SH2-containing Inositol Phosphatase (SHIP) is a 145 kD protein expressed in hematopoietic cells. SHIP is phosphorylated on tyrosine after receptor binding by several cytokines and has a negative role in hematopoiesis. We cloned a murine complementary DNA (cDNA) sequence for an isoform of SHIP with an internal 183 nucleotide deletion, encoding a protein 61 amino acids shorter than 145 kD SHIP. This deletion eliminates potential SH3-domain binding regions and a potential binding site for the p85 subunit of Phosphatidylinositol 3-Kinase. Using polyclonal anti-SHIP antibodies, we and others have previously observed a 135 kD SHIP isoform that is coexpressed with 145 kD SHIP. Here, we used monoclonal antibodies raised against the region deleted in the spliced form to show that the product of the novel spliced SHIP cDNA is antigenically identical to the 135 kD SHIP isoform. Like 145 kD SHIP, 135 kD SHIP expression was induced on differentiation of bone marrow cells. After macrophage colony-stimulating factor (M-CSF) stimulation of FDC-P1(Fms) myeloid cells, both 145 and 135 kD SHIP forms were tyrosine phosphorylated and could be coimmunoprecipitated with antibodies to Shc and Grb2. However, experiments showed only a weak association of 135 kD SHIP with p85. A potentially analogous 135 kD SHIP species also appears in human differentiated leukocytes.
PRINCIPAL FACTORS THAT mediate the survival and differentiation of hematopoietic progenitor cells include colony stimulating factors (CSF), interleukins (IL), erythropoietin (Epo), and stem cell factor (SCF).1 These receptor-ligand systems have been used as models to investigate signal transduction governing hematopoietic cell proliferation and differentiation, and several molecules that become phosphorylated when these cytokines bind their cognate receptors have been identified. One recent example is the SH2-containing Inositol Phosphatase (SHIP).2,3 SHIP is a signaling protein expressed in most, if not all, hematopoietic cells, and is a negative regulator in the process of hematopoietic cell development and function.2-7 SHIP contains an N-terminal SH2 domain, a central inositol polyphosphate-5-phosphatase catalytic region, two recognition sequences for phosphotyrosine binding (PTB) domains, and several proline-rich potential SH3 domain-binding regions near the C-terminus. We and others2-6,8-12 have shown previously that SHIP is phosphorylated on tyrosine and associates with the signaling molecule Shc after treatment of cells with a variety of cytokines and growth factors including IL-3, macrophage CSF (M-CSF), granulocyte macrophage CSF (GM-CSF), Epo, and SCF, or on activation of several immunological receptors such as Fc receptors and the B-cell–receptor Ig-α and -β chains. Recently, Helgason et al7 produced mice with a targeted disruption in bothship alleles. These mice, although viable, have a decreased lifespan associated with multiple abnormalities in hematopoietic cell development and cytokine responsiveness.7 Together, these studies indicate a broad role for SHIP in hematopoiesis.
The protein interaction domains of SHIP potentially allow association with a variety of different molecules. SHIP interaction with Shc occurs via the PTB domain of Shc, which recognizes the tyrosine-phosphorylated amino acid consensus sequence NPXY on SHIP.2,13,14 Two NPXY motifs are present in the carboxy terminal region of SHIP. The tyrosine in the first of these is followed by the amino acids IGM, which constitutes a potential recognition site for SH2-mediated binding of the p85 subunit of Phosphatidylinositol 3-Kinase (PI3K).15Several studies have also shown a strong role for the SH2 domain of SHIP in protein-protein interactions. Ono et al5 and Osborne et al16 showed functional interactions of the SHIP SH2 domain with immunoreceptor tyrosine-based activation or inhibitory motifs (ITAMs or ITIMs), present in the cytoplasmic regions of many receptors. Additionally, reports by Liu et al17 and Sattler et al18 showed that SHIP interacts with the protein phosphatase SHP-2 through the SH2 domain of SHIP, and also that the SH2 domain is important for SHIP tyrosine phosphorylation and interaction with Shc.19 Proline-rich stretches, present in the C-terminal region of SHIP, may be recognized by proteins with SH3 domains.20 Earlier studies from several laboratories showed that SHIP was found in a complex with Grb2,8,9 an interaction that may be governed by these polyproline motifs. Damen et all3 and Osborne et al16 have shown specific interactions of SHIP with the SH3 domains of PLCγ or Grb2 in vitro. Evidence of an in vivo interaction was supplied by Odai et al,12 who showed binding of SHIP with Grb2 in human leukemia cells.
The existence of numerous interaction partners as well as activating factors suggests multiple functions and regulatory mechanisms for SHIP in vivo. Using immunoblot assays with anti-SHIP antibodies, we and others2,5,7,21,26,30 have previously detected isoforms of SHIP in addition to 145 kD SHIP that may be important in the multiple activities and associations of this molecule.* Kavanaugh et al21 reported that three SHIP isoforms (SIP-110, -130, and -145) in human B cells are due to alternative splicing of a single gene, and showed that the differences between the forms are due to loss of complementary DNA (cDNA) sequence encoding the N-terminal SH2 domain. In contrast, Damen et al30 recently reported that isoforms of SHIP observed in the murine hematopoietic cell line DA-ER are the result of C-terminal proteolytic cleavages of 145 kD SHIP, possibly by calpain. In the following report, we identify a cDNA sequence encoding an approximately 135 kD SHIP isoform that is coexpressed with 145 kD SHIP. However, unlike the SIP proteins described by Kavanaugh or the SHIP cleavage products described by Damen, this 135 kD version of SHIP results from a specific internal deletion that removes a polyproline-containing stretch between the two NPXY motifs. This 135 kD SHIP protein is expressed under the same conditions as 145 kD SHIP, and appears to be the result of alternative splicing of SHIP messenger RNA (mRNA). Our studies also indicate that a 135 kD form of SHIP is produced in a human myeloid leukemia cell line and human peripheral white blood cells, and that a similar deletion in the SHIP message may be responsible for this isoform as well. We further show that the murine 135 kD isoform is phosphorylated in vivo and retains its ability to associate with the signaling proteins Shc and Grb2 after cytokine stimulation. However, our experiments showed only a weak association of 135 kD SHIP with the C-terminal SH2 domain of p85 compared with the full-length 145 kD form. This 135 kD SHIP isoform may therefore be involved in alternative protein associations or functions of SHIP.
MATERIALS AND METHODS
Cell culture.
The WEHI-3 monocyte cell line was obtained from the American Type Culture Collection (ATCC, Rockville, MD) and was maintained in RPMI 1640 medium supplemented with penicillin and streptomycin, plus 10% fetal bovine serum (FBS) (Summit Labs, Fort Collins, CO). FDC-P1 myeloid cells expressing the murine M-CSF receptor Fms (Genbank accession number FMSCR) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS, plus 10% WEHI-3 conditioned media as a source of IL-3, as described.2 For M-CSF stimulation, cells were first incubated for 5 hours in 1% FBS without IL-3. Cells were pelleted by centrifugation and resuspended in one half the pellet volume of PBS (control) or of M-CSF (70,000 U/mL final concentration). Cells were incubated for 2 minutes at room temperature, then lysed in cold NP-40 lysis buffer [50 mmol/L NaCl, 100 mmol/L HEPES, 30 mmol/L Na4P2O7, 50 mmol/L NaF, 5 μmol/L ZnCl2, 0.5% NP-40, 1 mmol/L Na3VO4(all from Sigma, St Louis, MO), pH 7.0]. 293T and Rat2 fibroblast cell lines were also maintained in DMEM, plus 5% FBS and 5% calf serum (Hyclone, Logan, UT). Bone marrow cells (BMCs) were obtained from the femurs of mature C57-BL6 mice and plated in DMEM plus 10% FBS for 6 hours. At this point, nonadherent cells were recovered and either lysed in NP-40 lysis buffer or were replated and incubated for 7 days in DMEM with 10% FBS, IL-3, and M-CSF (2000 U/mL). After 4 days, nonadherent cells were removed by washing with PBS and the adherent BM-derived macrophages (BMM) were lysed in NP-40 lysis buffer. The ML-1 cell line was cultured in RPMI 1640 plus 10% FBS as described.26Normal human peripheral white blood cells were purified from whole blood using Lymphoprep (Nycomed, Oslo, Norway) according to the manufacturer’s instructions.
Reverse transcription-polymerase chain reaction (RT-PCR).
Total RNA was isolated using TRIzol reagent (GIBCO-BRL, Grand Island, NY) according to the manufacturer’s instructions. To ensure that there was no genomic DNA contamination, the RNA preparation was treated with 10 U RNase-free DNase I (Boehringer Mannheim, Indianapolis, IN) at room temperature for 20 minutes in the presence of 3.75 mmol/L Tris-HCl (pH 7.5), 10 mmol/L MgCl2, 1 mmol/L dithiothreitol (DTT). This reaction was terminated with 5 μL of 0.5 mol/L EDTA and the RNA was extracted with chloroform and reprecipitated. RT-PCR assays were performed as previously described.22 Briefly, single-stranded cDNA was synthesized from 10 μg purified total RNA after priming with 10 pmol oligo dT15, using 200 U Superscript (GIBCO BRL) according to the manufacturer’s instructions. The final reaction volume was diluted to 100 μL in water, and 2 μL of the cDNA sample was used as a template in each 40 μL PCR. The amplification conditions were: 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C, for varying cycles. These conditions were also used to perform competitive PCR experiments with the 2666 to 3247 primer set. Primer sequences used were as follows: numbers refer to the murine SHIP cDNA sequence, Genbank accession number U51742: 73 (sense): 5′-GACCCAGTCCAGGAGACCC-3′; 800 (antisense): 5′-GCAGAGACTCCAGGGATG-3′; 2042 (sense): 5′-CTGACCCGGGACAAGTATGC-3′; 2703 (antisense): 5′-GGATTCATCCCGCTCTGTCT-3′; 2666 (sense): 5′-CAGGGCAAGATGAGGGAGAA-3′; 3247 (antisense): 5′-CGATGCTGGGTGATGAGATT-3′. Primer sequences for human SHIP were as follows; numbers refer to SHIP cDNA sequence, Genbank accession number U84400: 2615 (sense): 5′-GCCACTTCCAGGGGGAGATCA-3′; 3212 (antisense): 5′-GCAGGGCGGGGGTTCCTTCC-3′.
Genomic DNA sequencing.
A murine genomic DNA library (using Lambda DASH II; Stratagene, La Jolla, CA) was constructed by Dr Zhi Chen and was the kind gift of Dr Phil Soriano, Fred Hutchinson Cancer Research Center. Approximately 1.5 × 106 phage plaques were screened using full-length SHIP cDNA labeled with [α-32P] dCTP (RTS RadPrime DNA Labeling System, GIBCO BRL). Thirty positive clones were isolated and rescreened with the 581 bp product of the 2666-3247 PCR primer set, using a slot-blot apparatus (Schleicher & Schuell, Keene, NH). Positive clones were digested with EcoRI and/or BamHI and subjected to southern analysis with the same probe. Bands that hybridized were purified, subcloned into pBluescript (Stratagene), and sequenced using an automated sequencer (Perkin-Elmer, Norwalk, CT).
SHIP cDNA cloning.
A cDNA library was constructed from FDC-P1 cells,2 using the Lambda-ZAP kit (Stratagene). This library was probed using full-length SHIP cDNA labeled with [α-32P] dCTP. Thirty-four positive clones were obtained, and these were reprobed using [α-32P] dCTP–labeled cDNA representing nucleotides 38-502 of the SHIP cDNA sequence. Clones that were positive for this probe (5′ region of SHIP) were identified, and the membrane was stripped and rehybridized using a probe representing nucleotides 2842-3019 (183 nucleotide deletion) of SHIP cDNA. Clones that hybridized with both probes, as well as clones that hybridized to the first probe and not the second probe, were treated as described in the manufacturer’s instructions to excise the plasmid pBK-CMV containing the insert.
Transfections.
pBK-CMV containing SHIP cDNA or Δ183 SHIP cDNA was transfected into 293T fibroblast cells using a calcium phosphate precipitation protocol.23 Briefly, 10 μg of DNA was diluted in 500 μL of 250 mmol/L CaCl2 before being mixed with an equal volume of a 2× HeBS phosphate buffer (280 mmol/L NaCl, 10 mmol/L KCl, 1.5 mmol/L Na2 HPO4, 12 mmol/L glucose, 50 mmol/L HEPES, pH 7.06). This solution was incubated at room temperature for 5 minutes, then added to a 150 mm plate of 293T fibroblast cells at approximately 75% confluence. The cells were incubated for 48 hours before being washed and processed for immunoblotting.
Antibodies.
The anti-SHIP polyclonal antibody #5340 was made using SHIP amino acids 670-868, and the #5369 antibody was made using amino acids 889-1046.2 The anti-Shc polyclonal antibody was produced using a GST fusion protein with the SH2 domain of Shc, amino acids 374-469 (Genbank accession number U15784). The anti-p85 polyclonal antibody #06-195 was purchased from Upstate Biotechnology (Lake Placid, NY). Polyclonal antibodies were used at a dilution of 1:5000. Monoclonal antibodies (MoAbs) to SHIP were produced using BALB/C mice immunized with histidine-tagged proteins representing either amino acids 4-126 of SHIP (P6H8), or amino acids 866-1020 of SHIP (P2C6, P1D7, and P1C1). The 866-1020 peptide includes both NPXY motifs and the intervening polyproline stretch, as well as the region deleted in the Δ183 SHIP cDNA sequence (amino acids 920-980). Immunizations and fusions were performed by Dr Elizabeth Wayner, Fred Hutchinson Hybridoma Shared Resource. These clonal hybridoma supernatants were used at a dilution of 1:100. The antiphosphotyrosine MoAb 4G10 (the kind gift of Dr Brian Druker, Dana Farber Cancer Institute, Boston, MA) was used at a dilution of 1:10,000.
SDS-PAGE, immunoprecipitations, and immunoblots.
These assays were performed as described by Lioubin et al9and Carlberg et al.24 All cells were lysed in NP-40 lysis buffer containing 1 mmol/L Na3VO4, and cell debris was removed by centrifugation. Total protein amount was measured by spectrophotometry using Protein Assay Reagent (BioRad, Hercules, CA), and equal amounts of protein were loaded into each lane. SDS-PAGE was run using a 6% acrylamide gel, and protein molecular weights were compared using unstained Perfect Protein Markers (Novagen, Madison, WI). For immunoprecipitations, 400 μg of total protein was used in each reaction along with 40 μL of Protein A (or Protein G for the 4G10 MoAb) conjugated to sepharose beads (Pharmacia, Piscataway, NJ). Proteins were precipitated with 2 μL of anti-Shc, 1 μL of antiphosphotyrosine, or 2 μL of #5340 polyclonal anti-SHIP antibody. The lysate was mixed at 4°C for 6 hours, and beads were washed five times in cold NP-40 buffer. 2× sample loading buffer (100 mmol/L Tris pH 6.8, 200 mmol/L DTT, 4% SDS, 20% glycerol, 0.1% bromophenol blue) was added, and the sample was heated for 2 minutes at 100°C. SDS-PAGE, transfer, and detection were performed as described by Carlberg.24 For GST pull-down experiments, cDNA fragments encoding either the N-terminal or C-terminal SH2 domains from p85 were cloned into the pGEX-3X vector (Pharmacia), and protein was expressed and purified as described by Carlberg.24 Fifty μg of purified GST-SH2 protein was used in each reaction, plus 40 μL of a 50% slurry of GST-agarose beads (Sigma). The experiments were performed identically to the immunoprecipitations, except that the binding and washing buffer composition was as described in Joos et al.28
RESULTS
Identification of Δ183 SHIP.
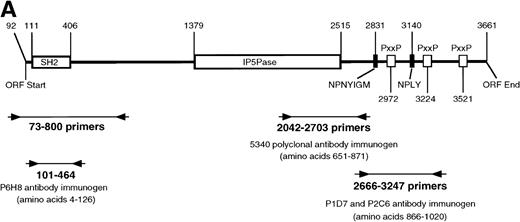
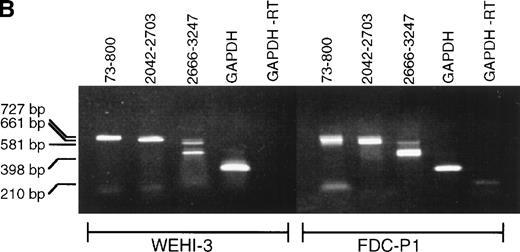
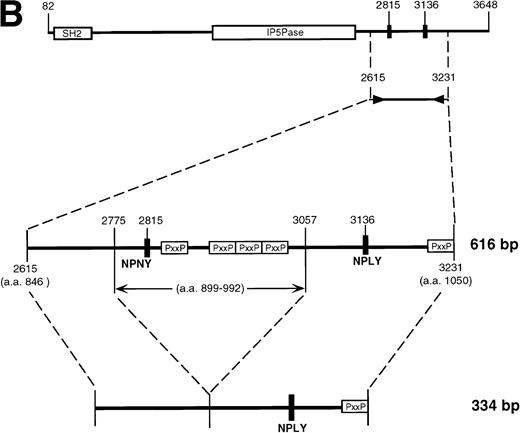
Previous immunoblot data from our laboratory and others2,5,7,21,26 showed the expression of several proteins that react with anti-SHIP polyclonal antibodies. The polyclonal anti–SHIP antibodies #5340 and #5369 detect the 145 kD SHIP protein in the myeloid cell line FDC-P1. These antibodies also react with a larger SHIP–related form, approximately 160 kD, two smaller proteins of approximately 135 and 130 kD, and under certain conditions a 110 kD protein.26 We performed RT-PCR analysis to detect alternative SHIP transcripts that might produce these additional protein forms. We generated oligonucleotide primers that annealed to several regions of SHIP sequence (Fig 1A) and performed RT-PCR using RNA from either WEHI-3 or FDC-P1 myeloid cell lines, which produce identical expression patterns of SHIP isoforms or SHIP-like proteins in immunoblot assays. A primer set flanking the region of SHIP cDNA encoding the SH2 domain (nucleotides 73-800, amino acids 1-237), as well as a primer set flanking the region encoding the epitope for antibody #5340 (nucleotides 2042-2703, amino acids 651-871) resulted in amplification of the predicted product sizes only. However, primers flanking nucleotides 2666-3247 (amino acids 866-1059) amplified a product at the predicted size of approximately 600 nucleotides, plus a smaller product of approximately 400 nucleotides (Fig 1B). A larger product of approximately 700 nucleotides was also visible in some experiments.
SHIP RT-PCR. (A) The 3,570 nucleotide ORF of murine SHIP cDNA is shown with structural and potential functional domains labeled. The NPXY and YIGM motifs are depicted (▪), as well as the proline-rich potential SH3 domain binding regions (□). Also shown are the binding sites for the PCR primer sets used. Nucleotide numbering refers to the cDNA sequence of murine SHIP, Genbank accession numberU51742. (B) RT-PCR was conducted using RNA from the WEHI-3 and FDC-P1 myeloid cell lines and the primer sets shown in Fig 1A. The products were separated by electrophoresis in a 1% agarose gel and stained with ethidium bromide. Amplification of cDNA with primers to GAPDH was included to verify cDNA quality.
SHIP RT-PCR. (A) The 3,570 nucleotide ORF of murine SHIP cDNA is shown with structural and potential functional domains labeled. The NPXY and YIGM motifs are depicted (▪), as well as the proline-rich potential SH3 domain binding regions (□). Also shown are the binding sites for the PCR primer sets used. Nucleotide numbering refers to the cDNA sequence of murine SHIP, Genbank accession numberU51742. (B) RT-PCR was conducted using RNA from the WEHI-3 and FDC-P1 myeloid cell lines and the primer sets shown in Fig 1A. The products were separated by electrophoresis in a 1% agarose gel and stained with ethidium bromide. Amplification of cDNA with primers to GAPDH was included to verify cDNA quality.
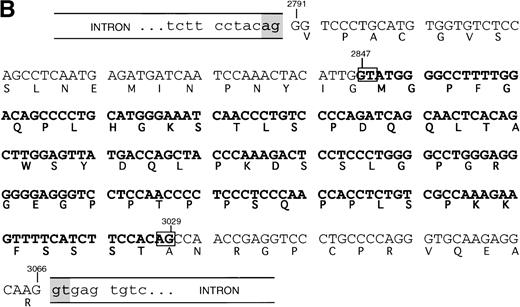
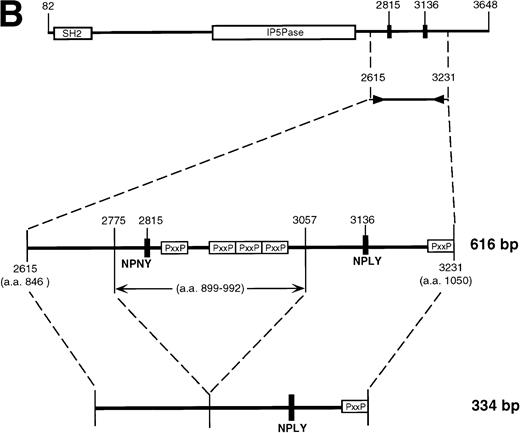
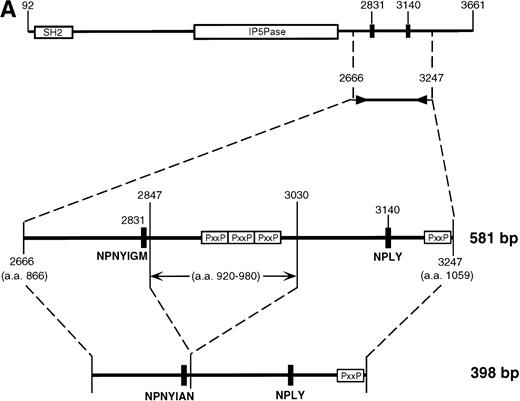
Subcloning and sequencing of the two major bands produced by the 2666-3247 primers showed the known full-length SHIP cDNA and SHIP cDNA with a deletion of 183 nucleotides (Fig 2A). This deletion occurs within the region amplified by the 2666-3247 primers; the primer binding sites are unaffected, and thus the amplification of this smaller band was not due to improper annealing of the primers. At the protein level, this “Δ183” sequence predicts an in-frame deletion of 61 amino acids in the proline-rich C-terminal end of SHIP producing an isoform with a calculated size of 127 kD.
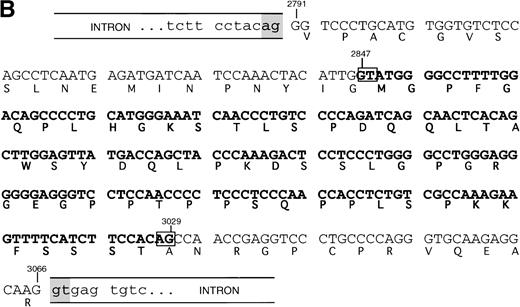
SHIP deletion. (A) The two bands produced using the 2666-3247 primer set, shown in Fig 1B, were subcloned and sequenced. The larger (581 bp) band corresponds to the published SHIP sequence. The smaller (398 bp) band has a deletion of 183 nucleotides at the site shown, but is otherwise identical. (B) SHIP genomic DNA was isolated from a lambda genomic DNA library. Phage clones which hybridized with the 581 bp product of the 2666-3247 PCR primers were sequenced to show the NPXY-containing exon. Exon sequence is in upper case letters, and the sequence of the ▵183 deletion is in bold type. The verified splice acceptor and donor are shown in shaded boxes, and the potential splice sites that would produce the ▵183 deletion as an intron are shown in open boxes. Numbering refers to the SHIP cDNA sequence as in Fig 1.
SHIP deletion. (A) The two bands produced using the 2666-3247 primer set, shown in Fig 1B, were subcloned and sequenced. The larger (581 bp) band corresponds to the published SHIP sequence. The smaller (398 bp) band has a deletion of 183 nucleotides at the site shown, but is otherwise identical. (B) SHIP genomic DNA was isolated from a lambda genomic DNA library. Phage clones which hybridized with the 581 bp product of the 2666-3247 PCR primers were sequenced to show the NPXY-containing exon. Exon sequence is in upper case letters, and the sequence of the ▵183 deletion is in bold type. The verified splice acceptor and donor are shown in shaded boxes, and the potential splice sites that would produce the ▵183 deletion as an intron are shown in open boxes. Numbering refers to the SHIP cDNA sequence as in Fig 1.
This deleted form of SHIP cDNA could be the result of alternative RNA splicing. We therefore sequenced murine genomic DNA to map the intron/exon boundaries and investigate this possibility. Our genomic sequencing of ship showed that the deleted 183 nucleotide section occurs entirely within a single 276 nucleotide exon. However, the 183 nucleotide segment contains a consensus splice donor and acceptor site at either end, and thus could act as an intron (Fig 2B).
Δ183 SHIP mRNA expression.
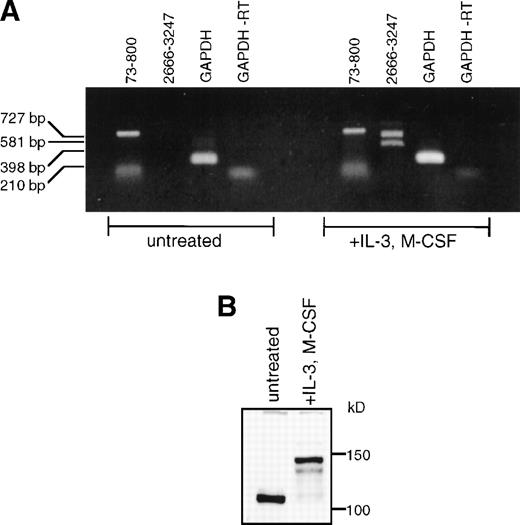
It has previously been shown that SHIP message is present in a wide variety of hematopoietic cells.2,10 Using RT-PCR with the 2666-3247 primer set, we have detected Δ183 SHIP message along with full-length SHIP message in the macrophage cell lines WEHI-3 and RAW 264.7, the B-cell progenitor line BaF3, and the T-cell line CTLL-2 (not shown), in addition to the myeloid progenitor cell line FDC-P1 from which Δ183 SHIP was cloned. No SHIP mRNA was detected in the fibroblast cell lines NIH 3T3 (mouse), Rat2 (rat), or 293T (human) (not shown). We were unable to detect either full-length or Δ183 SHIP message in untreated murine BMC RNA by RT-PCR using the primers to nucleotides 2666-3247. However, using primers to nucleotides 73-800, we detected a band of the predicted size for SHIP cDNA (Fig 3A). When BMCs were differentiated to macrophages using M-CSF, we readily detected both full-length and Δ183 SHIP mRNA using the 2666-3247 primer set, and full-length SHIP using the 73-800 primer set. This finding correlates with our immunoblot analysis using untreated BMC lysates or IL-3 plus M-CSF–differentiated BMM shown in Fig 3B. In untreated BMC lysates, a SHIP-like protein of approximately 110 kD was expressed as reported by us previously,26 but full-length SHIP was not detected (Fig 3B). Within 4 days of incubation in media containing IL-3 and M-CSF, adherent BMM cells expressed the 145 kD and 135 kD SHIP proteins. The band produced using the 73-800 primer set in RT-PCR analysis of untreated BM may represent this approximately 110 kD SHIP protein form, which may lack part of the region encoded by the nucleotides 2666-3247 and thus is not recognized by our MoAbs.
SHIP expression in BM and BMM. (A) Murine BM was isolated and treated as described in Materials and Methods. Total RNA was purified from undifferentiated BM or from BMM. RT-PCR was performed using the primer sets shown in Fig 1A. (B) Untreated murine BMCs or BMM were lysed in NP-40 lysis buffer. Equal amounts (40 μg) of total protein were loaded into each lane of a 6% polyacrylamide gel. The gels were processed as described and protein was detected with the anti-SHIP polyclonal antibody #5340.
SHIP expression in BM and BMM. (A) Murine BM was isolated and treated as described in Materials and Methods. Total RNA was purified from undifferentiated BM or from BMM. RT-PCR was performed using the primer sets shown in Fig 1A. (B) Untreated murine BMCs or BMM were lysed in NP-40 lysis buffer. Equal amounts (40 μg) of total protein were loaded into each lane of a 6% polyacrylamide gel. The gels were processed as described and protein was detected with the anti-SHIP polyclonal antibody #5340.
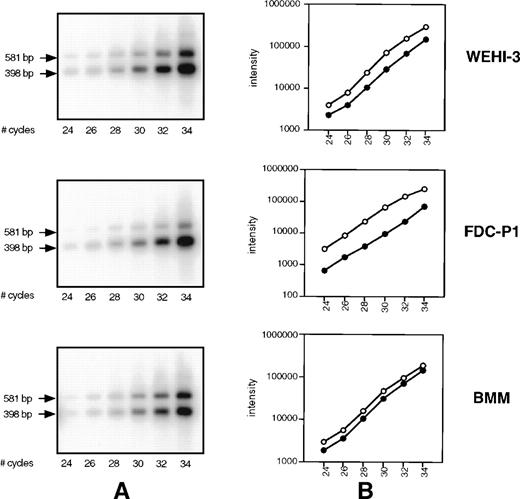
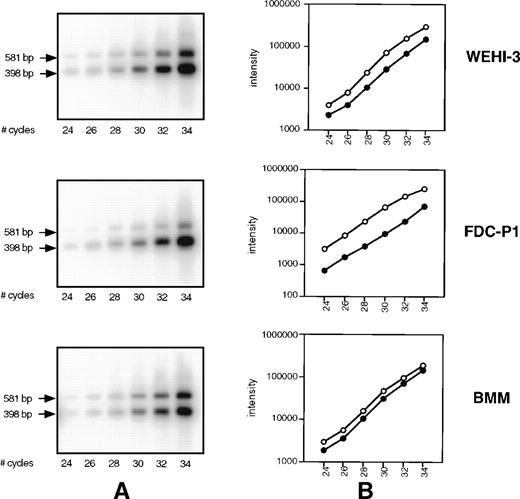
We hypothesized that the smaller PCR product of the primer set 2666-3247, observed in Fig 1B and 3A, may in fact be a minor component of SHIP message detected only because of the sensitivity of RT-PCR. To address this, we performed competitive PCR analysis to compare the rate of amplification of the two products, as well as the level of Δ183 SHIP cDNA relative to the full-length product. Using the 2666-3247 primer set, cDNA from WEHI-3 cells, FDC-P1 cells, and BMM was amplified for varying cycles under the conditions described in Materials and Methods. The resulting PCR products were transferred to a membrane and hybridized to the radiolabeled probe made from SHIP cDNA (nucleotides 2666-2795) that anneals to a region common to both products. The signal of each band was measured using a Phosphorimager and plotted against cycle number (Fig 4A). As can be observed, the rate of amplification of the two products is similar; no preferential amplification of either product is detected. We then compared the lower (Δ183 SHIP) band with the upper (full-length SHIP) band in the same lane in each cell type. Within the linear range of amplification (cycles 26 through 32), Δ183 SHIP cDNA was present in amounts 2 to 10 times greater than full-length SHIP cDNA (Fig 4B).
SHIP competitive PCR. (A) cDNA from WEHI-3 cells, FDC-P1 cells, and BMM was amplified for the specified number of cycles using the 2666-3247 primer set. Products were transferred to a membrane and hybridized using a 32P-labeled SHIP cDNA sequence common to both. (B) The signal of each band was measured using a Phosphorimager and plotted against cycle number. Within the linear range of amplification, cycles 26 through 32, the 398 bp ▵183 SHIP band (○) from each cDNA type was compared with the 581 bp full-length SHIP band (•) in the same lane.
SHIP competitive PCR. (A) cDNA from WEHI-3 cells, FDC-P1 cells, and BMM was amplified for the specified number of cycles using the 2666-3247 primer set. Products were transferred to a membrane and hybridized using a 32P-labeled SHIP cDNA sequence common to both. (B) The signal of each band was measured using a Phosphorimager and plotted against cycle number. Within the linear range of amplification, cycles 26 through 32, the 398 bp ▵183 SHIP band (○) from each cDNA type was compared with the 581 bp full-length SHIP band (•) in the same lane.
SHIP cDNA cloning.
We used full-length SHIP cDNA to probe a library constructed from FDC-P1 cell mRNA. Positive clones were analyzed on a slot blot, using a probe to the 5′ untranslated region of SHIP cDNA (nucleotides 38-502), to determine whether they contained the beginning of the open reading frame (ORF) of 145 kD SHIP protein. This slot blot was also probed with SHIP cDNA representing the 183 nucleotides deleted in the PCR products (nucleotides 2842-3019). Clones that hybridized to both probes, as well as clones that hybridized to the 5′ probe but not to the Δ183 probe were subjected to sequence analysis. In this screen, we obtained two complete full-length SHIP cDNA clones and three complete Δ183 SHIP cDNA clones. These Δ183 SHIP clones were identical to full-length SHIP cDNA except for the 183 nucleotide deletion described.
Δ183 SHIP protein expression.
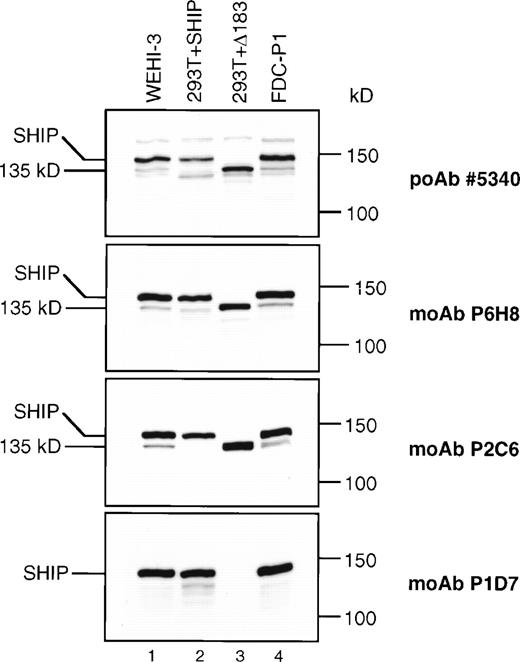
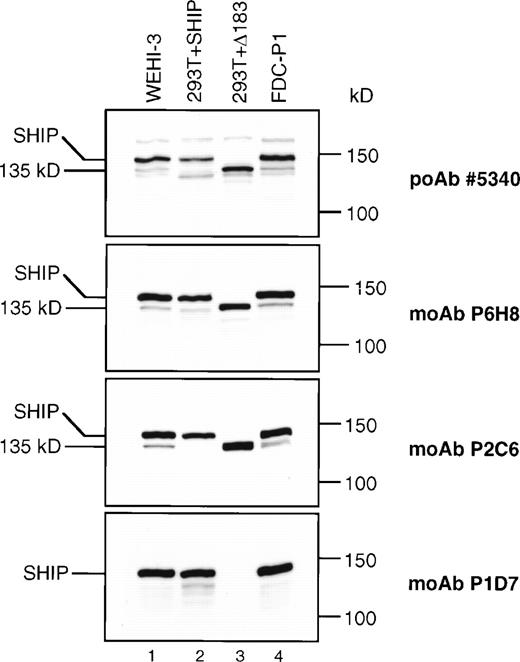
To more precisely identify SHIP and its potential isoforms, we produced MoAbs to SHIP. The first immunogen used included SHIP amino acids 866-1020, encompassing the region eliminated in the Δ183 SHIP cDNA (Fig 1) and resulted in the MoAbs P2C6, P1D7, and P1C1. The second immunogen included amino acids 4-126, encompassing the N-terminal SH2 domain, and resulted in the MoAb P6H8. Immunoprecipitation and immunoblot analyses showed that the 145 kD band recognized by each of these anti-SHIP MoAbs was also recognized by the anti-SHIP polyclonal antibodies #5340 and #5369 previously produced, and thus is a closely related if not identical protein (Fig 5). To examine the products of the SHIP cDNA clones that were isolated, 293T fibroblast cells were transfected with an expression vector containing either full-length or Δ183 SHIP cDNA. Lysates from these cells, along with lysates from WEHI-3 and FDC-P1 myeloid cells, were then analyzed by immunoblot assay (Fig 5). Although untransfected 293T fibroblast cells showed no bands using any of the anti-SHIP antibodies (not shown), 293T cells transfected with either full-length or Δ183 SHIP cDNA produced bands of the predicted size that were recognized by the anti-SHIP polyclonal antibody #5340 and the anti-SHIP MoAbs P2C6 and P6H8 (Fig 5, lanes 2 and 3). The MoAb P1D7 did not recognize the product of transfected Δ183 SHIP cDNA, nor did it recognize the 135 kD protein in either WEHI-3 or FDC-P1 myeloid cells. However, MoAb P1D7 recognized the 145 kD SHIP protein in the hematopoietic cells and in 293T cells transfected with full-length SHIP cDNA. This result shows that the epitope recognized by the P1D7 antibody exists within the 61 amino acids eliminated in Δ183 SHIP. Furthermore, the migration characteristics of 135 kD SHIP and the translated product of Δ183 SHIP cDNA were indistinguishable in higher resolution SDS-PAGE experiments. The P1C1 antibody recognition pattern was the same as that of the P2C6 and P6H8 antibodies (not shown). Taken together, these data provide evidence that Δ183 SHIP and the 135 kD protein observed in hematopoietic cells are identical, and that this protein differs from full-length SHIP in the region encoded by the deleted 183 nucleotides.
SHIP immunoblots. Equal amounts (40 μg) of total protein from the cell type indicated was loaded into each lane of four identical 6% polyacrylamide gels. The blots were processed as described and incubated with the anti-SHIP polyclonal antibody #5340 or the anti-SHIP MoAbs as indicated.
SHIP immunoblots. Equal amounts (40 μg) of total protein from the cell type indicated was loaded into each lane of four identical 6% polyacrylamide gels. The blots were processed as described and incubated with the anti-SHIP polyclonal antibody #5340 or the anti-SHIP MoAbs as indicated.
In both 293T fibroblasts (Fig 5) and Rat2 fibroblasts2transfected with full-length SHIP cDNA, we observed a SHIP antibody-reactive protein of approximately 130 kD (Fig 5, lane 2). This band was present in fibroblasts that were transfected with only full-length SHIP cDNA. The Δ183 SHIP sequence could not be detected in these cells by RT-PCR analysis, although full-length SHIP cDNA is readily amplified (not shown). Unlike the translated product of Δ183 SHIP cDNA, shown in lane 3 of Fig 5, this approximately 130 kD band in full-length SHIP–transfected fibroblasts is recognized by all our anti–SHIP antibodies and also migrates slightly faster than the hematopoietic 135 kD SHIP protein on SDS-PAGE. In the WEHI-3 and FDC-P1 cells we also detected additional smaller bands including the 130 kD band observed in SHIP-transfected fibroblasts (Fig 5, #5340 panel), but these bands were inconsistently detected and none shared the size or antibody recognition pattern of Δ183 SHIP.
Human hematopoietic cells produce a 135 kD isoform of SHIP.
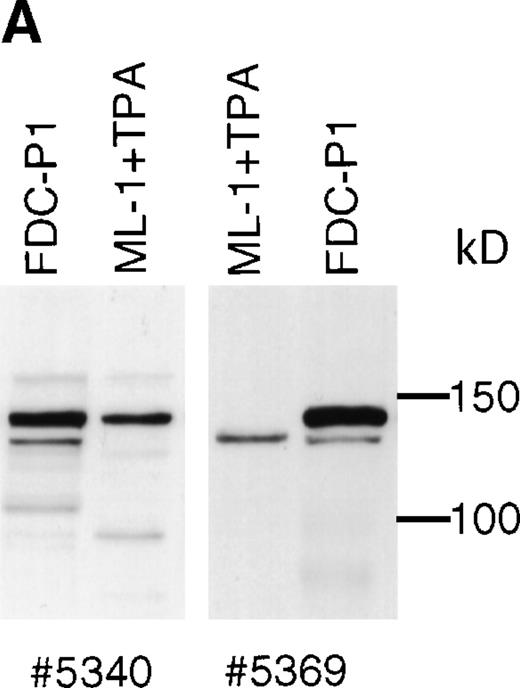
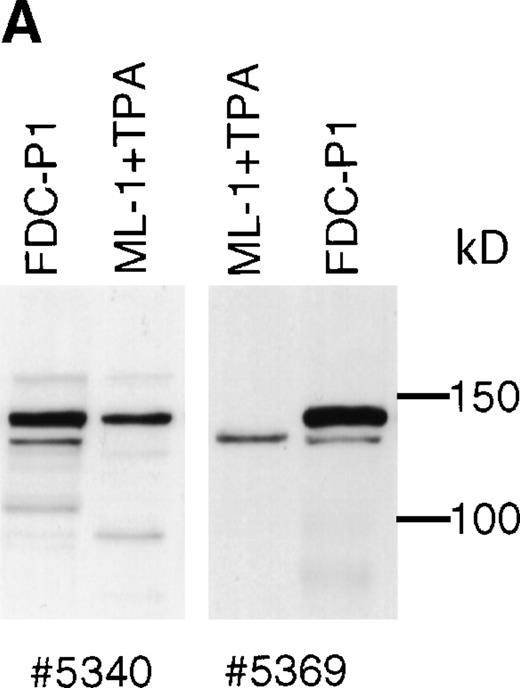
Western blotting experiments of tetradecanoyl phorbol acetate (TPA)-treated ML-1 cells using polyclonal SHIP antibodies #5340 and #5369 show that a protein of 135 kD is produced in addition to 145 kD SHIP (Fig 6A). This 135 kD protein is also detectable in human peripheral white blood cells, but at a very low level. Human SHIP is recognized poorly by P2C6, P1D7, and P6H8. The P1C1 MoAb, produced using the same immunogen as for P1D7 and P2C6, recognizes human 145 kD SHIP well, but does not recognize human 135 kD SHIP (not shown).
Human SHIP 135 kD protein. (A) ML-1 human monocytic leukemia cells were treated for 2 days with TPA, and adherent cells were lysed and subjected to SDS-PAGE. Blots were incubated with SHIP polyclonal antibodies #5340 or #5369. (B) RNA from ML-1 cells with the same TPA treatment was analyzed by RT-PCR using primers to human SHIP 2615-3231 (Genbank accession number U84400). Two bands were detected at approximately 600 and 350 bp. These were purified and subjected to DNA sequencing, and the results are depicted as in Fig 2A.
Human SHIP 135 kD protein. (A) ML-1 human monocytic leukemia cells were treated for 2 days with TPA, and adherent cells were lysed and subjected to SDS-PAGE. Blots were incubated with SHIP polyclonal antibodies #5340 or #5369. (B) RNA from ML-1 cells with the same TPA treatment was analyzed by RT-PCR using primers to human SHIP 2615-3231 (Genbank accession number U84400). Two bands were detected at approximately 600 and 350 bp. These were purified and subjected to DNA sequencing, and the results are depicted as in Fig 2A.
RT-PCR experiments using human hematopoietic cells detected a smaller product in addition to the full-length band, but the level was significantly lower than Δ183 SHIP cDNA expression in murine cells and was not easily detected with ethidium bromide staining. This 334 bp product was amplified from both TPA-treated ML-1 cells and normal human peripheral white blood cells, using primers to the human SHIP sequence analogous to the murine SHIP primers 2666-3247 (Fig 6B). Sequence analysis showed the same in-frame 282 bp deletion in samples from both ML-1 cells and peripheral white blood cells, encoding 94 amino acids with a calculated molecular weight of 10 kD. The human Δ282 deletion is in a region of SHIP overlapping the murine Δ183 deletion region, but is not identical. However, both ends of the deleted segment in human SHIP correspond perfectly to intron-exon boundaries we detected in murine genomic DNA, and are preceded by the required consensus splice acceptor nucleotides AG.
Δ183 SHIP is tyrosine phosphorylated and interacts with Shc and Grb2 in response to M-CSF.
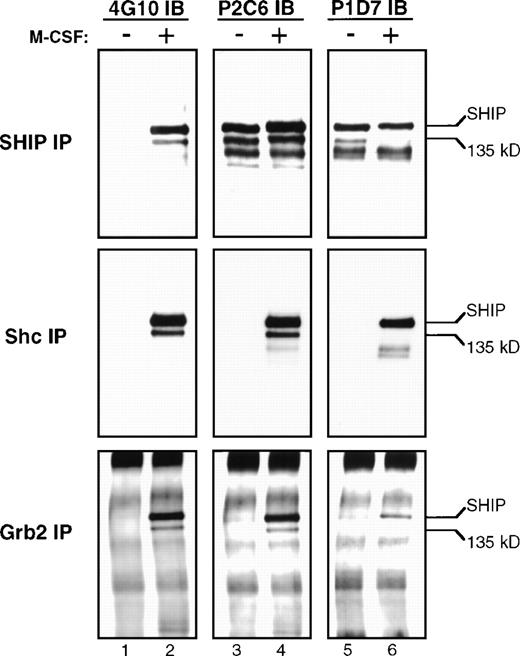
After 5 hours of incubation in media containing 1% serum without IL-3, FDC-P1 cells were centrifuged and either left untreated or were treated with M-CSF for 2 minutes. Lysates from these cells were immunoprecipitated with #5340 anti-SHIP antibody, and one third of the resulting supernatant was loaded onto each of four identical polyacrylamide gels for SDS-PAGE as described. The blots were incubated with the phosphotyrosine-specific antibody 4G10 or the anti-SHIP MoAbs P2C6 or P1D7 as indicated (Fig 7). As can be observed in the SHIP IP lanes 3 and 4, comparable amounts of all SHIP proteins were immunoprecipitated from untreated or M-CSF–treated lysates using the polyclonal anti-SHIP antibody #5340. The 135 kD band was readily observed with MoAb P2C6 as expected, and was not visible using MoAb P1D7 (SHIP IP lanes 5 and 6). In this experiment we also detected a band at 130 kD (SHIP IP lane 5), but its recognition by the P1D7 antibody shows that it is distinct from the Δ183 SHIP isoform and likely represents proteolytic degradation of 145 kD SHIP. Both the 145 kD and the 135 kD bands are detected by the 4G10 antibody in the SHIP immunoprecipitations using lysates from M-CSF–treated cells, showing they are both tyrosine phosphorylated in response to M-CSF treatment (SHIP IP lane 2).
Immunoprecipitations. Lysates from FDC-P1 cells, either untreated (−) or treated with M-CSF (+), were mixed with protein A sepharose beads and each of the antibodies indicated on the left (IP). Immunoprecipitation and immunoblotting analyses were performed as described, and three identical gels were prepared. These gels were incubated with the antiphosphotyrosine antibody 4G10, or the SHIP MoAbs P2C6 or P1D7 (IB). The positions of 145 kD and 135 kD SHIP are indicted with arrows.
Immunoprecipitations. Lysates from FDC-P1 cells, either untreated (−) or treated with M-CSF (+), were mixed with protein A sepharose beads and each of the antibodies indicated on the left (IP). Immunoprecipitation and immunoblotting analyses were performed as described, and three identical gels were prepared. These gels were incubated with the antiphosphotyrosine antibody 4G10, or the SHIP MoAbs P2C6 or P1D7 (IB). The positions of 145 kD and 135 kD SHIP are indicted with arrows.
As can be observed in the Shc IP lane 2, immunoprecipitations using the anti-Shc antibody contain tyrosine-phosphorylated proteins of 145 and 135 kD in lysates from M-CSF–treated FDC-P1 cells. These proteins comigrate exactly with the SHIP isoforms and are recognized by the MoAb P2C6. The 145 kD, but not the 135 kD, isoform is also detected with P1D7 antibody, as was shown in Fig 5. As we and others have previously described,8-10 145 kD SHIP is found in a complex with both Shc and Grb2. To address whether 135 kD SHIP is also involved in this interaction, we performed immunoprecipitations using a polyclonal anti-Grb2 antibody and lysates from the untreated or M-CSF–treated FDC-P1 cells. As can be observed in the Grb2 IP lane 2, tyrosine-phosphorylated proteins are present in Grb2 immunoprecipitates that migrate identically to 145 kD and 135 kD SHIP. Additionally, both bands are recognized by the MoAb P2C6, but the 135 kD band is not recognized by the antibody P1D7 (Grb2 IP lanes 3-6). The Grb2 immunoprecipitation experiments required longer exposure times than the Shc or SHIP experiments, suggesting a weaker interaction of Grb2 with SHIP.
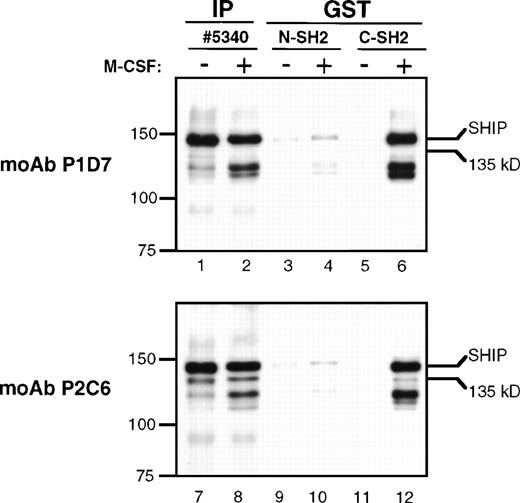
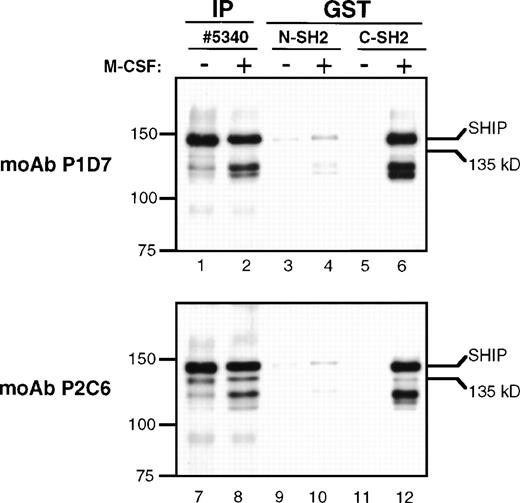
The presence of the YIXM motif in SHIP suggests that it may bind the SH2 domain of p85, the regulatory subunit of PI3K.27Previous experiments in our laboratory showed that immunoprecipitates using anti-p85 polyclonal antibody contained SHIP, but it was not clear whether this was a direct interaction or the result of a larger complex formation.24 To more specifically detect p85-SHIP interactions, we performed pull-down experiments using either the N-terminal or the C-terminal SH2 domains of p85 fused to GST (Fig 8). An immunoblot of the GST-SH2 domain pull-down eluates, incubated with the anti-SHIP MoAb P1D7, showed a clear phosphorylation-dependent interaction of the p85 C-terminal SH2 domain with 145 kD SHIP (Fig 8, lanes 5 and 6). Identical immunoblots incubated with the monoclonal anti-SHIP antibody P2C6 also indicated an interaction of the p85 C-terminal SH2 domain with 145 kD SHIP, and a much weaker interaction of the SH2 domain with Δ183 SHIP (lanes 11 and 12). Comparisons of 145 kD SHIP and Δ183 kD SHIP bands in GST pull-downs (lanes 11 and 12) and SHIP immunoprecipitations (lanes 7 and 8) showed that the ratio of 145 kD SHIP to Δ183 SHIP was approximately 3:1 in the SHIP immunoprecipitates, but was at least 10:1 in the SH2 domain GST pull-down experiments. In each case, the p85 N-terminal SH2 domain showed only a faint SHIP antibody-reactive band at 145 kD, which was similar in the unstimulated and the M-CSF–stimulated lysates (lanes 3, 4, 9, and 10). This may indicate a weak phosphorylation-independent interaction between SHIP and the N-terminal SH2 domain.
GST pull-downs. Lysates from FDC-P1 cells were either untreated (−) or treated with M-CSF (+) as for Fig 7. Lysates were immunoprecipitated with anti-p85 or anti-SHIP #5340 polyclonal antibodies (IP), or were mixed with GST-agarose beads and purified peptides representing either the N-terminal or C-terminal SH2 domains of p85 fused to GST (GST). Beads were incubated and washed and the eluted proteins subjected to immunoblotting with MoAbs P1D7 or P2C6.
GST pull-downs. Lysates from FDC-P1 cells were either untreated (−) or treated with M-CSF (+) as for Fig 7. Lysates were immunoprecipitated with anti-p85 or anti-SHIP #5340 polyclonal antibodies (IP), or were mixed with GST-agarose beads and purified peptides representing either the N-terminal or C-terminal SH2 domains of p85 fused to GST (GST). Beads were incubated and washed and the eluted proteins subjected to immunoblotting with MoAbs P1D7 or P2C6.
DISCUSSION
In this report, we describe a novel spliced form of the 145 kD inositol phosphatase SHIP that is expressed in hematopoietic cells. Our experiments show that this 135 kD form, Δ183 SHIP, is produced along with 145 kD SHIP at both the mRNA and protein levels in M-CSF–differentiated BMCs and in various hematopoietic cell lines. In the report by Helgason et al,7 a targeted disruption ofship in mice eliminated both the 135 kD and the 145 kD forms of SHIP protein, showing that these two isoforms are the product of a single gene. Therefore, Δ183 SHIP expression is not likely to be the product of a separate allele or transcriptional regulatory mechanism. Also, the 183 nucleotide region deleted in Δ183 SHIP cDNA is flanked by consensus intron splice donor and acceptor sequences, and the deletion results in the elimination of 61 amino acids from within the protein sequence. This finding suggests that the appearance of this 135 kD form is the result of alternative mRNA splicing and is not the result of proteolytic cleavage or other post-translational modification of SHIP protein.
To further study the SHIP isoforms, we developed several MoAbs. In the hybridoma screening process, MoAbs were selected that recognized only 145 kD SHIP (P1D7), but we also obtained antibodies that recognized both 145 kD SHIP and a 135 kD form (P2C6, P1C1, and P6H8), and these specificities were not separable by repeated cloning of the hybridomas. The recognition of both 145 and 135 kD SHIP bands by the MoAbs P2C6, P1C1, and P6H8, as well as both the anti-SHIP polyclonal antibodies, shows that the two species are closely related. Furthermore, the 135 kD form of SHIP lacks the epitope recognized by the MoAb P1D7. Our comparison of SHIP isoforms using these antibodies, together with our RT-PCR analyses, provides a powerful means of detecting specific differences in the various SHIP proteins.
Initially, we considered that the 135 kD band recognized by our anti-SHIP antibodies may be a proteolytic cleavage product of 145 kD SHIP, as was recently described by Damen et al.30 However, treatment of cell lysates with protease inhibitors or lysing cells directly into SDS-PAGE sample loading buffer did not affect the appearance of the 135 kD band in immunoblotting experiments. Furthermore, our data showed that the epitope recognized by the MoAb P1D7 is absent in the 135 kD isoform. To remove this epitope by normal proteolysis, more than 160 amino acids (about 19 kD) must be deleted from the C terminus. Our SDS-PAGE size estimates show that full-length and 135 kD SHIP differ by less than 10 kD, making it unlikely that the epitope recognized by P1D7 is eliminated by proteolytic degradation.
We obtained complete cDNA sequences for both the 145 kD and the 135 kD forms of SHIP using an FDC-P1 murine cDNA library. 293T fibroblasts transfected with this full-length SHIP cDNA in an expression vector produce SHIP protein of the expected size and also a protein approximately 15 kD smaller (Fig 5, lane 2). However, this smaller (130 kD) band in the full-length SHIP transfectants is recognized using the MoAb P1D7 that does not recognize either the translated product of Δ183 SHIP in the same cell lines, or the 135 kD SHIP in hematopoietic cells. RT-PCR analysis of full-length SHIP–transfected 293T cells using the 2666-3247 primer set readily detects full-length SHIP cDNA as expected, but is unable to detect Δ183 SHIP cDNA. These results suggest that the smaller bands observed in fibroblasts transfected with SHIP cDNA are the result of proteolysis or other modification of SHIP perhaps as described by Damen et al,30 but that these modifications are not responsible for the appearance of Δ183 SHIP/135 kD protein. We also observe the 130 kD protein in some, but not all, preparations of hematopoietic cells (Fig 7, SHIP IP, lane 5). Considering the specificity of the MoAbs, this 130 kD SHIP protein must differ from full-length SHIP in a region other than that deleted in Δ183 SHIP/135 kD protein.
Our competitive PCR experiments have shown that in the cell types we have tested, Δ183 SHIP cDNA is present in higher abundance than full-length SHIP cDNA. This is not the result of preferential amplification of the shorter product, because as can be observed in Fig4, the rate of amplification for both products is nearly identical. In contrast, our immunoblot assays show that 145 kD SHIP protein appears to be present in at least three-fold greater abundance than Δ183 SHIP protein. Reasons for this difference may include lesser affinities of the anti-SHIP antibodies for Δ183 SHIP, an increased rate of protein degradation, or a decreased efficiency of translation for the Δ183 SHIP isoform. We are conducting experiments to distinguish between these or other possibilities. As we have observed in murine BMCs, the expression pattern of SHIP or SHIP isoforms appears to be based on the maturational stage of the cell, and further experimentation is necessary to determine the details of this change in regulation. In future studies, the competitive PCR strategy we describe will allow us to compare ratios of Δ183 SHIP to full-length SHIP message in the course of hematopoietic cell development to determine if there is a difference in abundance related to cell differentiation.
The human myeloid leukemia cell line ML-1 produces a SHIP isoform of approximately 110 kD. When these cells are treated with TPA to induce differentiation toward a macrophage phenotype, SHIP forms of approximately 135 and 145 kD are expressed and production of the 110 kD form is diminished26 (Fig 6A). This pattern parallels what we have observed in this study using murine BMCs treated with IL-3 plus M-CSF, and suggests that human cells may produce SHIP splice forms as well. Our experiments also showed that normal human peripheral white blood cells express a 135 kD SHIP protein in addition to 145 kD SHIP, but the expression of this form is low relative to the 145 kD form. Using RT-PCR, we detected a shorter species of SHIP cDNA, as well as full-length SHIP cDNA, in both TPA-treated ML-1 cells and in normal human peripheral white blood cells. The smaller cDNA species was amplified in addition to the expected product using primers to the human SHIP sequence analogous to the murine SHIP 2666-3247 primers, and may represent a human SHIP splice form similar to Δ183 SHIP. Our genomic and cDNA sequencing results show that the 282 bp deletion in this potential splice form overlaps the site of the Δ183 SHIP deletion. Also, the segment deleted is preceded on each end by a consensus splice acceptor, and corresponds perfectly to intron-exon boundaries we determined for murine genomic DNA. Thus, Δ282 SHIP is likely to arise from mRNA splicing. The deleted segment encodes 10 kD, and may be responsible for the size difference between the 145 and 135 kD forms observed in our immunoblotting experiments. Interestingly, the deletion in the human SHIP cDNA has the same result as the Δ183 SHIP deletion, which is to either eliminate or alter the specificity of the proximal NPXY motif and to remove potential SH3-binding motifs from the SHIP protein.
Kavanaugh et al21 have identified SHIP isoforms of predicted molecular weight 110, 130, and 145 kD (SIP-110, SIP-130, and SIP-145) in human B cells. According to this study, these SIP/SHIP species vary from each other in the N-terminal region and do not contain a deletion in the C-terminal region. Also, our P6H8 MoAb, made to the SH2 domain of SHIP, recognizes both the 145 and 135 kD forms of SHIP (Fig 5). Thus, Δ183 SHIP and the murine homologs of the human SIP isoforms, if present in these myeloid cells, are distinct. Similarly, a recent report from Pesesse et al25 describes an additional member of the SHIP family in human cells, SHIP2. We do not believe that the 135 kD SHIP protein we have examined is the murine version of SHIP2. Our results show that the 135 kD band observed in SHIP immunoblots of myeloid cells is identical to the predicted character of translated Δ183 SHIP cDNA, which in turn is identical in sequence to full-length SHIP except for the 183 nucleotide deletion. SHIP2, in contrast, is less than 50% identical to human 145 kD SHIP by amino acid comparison, and is larger than SHIP by approximately 10 kD.
In some instances, our results contradict the recent findings of Damen et al,30 who showed that a 135 kD protein, as well as smaller forms, are produced by C-terminal cleavage of full-length SHIP. For example, they show that 145 kD, but not 135 kD, SHIP associates with Shc after IL-3 treatment of DA-ER cells. It is possible that the phosphorylation pattern of SHIP isoforms differs between FDC-P1 and DA-ER cells, or that IL-3 treatment of DA-ER cells causes the phosphorylation of 145 kD SHIP and not of 135 kD SHIP. Another explanation is that our antibodies detect a different SHIP form than the 135 kD cleavage product observed by Damen et al. As discussed above, we also observe a separate SHIP form at approximately 130 kD in some experiments that is distinct from 135 kD SHIP; this may be the same form described by Damen et al. Additionally, the 135 kD protein observed by Damen et al could be the product of mRNA splicing as for Δ183 SHIP cDNA. Their results show that 135 kD SHIP is recognized by their C-terminal polyclonal antibody. The immunogen used to produce this antibody includes the amino acids 1157-1178. Therefore, this antibody could not recognize a proteolytic cleavage product of 145 kD SHIP lacking more than 4 kD from the C-terminal end. Their results, however, are consistent with our data showing that a deletion occurs internally and not at the C-terminal end of SHIP. This explanation would require splicing of message from the transfected SHIP cDNA, which we did not find in our experiments using Rat2 fibroblasts, but that may occur in murine hematopoietic cells. It should be determined whether DA-ER cells actually produce the message for Δ183 SHIP, and whether 135 kD SHIP in these cells has the same migration and antibody recognition pattern as the Δ183/135 kD SHIP we have described.
An important question raised by this work is the biological reason for expression and function of two similar forms of SHIP in the same cell types under the same conditions. It is likely that the 61 amino acid region deleted in Δ183 SHIP is critical in certain protein-protein interactions, and loss of this region alters SHIP association with other signaling intermediates while allowing normal expression of the SHIP SH2 and catalytic domains. We have shown in this report that like 145 kD SHIP, Δ183 SHIP is phosphorylated on tyrosine after M-CSF stimulation in the myeloid cell line FDC-P1. Furthermore, both 135 kD and 145 kD SHIP isoforms interact with Shc and Grb2 after M-CSF treatment in FDC-P1 cells; binding of these molecules to SHIP is not affected by the Δ183 deletion.
GST pull-down experiments using the two SH2 domains of the p85 regulatory subunit of PI3K showed a potentially important difference in the association of the two isoforms with PI3K. The deletion in Δ183 SHIP occurs within the amino acid sequence YIGM, which when phosphorylated has been shown to be the preferred binding target of p85.15 The Δ183 deletion eliminates the methionine at the +3 position, crucial for recognition by the p85 SH2 domain,27 and changes the sequence to YIAN. Although the 145 kD SHIP form bound well to the C-terminal SH2 domain of p85 in our experiments, Δ183 SHIP bound poorly. Thus, the Δ183 deletion appears to eliminate interaction of Δ183 SHIP with PI3K, possibly through this YIGM motif. PI3K is vital in phosphatidylinositol signaling, and is activated by many extracellular factors that also promote its recruitment to the cell membrane.15 PI3K has been shown to activate other protein tyrosine kinases, as well as to be involved in vesicular trafficking and actin cytoskeletal rearrangements. The enzymatic activities of SHIP and PI3K can affect the same phosphatidylinositol intermediates. Therefore, SHIP and PI3K might associate in a pathway and together regulate levels of specific phosphatidylinositols. Δ183 SHIP may escape this interaction with PI3K to alternately affect inositol levels. A likely target of this regulatory mechanism is the serine-threonine kinase Akt/protein kinase B, active in growth-factor mediated signal transduction.29Further experiments, including overexpression of Δ183 SHIP in hematopoietic cells, are under way to provide evidence for this hypothesis.
ACKNOWLEDGMENT
The authors thank all the members of the Rohrschneider laboratory for many helpful discussions and comments throughout this work. We would especially like to thank Dr Paul Algate who constructed the FDC-P1 cDNA library, Susan Geier and Carol Romero for expert technical assistance, and Dr Elizabeth Wayner in the FHCRC Hybridoma Shared Resource.
Supported by Public Health Service Grant No. CA20551 and Fred Hutchinson Cancer Research Center funding to L.R.R., and a National Research Service Award (F32 DK09774-01) to D.M.L.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
Full-length SHIP, with a calculated molecular weight of 133 kD, has been described previously as a 145 kD protein due to its slower migration in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses. To maintain consistency, we will refer to the apparent molecular weights of SHIP throughout this report.