the LIGAND program. Each point represents the average of duplicate measurements. The analyses clearly show two distinct affinities. (B) Cross-linking experiment using LO cells. LO cells were incubated with 5 nmol/L 125I-mOSM in the presence or absence of a 1,000-fold excess of unlabeled mOSM. After 4 hours of incubation at 4°C, cross-linked proteins were analyzed by SDS-PAGE.
(A) Scatchard plot analyses of mOSM binding to LO cells. LO cells were incubated with various concentrations of125I-labeled mOSM in the presence or absence of a 1,000-fold excess of unlabeled mOSM. After 3 hours of incubation at 4°C, free mOSM was washed out through a Whatman GF/C glass filter (Maidstone, UK), and the bound radioactivity was measured by a gamma counter. Specific binding was obtained by subtracting nonspecific binding from total binding. Data are plotted according to the Scatchard transformation using the LIGAND program. Each point represents the average of duplicate measurements. The analyses clearly show two distinct affinities. (B) Cross-linking experiment using LO cells. LO cells were incubated with 5 nmol/L 125I-mOSM in the presence or absence of a 1,000-fold excess of unlabeled mOSM. After 4 hours of incubation at 4°C, cross-linked proteins were analyzed by SDS-PAGE.
RT-PCR cloning using degenerated oligonucleotide primers.
Although we attempted to clone the mOSMRβ gene by standard low-stringency hybridization using the hOSMRβ cDNA as a probe, we were unable to isolate any clones with a homologous sequence. Therefore, we employed a strategy using RT-PCR with degenerated oligonucleotide primers. The failure to isolate the putative mOSMRβ by cross-hybridization with hOSMRβ implied a low overall sequence homology between the two; however, we expected that there may be some conserved regions between them. Since the hOSMRβ and LIFRβ are structurally related,21 we searched for regions of homology among hOSMRβ, hLIFRβ, and mLIFRβ by comparing the amino acid sequences and nucleotide sequences of the three genes. Although the amino acid similarity between hOSMRβ and hLIFRβ was relatively low (32%), their nucleotide sequences showed significantly higher homology (50%). Therefore, we designed a pair of degenerated oligonucleotide primers that correspond to two of the most conserved regions (Fig2B). The RT-PCR using polyA+ RNA derived from LO cells generated a product of the expected size. After subcloning of the DNA fragments, one clone (ORP3-5) was found to encode a novel amino acid sequence with 58% identity to the corresponding amino acid sequence of hOSMRβ.
(A) Schematic representation of the structure of mOSMRβ cDNA. The 5′ and 3′ UTRs (solid line) and the coding region (boxed region) containing the predicted signal sequence (hatched box) and the transmembrane domain (filled box) are shown. The position of cysteines and WS motifs that are conserved among the cytokine receptor superfamily is marked. The location of three overlapping cDNA clones isolated is also indicated. (B) Nucleotide and predicted amino acid sequence of mOSMRβ. Amino acids are shown by the one-letter code. Conserved cysteines and WS motifs are shaded. Potential asparagine-linked glycosylation sites (NXS/T) are underlined. The putative signal sequence and transmembrane domain are shown by a broken line and a double underline, respectively. Primers used to clone the cDNA are also shown as an underline with an arrow. YXXQ motifs are boxed. (C) Comparison of amino acid sequences between mOSMRβ and hOSMRβ. Identical amino acid residues are shown as bold letters.
(A) Schematic representation of the structure of mOSMRβ cDNA. The 5′ and 3′ UTRs (solid line) and the coding region (boxed region) containing the predicted signal sequence (hatched box) and the transmembrane domain (filled box) are shown. The position of cysteines and WS motifs that are conserved among the cytokine receptor superfamily is marked. The location of three overlapping cDNA clones isolated is also indicated. (B) Nucleotide and predicted amino acid sequence of mOSMRβ. Amino acids are shown by the one-letter code. Conserved cysteines and WS motifs are shaded. Potential asparagine-linked glycosylation sites (NXS/T) are underlined. The putative signal sequence and transmembrane domain are shown by a broken line and a double underline, respectively. Primers used to clone the cDNA are also shown as an underline with an arrow. YXXQ motifs are boxed. (C) Comparison of amino acid sequences between mOSMRβ and hOSMRβ. Identical amino acid residues are shown as bold letters.
Cloning of a full-length cDNA encoding mOSMRβ.
To isolate a full-length cDNA clone, LO cell cDNA libraries were screened with ORP3-5 as a probe. Two positive clones (OR1C5 and OR4C6) were obtained and their sequences determined (Fig 2B). OR1C5 and OR4C6 carried two distinct cDNAs of 2.2 kb. These two sequences were overlapped to yield one large single open reading frame (ORF) (Fig2A). We further cloned a 5′ untranslated region (UTR) of this cDNA by the 5′-RACE method. The ORF of the combined cDNA (2,913 bp) is predicted to encode a polypeptide of 970 amino acid residues with a calculated molecular weight of 110 kD (Fig 2B). There are 20 potential N-linked glycosylation sites in the extracellular domain. The deduced protein is a member of the class I cytokine receptor superfamily,40,41 and it shows 55.5% and 27.8% identity at the amino acid level with hOSMRβ and mLIFRβ, respectively (Fig2C). In the extracellular domain, there are two modified Trp-Ser-X-Trp-Ser (WSXWS) motifs characteristic of this family42: one is WGNWS and the other is WSDWT. Two pairs of cysteine residues were also conserved (Fig 2B). The cytoplasmic domain of hematopoietin receptors is also characterized by Box1 and Box2 regions, which are critical for generating proliferation signals43-46 and are involved in the activation of the Janus kinase (Jak) family.47-49 Box1 and Box2 regions, as well as a Tyr-X-X-Gln (YXXQ) motif critical for the activation of STAT3,44,46 50 are all conserved in the cytoplasmic domain of the cloned cDNA (Fig 2B). Therefore, we considered this novel cDNA (clone OR1C5/OR4C6) to be a strong candidate for mOSMRβ.
Functional reconstitution of mOSMR in Ba/F3 cell transfectant.
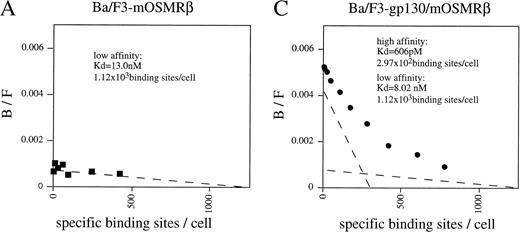
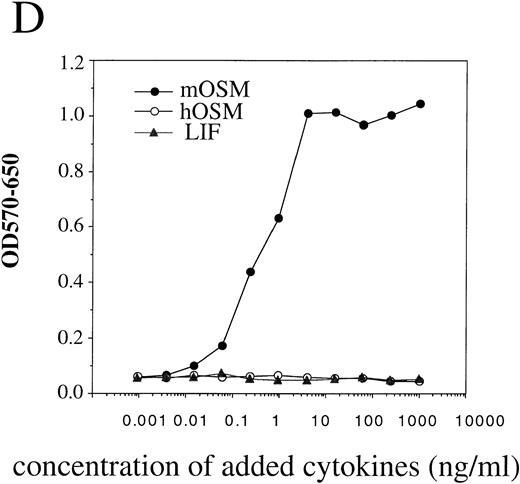
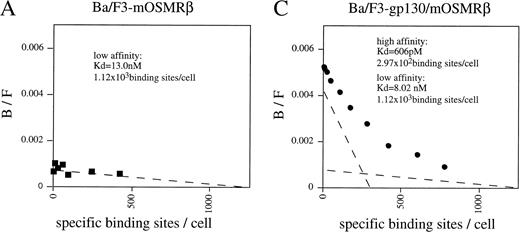
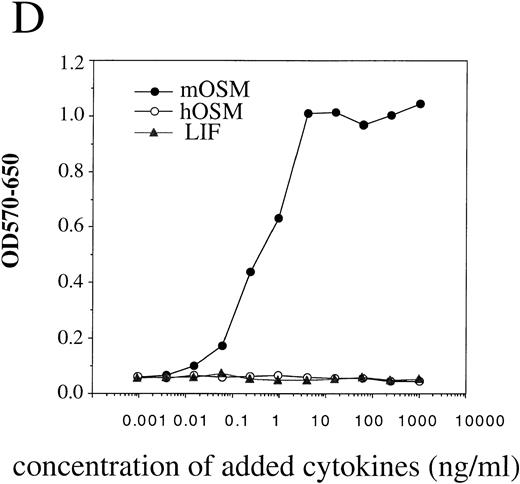
To confirm that the OR1C5/OR4C6 cDNA encodes a functional mOSMRβ subunit, we generated Ba/F3 transfectants expressing both OR1C5/OR4C6 and gp130 or OR1C5/OR4C6 alone. Expression of both proteins on the cell surface was confirmed by flow cytometry using monoclonal antibodies against gp130 and OR1C5/OR4C6, respectively (data not shown). Scatchard analysis of these transfectants demonstrated that mOSM bound to the Ba/F3 transfectant expressing mOSMRβ alone with low affinity (kd = 13.0 nmol/L; Fig 3A), whereas it was able to bind to the double transfectant with high affinity (kd = 606 pmol/L; Fig 3C). The direct binding of mOSM to mOSMRβ was confirmed by chemical cross-linking experiments using Ba/F3 transfectants (Fig 3B). Furthermore, the double transfectant proliferated in response to mOSM, but not to hOSM and mLIF, in a dose-dependent manner (Fig 3D). On the other hand, transfectants expressing either mOSMRβ or gp130 alone never responded to mOSM even in the presence of high concentrations (> 100 ng/mL) of mOSM (data not shown). These results confirmed that the OR1C5/OR4C6 cDNA encodes mOSMRβ and the functional mOSMR consists of gp130 and mOSMRβ.
Reconstitution of OSM receptor in Ba/F3 cells. (A) Scatchard plot analysis of Ba/F3 transfectant expressing only mOSMRβ; mOSM binds only to mOSMRβ with low affinity. (B) Chemical cross-linking experiment using Ba/F3 transfectant expressing only mOSMRβ; 125I-mOSM binds to mOSMRβ specifically. (C) Scatchard plot analysis of Ba/F3 transfectant expressing mOSMRβ and gp130; double transfectant exhibits high- and low-affinity binding sites. (D) Growth stimulation of transfectant expressing mOSMRβ and gp130 responding to various ligands. Double transfectant proliferates in a mOSM-dependent manner, while neither hOSM nor mLIF stimulate its growth.
Reconstitution of OSM receptor in Ba/F3 cells. (A) Scatchard plot analysis of Ba/F3 transfectant expressing only mOSMRβ; mOSM binds only to mOSMRβ with low affinity. (B) Chemical cross-linking experiment using Ba/F3 transfectant expressing only mOSMRβ; 125I-mOSM binds to mOSMRβ specifically. (C) Scatchard plot analysis of Ba/F3 transfectant expressing mOSMRβ and gp130; double transfectant exhibits high- and low-affinity binding sites. (D) Growth stimulation of transfectant expressing mOSMRβ and gp130 responding to various ligands. Double transfectant proliferates in a mOSM-dependent manner, while neither hOSM nor mLIF stimulate its growth.
Expression of mOSMRβ mRNA.
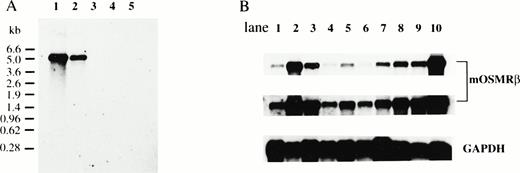
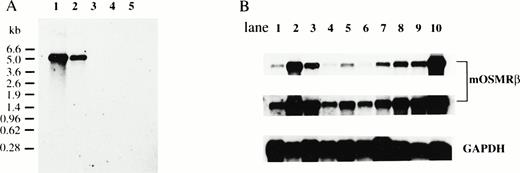
The distribution of mOSMRβ mRNA was analyzed by Northern blotting. A single band of 5.2 kb corresponding to mOSMRβ mRNA was detected in LO cells and NIH3T3 cells; no such mRNA was present in Ba/F3 cells, M1 cells, and CCE embryonic stem cells (Fig4A). Next, we examined mOSMRβ mRNA expression in various tissues of adult mice by Northern blotting. We found that mOSMRβ was widely distributed in all tissues examined. The expression level was highest in the lung, heart, thymus, and spleen (Fig 4B). However, mOSM expression is undetectable in the liver, lung, small intestine, kidney, and brain, and is inducible by cytokines in hematopoietic cells.25 Therefore, in these tissues of adult mice, it is likely that the mOSMRβ responds to OSM produced during an inflammatory process.
Expression of mOSMRβ mRNA. (A) Northern blot analyses of mOSMRβ mRNA in various cell lines. PolyA+ RNAs were prepared from various cell lines, and 1 μg of each sample was subjected to Northern blot analysis: lane 1, LO cells; lane 2, NIH3T3 cells; lane 3, Ba/F3 cells; lane 4, CCE cells; lane 5, M1 cells. (B) Northern blot analyses of mOSMRβ mRNA in various adult tissues; 1 μg of each sample was subjected to Northern blot analyses: lane 1, brain; lane 2, lung; lane 3, heart; lane 4, liver; lane 5, kidney; lane 6, small intestine; lane 7, muscle; lane 8, thymus; lane 9, spleen; lane 10, LO cells (upper panel). The middle panel shows the long exposure of the upper panel. The lower panel demonstrates equal loading by rehybridization with the GAPDH probe.
Expression of mOSMRβ mRNA. (A) Northern blot analyses of mOSMRβ mRNA in various cell lines. PolyA+ RNAs were prepared from various cell lines, and 1 μg of each sample was subjected to Northern blot analysis: lane 1, LO cells; lane 2, NIH3T3 cells; lane 3, Ba/F3 cells; lane 4, CCE cells; lane 5, M1 cells. (B) Northern blot analyses of mOSMRβ mRNA in various adult tissues; 1 μg of each sample was subjected to Northern blot analyses: lane 1, brain; lane 2, lung; lane 3, heart; lane 4, liver; lane 5, kidney; lane 6, small intestine; lane 7, muscle; lane 8, thymus; lane 9, spleen; lane 10, LO cells (upper panel). The middle panel shows the long exposure of the upper panel. The lower panel demonstrates equal loading by rehybridization with the GAPDH probe.
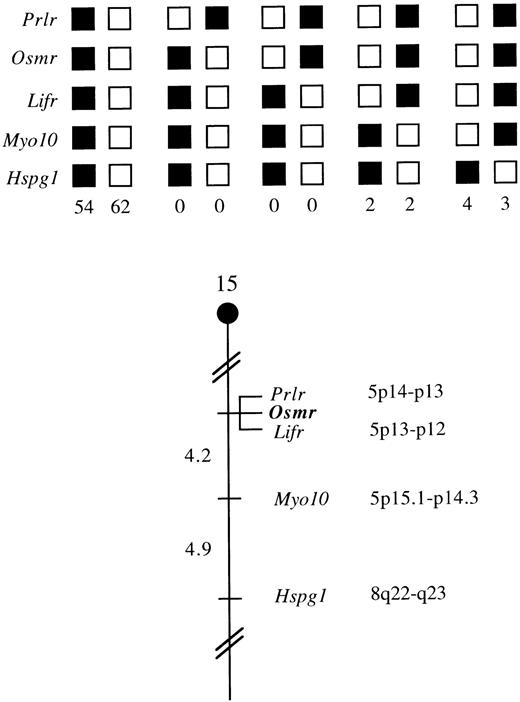
Chromosomal mapping of the OSMRβ gene.
The mouse chromosomal location of Osmr was determined by interspecific backcross analysis using progeny derived from mice matings ([C57BL/6 × M. spretus] F1 × C57BL/6J). This interspecific backcross mapping panel has been typed for over 2,600 loci that are well distributed among all autosomes, as well as the X chromosome.37 C57BL/6J and M. spretus DNAs were digested with several enzymes and analyzed by Southern blot hybridization for informative RFLPs using a mouse cDNA probe. The 7.5-, 7.1-, and 3.6-kb BglI M. spretus RFLPs were used to follow the segregation of the Osmr locus in backcross mice. The mapping results indicated that Osmr is located in the proximal region of mouse chromosome 15 linked to Prlr, Lifr, Myo10, andHspg1. Although 127 mice were analyzed for every marker and are shown in the segregation analysis (Fig 5), up to 174 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. The ratio for the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are as follows: centromere—Prlr-0/174-Osmr-0/172-Lifr-7/168-Myo10-7/144-Hspg1.The recombination frequencies (expressed as genetic distance in centimorgans [cM] ± SE) are as follows: [Prlr, Osmr, Lifr] 4.2 ± 1.5-Myo10-4.9 ± 1.8-Hspg1. No recombinants were detected between Prlr and Osmr in 174 animals typed in common and between Osmr and Lifr in 172 animals typed in common, suggesting that the two loci within each pair are within 1.7 cM of each other (upper 95% confidence limit).
Osmr maps in the proximal region of mouse chromosome 15. Osmr was placed on mouse chromosome 15 by interspecific backcross analysis. The segregation patterns ofOsmr and flanking genes in 127 backcross animals that were typed for all loci are shown at the top. For individual pairs of loci, >127 animals were typed. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus) F1 parent. Shaded boxes represent the presence of a C57BL/6J allele, and white boxes represent the presence of a M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 15 linkage map showing the location ofOsmr in relation to linked genes is shown at the bottom. Recombination distances between loci (in centimorgans) are shown to the left of the chromosome, and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
Osmr maps in the proximal region of mouse chromosome 15. Osmr was placed on mouse chromosome 15 by interspecific backcross analysis. The segregation patterns ofOsmr and flanking genes in 127 backcross animals that were typed for all loci are shown at the top. For individual pairs of loci, >127 animals were typed. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus) F1 parent. Shaded boxes represent the presence of a C57BL/6J allele, and white boxes represent the presence of a M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 15 linkage map showing the location ofOsmr in relation to linked genes is shown at the bottom. Recombination distances between loci (in centimorgans) are shown to the left of the chromosome, and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
DISCUSSION
The hOSM gene was isolated almost a decade ago.51 However, the molecular characteristics and unique biologic properties of OSM were not extensively studied because OSM had long been considered as just another LIF, a cytokine with close structural and biologic similarity. In addition, since mOSM remained molecularly unidentified until recently, characterization of the receptor was not possible. Cloning of the mOSM cDNA allowed us to examine its biologic functions and led us to discover various unique activities.25 These include stimulation of definitive hematopoiesis in the embryo,26 maturation of hepatocytes (Kamiya et al, submitted), and proliferation of neonatal Sertoli cells.28 None of these activities were exhibited by LIF and hOSM. In addition, we found various functions that are mediated by mOSM but not by LIF and hOSM and vice versa, as described. As cytokine functions are generally conserved between mouse and human, the functional differences between mOSM and hOSM are unusual. Based on the functions of mOSM and hOSM, as well as binding studies, we previously proposed that unlike hOSM, mOSM does not use the mouse LIF receptor and manifests its function only through the mOSM-specific receptor.29 However, the molecular nature of the mOSM-specific receptor remained unknown.
In this report, we describe molecular cloning of the mOSMRβ cDNA and demonstrate that the coexpression of mOSMRβ and gp130 results in the formation of a high-affinity mOSM receptor. Among IL-6 family members, OSM is unique in its ability to bind gp130 with low affinity.19,22 Interestingly, mOSM not only binds to gp130, it also binds to mOSMRβ with low affinity. In contrast, hOSM binds gp130 with low affinity but does not exhibit detectable binding to hOSMRβ.21 Since the activation of cytokine receptors is generally initiated by receptor dimerization,52 this direct binding of mOSM to both components will contribute to the formation of a stable heterodimer (Fig 6). The cytoplasmic domains of mouse and human OSMRβ contain Box1, Box2, and the YXXQ motifs important for signal transduction. As described previously,53 experiments using the G-CSFR-OSMRβ chimeric receptor revealed that homodimerization of the cytoplasmic domain of hOSMRβ was capable of activating STATs. The inability of the Ba/F3 transfectant expressing mOSMRβ alone to proliferate in response to mOSM suggested that mOSM does not form a homodimer of mOSMRβ, despite its ability to bind directly to mOSMRβ. On the other hand, IL-6 in combination with the soluble IL-6 receptor is able to induce homodimerization of gp130 and transduce signals through gp130 alone, indicating that gp130 itself is sufficient for generating signals.46 Alternatively, by analogy to the ability of CNTF to activate the LIFR/gp130 complex in association with its specific receptor subunit (CNTFRα),4 OSMRβ/gp130 might also be used by an unidentified cytokine in combination with its specific receptor subunit.
Proposed model of the functional mOSM receptor. Bold arrows, high-affinity binding; arrows, low-affinity binding.
Proposed model of the functional mOSM receptor. Bold arrows, high-affinity binding; arrows, low-affinity binding.
Studies on hOSMR-mediated signal transduction have revealed that hOSM activates both STAT3 and STAT5b through the type II receptor in the A375 cell line, whereas it phosphorylates STAT3 but not STAT5b through the type I receptor in the JAR cell line.54 On the other hand, the same set of STATs, ie, STAT1, STAT3, and STAT5b, are phosphorylated in Hep3B cells through both type I and type II OSMR.53 Thus, STATs and possibly other signaling molecules activated by hOSM are likely to depend on the cell type rather than the type of receptor. The distinct biologic actions of the gp130 family of cytokines are most simply explained by the specific expression of their receptors. For example, OSM transmits a growth promoting signal in LO cells and a growth suppressing signal in NIH3T3 cells through the type II receptor. NIH3T3 cells do not express the IL-6 receptor α chain and thus do not normally respond to IL-6. However, the combination of IL-6 and soluble IL-6 receptor is able to promote proliferation of LO cells and inhibit growth of NIH3T3 cells. OSM and LIF could therefore exhibit the same biologic activities if both LIFRβ and OSMRβ are expressed in the same cells. However, LIFRβ and OSMRβ are not always coexpressed in mouse cells, and this is probably the major reason that OSM and LIF exhibit their specific functions in mice.
Northern blot experiments revealed that mOSMRβ was widely distributed in adult mice. The expression level was highest in the lung, heart, thymus, and spleen. OSM is involved in inflammation. Consistently, in human lung-derived epithelial cells, hOSM, but not IL-6 or LIF, stimulates the synthesis of α1-proteinase inhibitor, which plays an important role in inflammation, and the type II receptor signaling pathway is critical for its synthesis.55 However, novel OSM-specific biologic activities have recently been noted in the AGM region, fetal liver, and neonatal testes as already described. In all of these cases, an OSM response is observed only in restricted cell populations and in a stage-specific manner during development. It would therefore be interesting to uncover the expression profiles of mOSMRβ and LIFRβ during development. Comparison of the transcriptional regulation of OSMRβ and LIFRβ expression would also be important for understanding the mechanism of mammalian embryogenesis and organogenesis.
We determined the chromosomal localization of the mOSMRβ gene. We have compared our interspecific map of chromosome 15 with a composite mouse linkage map that reports the map location of many uncloned mouse mutations (provided by the Mouse Genome Database, a computerized database maintained at The Jackson Laboratory, Bar Harbor, ME).Osmr mapped in a region of the composite map that lacks mouse mutations with a phenotype that might be expected for an alteration in this locus (data not shown). The proximal region of mouse chromosome 15 shares regions of homology with human chromosome 5p and 8p (Fig 5). In particular, Prlr and Lifr have been mapped to 5p14-p13 and 5p13-p12, respectively. The close linkage between Osmr andPrlr and Lifr in the mouse suggests that the human homolog of Osmr will map to 5p as well. Despite the difference for the receptor usage by OSM between human and mouse, mOSMRβ and hOSMRβ exhibit significant sequence conservation and both are structurally related to LIFRβ. Colocalization of the mOSMRβ and mLIFRβ genes strongly suggests that they were created by duplication during evolution.
During the preparation of this report, Lindberg et al56reported the cloning of mOSMRβ cDNA. Their nucleotide and amino acid sequences are identical to those for our cDNA, except the following positions. Nucleotides at the positions 1576 to 1578, which encodes a lysine residue, are deleted in our cDNA. In addition, a threonin residue at 505 is substituted with an alanine residue by a single nucleotide change. These differences might be due to the difference in mouse strains.
ACKNOWLEDGMENT
We thank Deborah B. Householder for excellent technical assistance. The nucleotide sequence data for this cDNA will appear in the DNA DATABANK of JAPAN/European Molecular Biology Laboratory/Genbank nucleotide sequence databases with accession no. AB015978.
Supported in part by a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists (M.T.), Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan, and grants from the Core Research for Evolutional Science and Technology, the Toray Research Foundation, the Uehara Memorial Foundation, Japan Science and Technology Corporation, and the National Institute, Department of Health and Human Services, under contract with ABL.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Atsushi Miyajima, PhD, Institute of Molecular and Cellular Biosciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan; e-mail:miyajima@ims.u-tokyo.ac.jp.