Activation of endothelial cells, important in processes such as angiogenesis, is regulated by cell surface receptors, including those in the tyrosine kinase (RTK) family. Receptor activity, in turn, can be modulated by phosphorylation, turnover, or proteolytic release of a soluble extracellular domain. Previously, we demonstrated that release of soluble tie-1 receptor from endothelial cells by phorbol myristate acetate (PMA) is mediated through protein kinase C and a Ca2+-dependent protease. In this study, the release of soluble tie-1 was shown to be stimulated by inflammatory cytokines and vascular endothelial growth factor (VEGF), but not by growth factors such as basic fibroblast growth factor (bFGF) or transforming growth factor (TGF). Release of soluble tie by tumor necrosis factor (TNF) or VEGF occurred within 10 minutes of stimulation and reached maximal levels within 60 minutes. Specificity was shown by fluorescence-activated cell sorting (FACS) analysis; endothelial cells exhibited a significant decrease in cell surface tie-1 expression in response to TNF, whereas expression of epidermal growth factor receptor (EGF-R) and CD31 was stable. In contrast, tie-1 expression on megakaryoblastic UT-7 cells was unaffected by PMA or TNF. Sequence analysis of the cleaved receptor indicated that tie-1 was proteolyzed at the E749/S750 peptide bond in the proximal transmembrane domain. Moreover, the hydroxamic acid derivative BB-24 demonstrated dose-dependent inhibition of cytokine-, PMA-, and VEGF-stimulated shedding, suggesting that the tie-1 protease was a metalloprotease. Protease activity in a tie-1 peptide cleavage assay was (1) associated with endothelial cell membranes, (2) specifically activated in TNF-treated cells, and (3) inhibited by BB-24. Additionally, proliferation of endothelial cells in response to VEGF, but not bFGF, was inhibited by BB-24, suggesting that the release of soluble tie-1 receptor plays a role in VEGF-mediated proliferation. This study demonstrated that the release of soluble tie-1 from endothelial cells is stimulated by inflammatory cytokines and VEGF through the activation of an endothelial membrane-associated metalloprotease.
SERVING BOTH BARRIER and conduit functions, the vasculature plays a critical role in maintaining normal organ function and exacerbating pathological processes. Endothelial cell activation, proliferation, migration, and matrix remodeling all are essential components of inflammation and angiogenesis, processes accompanying diseases such as cancer, diabetic retinopathy, and rheumatoid arthritis.1 2 The response of endothelial cells to the cytokines and growth factors that mediate these pathways is dependent on the expression and activity of their corresponding cell surface receptors. Consequently, the regulation of endothelial cell receptor function has become an area of expanding research focus and effort.
Receptor tyrosine kinases (RTK) play pivotal roles in the proliferation, activation, and differentiation of a spectrum of cell types.3 Endothelial cells express several members of the RTK family, including receptors for vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), and epidermal growth factor (EGF).4 The VEGF receptor (VEGF-R), Flk-1, stimulates endothelial cell proliferation, migration, and tubule formation in vitro.5In vivo, Flk-1 activation stimulates increased vascular permeability and induction of angiogenesis. FGF-R activation by basic FGF (bFGF) has also been implicated in angiogenesis by inducing endothelial cell migration, proliferation, and matrix reorganization.6 These studies suggest that the activation of RTKs initiates a cascade of signaling pathways leading to the functional and phenotypic changes associated with vascular remodeling. The importance of this process is underscored by evidence that the vasculature is largely quiescent in healthy adults.1Endothelial cell RTK activation is therefore more strongly associated with disease and its coordinate pathology. For example, in the absence of angiogenesis, solid tumor growth is severely limited, but tumors circumvent this restriction by expressing endothelial cell mitogens, including bFGF and VEGF.7 However, tumor growth in vivo is inhibited if RTK signaling is prevented, such as by dominant-negative mutations of the VEGF-R, Flk-1.8 Thus, endothelial cell RTK activation is an important component of disease progression and the regulation of RTKs increasingly a focal point for new approaches in drug development.
The tie-1 RTK is expressed on endothelial cells and a subset of hematopoietic progenitor cells.4 Tie-1 and tie-2 (tek) form a subfamily of RTKs possessing EGF-like repeats that distinguish it from other RTK subfamilies. Although tie-1 expression on CD34+ hematopoietic stem cells decreases during myeloid differentiation, in megakaryocyte differentiation a significant percentage of CD34− cells express tie-1.9Treatment of megakaryoblastic leukemia cells with phorbol myristate acetate (PMA) also upregulates tie-1 expression.10 During development, tie-1 expression is prominent in tissues that give rise to the vasculature. Tie-1 knockout mice fail to assemble an intact vasculature and die as a result of edema and hemorrhage between E13.5 and 14.5.11 In adults, tie expression increases during wound healing, in proliferating ovarian capillaries, and in the vasculature surrounding malignant glioblastoma and melanoma.12-14 Although the ligand for tie-1 has yet to be described, angiopoietin-1, the tie-2 ligand, was recently identified by Davis et al.15 Angiopoietin-1 appears to be involved in the regulation of cell-cell interactions and maintenance of the vascular architecture. In contrast to bFGF or VEGF, angiopoietin-1 does not directly stimulate endothelial cell growth. Angiopoietin-1 knock-out mice recapitulate the loss of vascular organization and embryonic lethality observed with tie-2 knock-outs,16,17and a mutated tie-2 has been linked with some forms of inherited venous malformations.18 Recently, adenoviral-mediated delivery of soluble tie-2 was shown to inhibit tumor growth and metastasis in vivo,19 suggesting that tie-2 is important in the development of tumor vasculature. These data suggest that the tie family of RTKs is critical in the organization and integrity of the vascular wall.
In this study, we show that the release of soluble tie-1 from endothelial cells is regulated by inflammatory cytokines and VEGF via the activation of a hydroxamic acid-sensitive metalloprotease. These data suggest that tie-1 plays a role in the modulation of cell-cell interactions and vascular permeability that accompany inflammation.
MATERIALS AND METHODS
Reagents.
All cytokines and growth factors were purchased from R&D Systems (Minneapolis, MN) unless stated otherwise. PMA and lipopolysaccharide (LPS) were obtained from Sigma (St Louis, MO). Phosphoramidon, Pefabloc, leupeptin, and aprotinin were purchased from Boehringer Mannheim (Indianapolis, IN). VEGF was purchased from PeproTech (Rocky Hill, NJ). Purified recombinant bFGF was provided by Dr Tsutomu Arakawa (Amgen, Thousand Oaks, CA). Purified recombinant soluble CD14 was provided by Dr Mike Kelley (Amgen). The hydroxamate-based metalloprotease inhibitor BB-24 was a kind gift from British Biotech Pharmaceuticals (Oxford, UK). Antibodies (Abs) to tie-1 and recombinant soluble tie have been described previously.20 Abs against CD31 and CD62E were purchased from Becton Dickenson (Franklin Lakes, NJ). Anti–EGF-R was from Oncogene Sciences (Uniondale, NY), and anti–FGF-R1 was from Upstate Biotechnology (Lake Placid, NY). The tie-1a peptide, DNP-NH-AEGPVQESRAAEEG-COOH, was synthesized and verified as previously described,21 lyophilized, and reconstituted to 10 mmol/L in dimethyl sulfoxide (DMSO) before use. Additionally, a truncated peptide (tie-1b) representing the correct cleavage product, DNP-NH-AEGPVQE-COOH, was also synthesized and run as a positive control in all peptide cleavage assays. Both peptides were synthesized with a dinitrophenyl (DNP) group at the N-terminus for detection at 350 nm in high-performance liquid chromatography (HPLC) assays.
Cells.
Human umbilical vein endothelial cells (HUVECs) and dermal microvascular endothelial cells (MDECs) were purchased from Cell Systems (Kirkland, WA). Lung microvascular endothelial cells were purchased from Clonetics Corp (San Diego, CA). Endothelial cells were grown in the media provided by the vendor and cultures were routinely used between passages 2 and 6. CHO-tie cells, CHO stable transfectants expressing full length tie-1 receptor, have been described in detail previously.20 CHO-tie cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% dialyzed fetal bovine serum (FBS). UT-7 cells, a megakaryoblastic leukemic cell line,22 were grown in Iscove’s medium containing 10% FBS and 2 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF).
Enzyme-linked immunosorbent assays (ELISAs).
Sandwich ELISAs for the detection of soluble tie-1 were performed as previously described.20 Aliquots of supernatants from endothelial cells treated with cytokines, growth factors, or control media were assayed in duplicate. Where indicated, cells were incubated for 10 minutes with 200 nmol/L GF109203X (Calbiochem, San Diego, CA) or 20 μmol/L PD098059 (New England BioLabs, Beverly, MA) before adding 10 ng/mL PMA, tumor necrosis factor α (TNFα), or 20 ng/mL VEGF for 2 hours. For inhibition experiments, cultures were preincubated with increasing concentrations of the metalloprotease inhibitor BB-24 for 15 minutes before the addition of the indicated cytokines for 1 hour. Plates were developed with the TMB substrate kit (KPL, Gaithersburg, MD), and absorbance was read at 450 nm. Results are expressed as the mean ± SD.
Fluorescence-activated cell sorting (FACS) analysis.
HUVECs treated with 10 ng/mL TNFα or PMA for 1 hour were washed with ice-cold phosphate-buffered saline (PBS) and harvested using versine-EDTA. In some experiments, cells were preincubated with 5 μmol/L BB-24 or DMSO (1:438 dilution) as a control for 10 minutes before treatment with TNFα. Cells were stained for 45 minutes on ice with the following Abs: monoclonal antibody (MoAb) 42G10 for tie-1, antihuman EGF-R, antihuman CD31, MoAb #9 for axl, antihuman CD62E (E-selectin), and antichicken FGF-R1. Appropriate isotype-matched Ab controls were used in each experiment. UT-7 cells were treated for 1 hour with 10 ng/mL PMA, TNFα, interleukin-1β (IL-1β), or VEGF before staining with tie-1 MoAb 42G10. Samples were processed and analyzed on a FACScan flow cytometer (Becton Dickinson, Milpitas, CA) as previously described.20
Tie-1 cleavage site analysis.
HUVECs or CHO-tie cells were grown in 100-mm dishes to confluency. Cultures were treated with 10 ng/mL TNFα or PMA for 1 hour, washed, and lysed as previously described.20 An affinity matrix for isolation of the C-terminal fragment of tie-1 was prepared by covalently cross-linking a C-terminal peptide antibody20 to protein A Sepharose (Pharmacia Biotech, Piscataway, NJ) with dimethylpimelimidate.23 Lysate was loaded onto the affinity matrix and bound material eluted with 0.1 mol/L glycine, 10 mmol/L CHAPS, pH 2.7. Fractions were immediately neutralized, and those containing the cleaved tie receptor were pooled, concentrated, and separated on a 14% sodium dodecyl sulfate (SDS)-polyacrylamide gel followed by transfer to polyvinylidene difluoride membrane (PVDF) for N-terminal microsequencing.
HUVEC membrane preparation.
HUVECs in 100-mm dishes were incubated in the presence or absence of 20 ng/mL TNFα for 1 hour before harvesting with 5 mmol/L EGTA/EDTA. Washed, pelleted cells were resuspended at 5 × 106/mL in lysis buffer (10 mmol/L NaPO4, pH 7.5, 1 mmol/L MgCl2, 30 mmol/L NaCl, 1 mmol/L Pefabloc, 200 μmol/L phosphoramidon) and homogenized on ice in a 5-mL Dounce until less than 20% of the cells remained intact (∼50 strokes). The homogenate was cleared of cell debris by centrifugation at 1,500 rpm for 5 minutes. The supernatant was then centrifuged at 200,000g for 1 hour at 4°C. The cytosolic 200,000g supernatant fraction was carefully transferred into a new tube and the membrane pellet was resuspended at approximately 1 mg/mL in lysis buffer (PBS containing 10% glycerol, 1 mmol/L Pefabloc, 200 μmol/L phosphoramidon, 1.3 μmol/L aprotinin, and 5 μmol/L leupeptin). Both fractions were stored at −80°C until analysis in the peptide cleavage assay.
Tie-1 peptide cleavage assays.
Fifty microliters (45 μg) of HUVEC cytosol or membrane extract was mixed with 500 μg/mL tie-1a peptide, DNP-NH-AEGPVQESRAAEEG-COOH, in PBS, pH 7.5, containing Ca2+ and Mg2+ and incubated for 1 hour at 37°C. In some experiments, 5 μmol/L BB-24 was added to the reaction mix. Reactions were quenched by the addition of 10 μL of 0.5 mol/L EDTA/EGTA. Samples were separated on a 33 × 4.6 mm Micra NPS C18 HPLC column and eluted with a linear gradient of 1% to 16% acetonitrile in 0.1% trifluoroacetic acid (TFA) over 30 minutes. Cleavage reaction products were followed by monitoring absorbance at 350 nm. The tie-1a peptide alone or a mixture of the tie-1a and tie-1b peptides were incubated in buffer and analyzed in parallel to facilitate identification of the cleavage products.
Proliferation assays.
HUVECs in growth media were plated into 96-well plates at 1 × 105/mL and incubated overnight before serum starvation in DMEM containing 0.5% FBS and 1% bovine serum albumin (BSA) for 24 hours. Where indicated, cells were pretreated with increasing concentrations of BB-24 for 1 hour before the addition of bFGF (1 ng/mL) or VEGF (20 ng/mL) and incubated overnight. The next day, BrdU was added to each well and incorporation was measured after 24 hours according to the manufacturer’s protocol (BrdU Cell Proliferation ELISA; Boehringer Mannheim, Indianapolis, IN). Plates were read at 450 nm. All data points were assayed in triplicate and results were expressed as the mean ± SD.
RESULTS
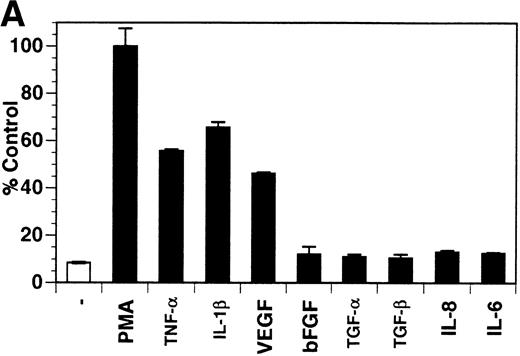
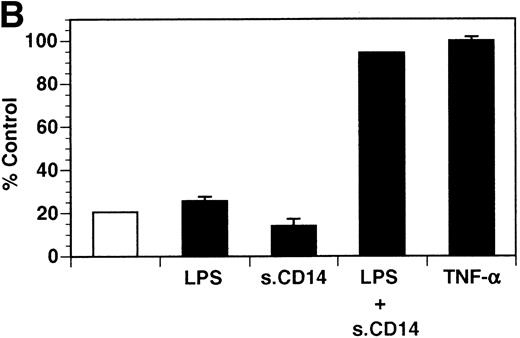
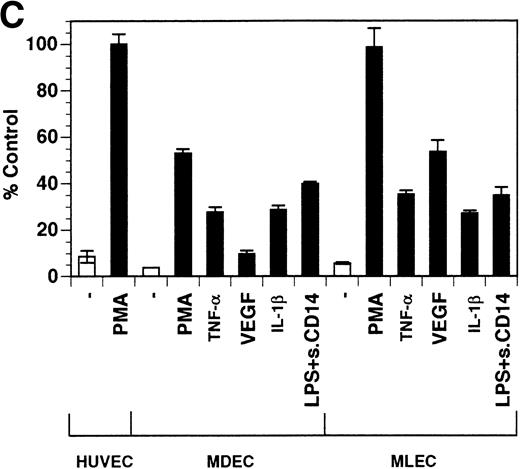
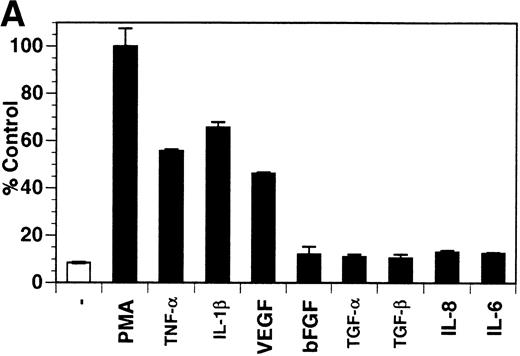
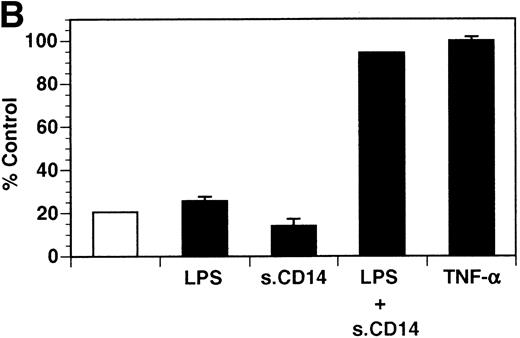
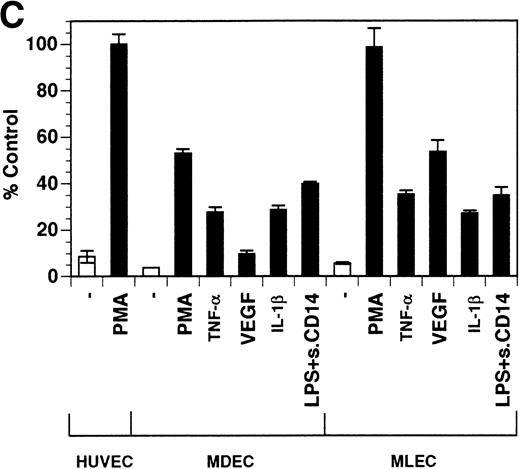
Previously, our lab demonstrated that treatment of endothelial cells with PMA stimulated the rapid release of soluble tie-1 receptor into the media.20 Because PMA is a biological response modifier with pleiotrophic effects on cell activation and function, we focused this study on soluble tie-1 release in response to specific cytokines and growth factors. Figure 1A shows that the inflammatory cytokines TNFα and IL-1β, as well as the growth/permeability factor VEGF, induced the significant release of soluble tie-1 from HUVECs. In contrast, bFGF, transforming growth factor α (TGFα), TGFβ, IL-8, and IL-6 did not stimulate soluble tie-1 release above background levels. Interestingly, soluble tie-1 release mediated by TNFα, IL-1β, or VEGF was approximately 50% of that seen in the presence of PMA. Soluble tie-1 was also released from HUVECs in response to LPS + soluble CD14, but not in response to LPS or soluble CD14 alone (Fig 1B). The release of soluble tie-1 was also evaluated in microvascular endothelial cells (Fig 1C). In dermal microvascular endothelial cells, tie-1 release was less pronounced overall than in HUVECs, especially in response to VEGF. However, in lung microvascular endothelial cells, the extent of tie-1 release in response to inflammatory cytokines and VEGF was quite similar to that observed in HUVECs. These data suggested that the release of soluble tie-1 from endothelial cells is specifically triggered by exposure to inflammatory cytokines and VEGF, but not by mitogenic growth factors such as bFGF or TGFα.
Soluble tie-1 is released from endothelial cells by inflammatory cytokines or VEGF. Cells were incubated with PMA or the indicated factors (all at 10 ng/mL except for VEGF at 100 ng/mL, LPS at 20 ng/mL, and soluble CD14 at 100 ng/mL) for 1 hour. After treatment, supernatants were collected and assayed for soluble tie-1 by sandwich ELISA as described in Materials and Methods. (A and B) HUVECs release soluble tie in response to PMA, TNF, IL-1β, VEGF, or the combination of LPS and soluble CD14. (C) Soluble tie release by HUVECs was compared with release by microvascular dermal endothelial cells (MDEC) and microvascular lung endothelial cells (MLEC). (□) The basal level of soluble tie release, cells incubated with media alone. Results are expressed as the percentage control (±SD), where 100% control represents soluble tie released by HUVECs in response to PMA (A and C) or TNF (B).
Soluble tie-1 is released from endothelial cells by inflammatory cytokines or VEGF. Cells were incubated with PMA or the indicated factors (all at 10 ng/mL except for VEGF at 100 ng/mL, LPS at 20 ng/mL, and soluble CD14 at 100 ng/mL) for 1 hour. After treatment, supernatants were collected and assayed for soluble tie-1 by sandwich ELISA as described in Materials and Methods. (A and B) HUVECs release soluble tie in response to PMA, TNF, IL-1β, VEGF, or the combination of LPS and soluble CD14. (C) Soluble tie release by HUVECs was compared with release by microvascular dermal endothelial cells (MDEC) and microvascular lung endothelial cells (MLEC). (□) The basal level of soluble tie release, cells incubated with media alone. Results are expressed as the percentage control (±SD), where 100% control represents soluble tie released by HUVECs in response to PMA (A and C) or TNF (B).
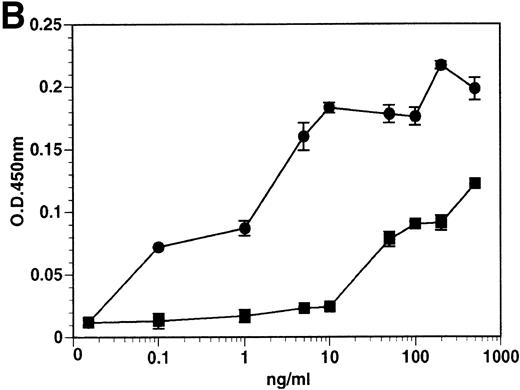
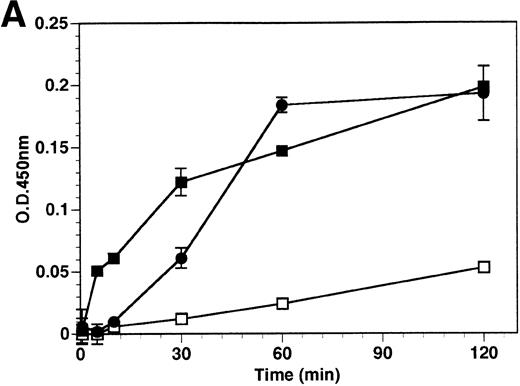
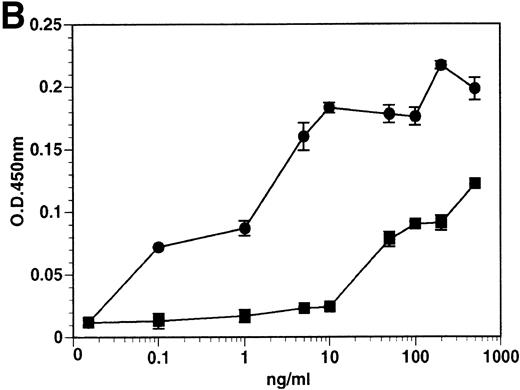
The kinetics of soluble tie-1 release in response to PMA are fairly rapid, with maximum release occurring within 30 minutes.20In comparison, the kinetics of tie-1 release in response to TNFα was somewhat slower; only minimal release was observed after 10 minutes and maximal release was not observed until 1 hour of exposure (Fig 2A). In contrast, VEGF stimulated a more rapid release of tie-1 from HUVECs. Significant release was detected after only 5 minutes of VEGF exposure. Release of soluble tie-1 slowed after 30 minutes and maximal release was reached only after about 1 hour. Figure 2B demonstrates the dose-response curve for soluble tie-1 release in the presence of TNFα and VEGF. For TNFα, soluble tie-1 release was evident at 0.1 ng/mL and peaked at 10 ng/mL. VEGF-mediated release occurred within a narrower dose range; only minimal tie-1 release was seen at 10 ng/mL VEGF, with maximal release seen at 100 ng/mL. These results suggested that the release of soluble tie-1 receptor by TNFα and VEGF was an immediate-early response and occurred at physiologically relevant concentrations.
Kinetics and dose-response of TNF- and VEGF-stimulated soluble tie-1 release in HUVECs. (A) HUVECs were incubated in the absence (□) or presence of 10 ng/mL TNF (•) or 100 ng/mL VEGF (▪) for the indicated times. (B) HUVECs were incubated for 1 hour with the indicated concentrations of TNF (•) or VEGF (▪). After treatment, supernatants were collected and assayed for soluble tie-1 release as described in Materials and Methods. Results are shown as the mean absorbance at 450 nm ± SD.
Kinetics and dose-response of TNF- and VEGF-stimulated soluble tie-1 release in HUVECs. (A) HUVECs were incubated in the absence (□) or presence of 10 ng/mL TNF (•) or 100 ng/mL VEGF (▪) for the indicated times. (B) HUVECs were incubated for 1 hour with the indicated concentrations of TNF (•) or VEGF (▪). After treatment, supernatants were collected and assayed for soluble tie-1 release as described in Materials and Methods. Results are shown as the mean absorbance at 450 nm ± SD.
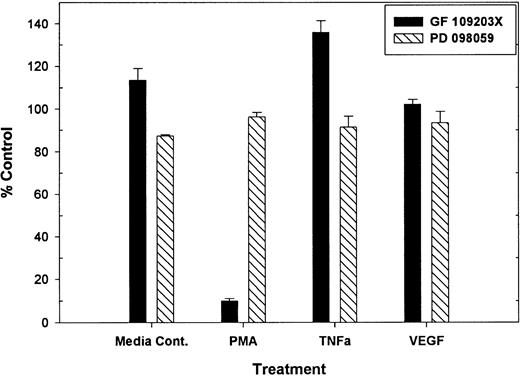
Previous work demonstrated that PMA-mediated tie-1 release is inhibited by GF109203X, indicating that activation of protein kinase C is critical in this pathway.20 To determine if protein kinase C was similarly involved in cytokine-mediated release, HUVECs were treated with TNFα or VEGF in the presence or absence of GF109203X and assayed for soluble tie-1 release (Fig 3). GF109203X did not inhibit tie-1 release in response to TNFα or VEGF, although, as expected, GF109203X completely inhibited PMA-dependent tie-1 release. In addition, soluble tie-1 release was evaluated in the presence of the MEK inhibitor, PD098059. As shown in Fig 3, inhibition of MAPK signaling did not inhibit the release of soluble tie-1 in response to PMA, TNFα, or VEGF. These results indicated that, although activation of protein kinase C modulates tie-1 release in response to PMA, tie-1 release in response to inflammatory cytokines or VEGF is modulated by a distinct set of signaling pathways.
PMA-mediated, but not TNF- or VEGF-mediated soluble tie-1 release is regulated by protein kinase C activation. HUVECs were incubated with 200 nmol/L GF109203X or 20 μmol/L PD098059 for 10 minutes before the addition of 10 ng/mL PMA, TNF, or 20 ng/mL VEGF for 2 hours. Supernatants were collected and assayed for soluble tie-1 release by sandwich ELISA as described in Materials and Methods. Results are expressed as the mean percentage of control ± SD, where 100% control represents soluble tie release in the absence of inhibitor.
PMA-mediated, but not TNF- or VEGF-mediated soluble tie-1 release is regulated by protein kinase C activation. HUVECs were incubated with 200 nmol/L GF109203X or 20 μmol/L PD098059 for 10 minutes before the addition of 10 ng/mL PMA, TNF, or 20 ng/mL VEGF for 2 hours. Supernatants were collected and assayed for soluble tie-1 release by sandwich ELISA as described in Materials and Methods. Results are expressed as the mean percentage of control ± SD, where 100% control represents soluble tie release in the absence of inhibitor.
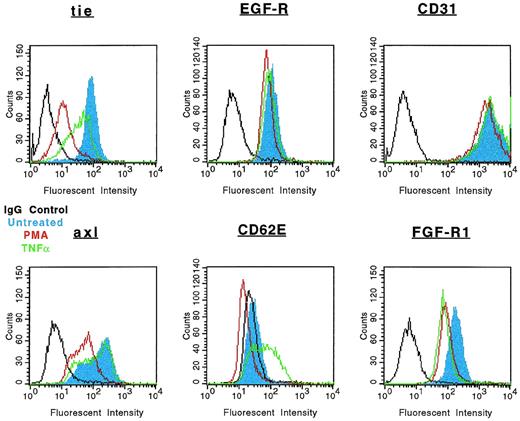
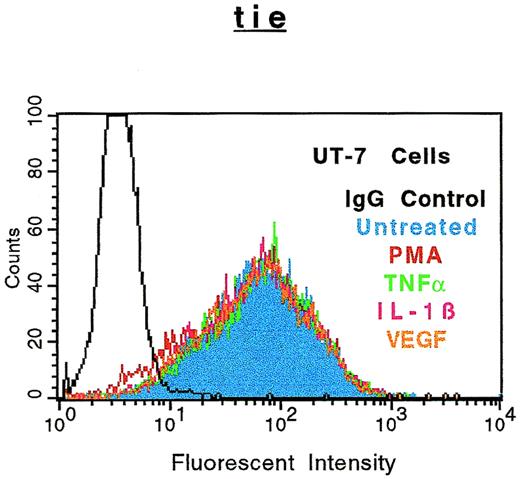
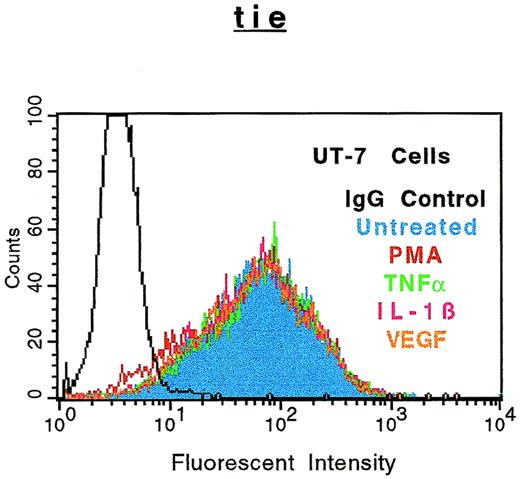
The specificity of soluble tie-1 release with regard to endothelial cell surface molecules and to hematopoietic cells expressing tie-1 was addressed by FACS analysis. First, HUVECs were treated with either TNFα or PMA for 1 hour and the expression of several cell surface markers was evaluated. Figure 4shows the expected loss of tie-1 expression in response to TNFα and PMA. In contrast, expression of the EGF-R, like tie an RTK, and CD31 were not affected by TNFα or PMA. The expression of several other cell surface molecules, including the IGF-1R, CD54, erbB2/Her2, and VLA-4, was also unaffected by TNFα or PMA (data not shown). Interestingly, the expression of axl, an RTK associated with myeloid leukemia cells,24 was also downregulated in response to PMA, but not TNFα. In addition, the expression of FGF-R1 decreased slightly in response to PMA and TNFα, although soluble FGF-R1 could not be detected in the media (data not shown). The upregulation of CD62E (E-selectin) expression after TNFα stimulation served as a positive control. Tie-1 expression was also evaluated on UT-7 megakaryoblastic leukemia cells stimulated with PMA, TNFα, IL-1β, or VEGF (Fig 5). In contrast to endothelial cells, UT-7 cells demonstrated no significant decrease in tie-1 expression in response to inflammatory cytokines, VEGF, or PMA. Control experiments confirmed that IL-1 and TNF receptors were expressed on UT-7 cells (data not shown). These experiments demonstrated that the release of soluble receptors from endothelial cells was not a universal response to TNFα or PMA stimulation. Moreover, soluble tie-1 release was also not a general response in hematopoietic cells expressing tie-1. These data suggested that soluble tie-1 is specifically released from endothelial cells in response to TNFα and PMA.
Specificity of tie-1 release in endothelial cells. HUVECs were incubated in the absence (blue) or presence of 10 ng/mL TNF (green) or PMA (red) for 1 hour before harvesting. Aliquots of cells were stained for 45 minutes on ice with Abs specific for tie-1, EGF-R, CD31 (PECAM), axl, CD62E (E-selectin), or FGF-R1. Control samples were stained with isotype-matched Abs (black). FACS analysis was performed as described in Materials and Methods.
Specificity of tie-1 release in endothelial cells. HUVECs were incubated in the absence (blue) or presence of 10 ng/mL TNF (green) or PMA (red) for 1 hour before harvesting. Aliquots of cells were stained for 45 minutes on ice with Abs specific for tie-1, EGF-R, CD31 (PECAM), axl, CD62E (E-selectin), or FGF-R1. Control samples were stained with isotype-matched Abs (black). FACS analysis was performed as described in Materials and Methods.
Soluble tie receptor is not released from megakaryocytic UT-7 cells. UT-7 cells were incubated in the absence (blue) or presence of 10 ng/mL PMA (red), TNF (green), IL-1β (pink), or 100 ng/mL VEGF (orange) for 1 hour. After treatment, cells were stained for 45 minutes on ice with phycoerythrin-coupled anti–tie-1 MoAb 42G10. An isotype matched IgG (black) was used as the negative control. FACS analysis was performed as described in Materials and Methods.
Soluble tie receptor is not released from megakaryocytic UT-7 cells. UT-7 cells were incubated in the absence (blue) or presence of 10 ng/mL PMA (red), TNF (green), IL-1β (pink), or 100 ng/mL VEGF (orange) for 1 hour. After treatment, cells were stained for 45 minutes on ice with phycoerythrin-coupled anti–tie-1 MoAb 42G10. An isotype matched IgG (black) was used as the negative control. FACS analysis was performed as described in Materials and Methods.
The specificity of soluble tie-1 release from endothelial cells in response to inflammatory cytokines suggested that tie-1 cleavage occurred at a discrete site. To determine the cleavage site, amino acid sequencing of the cleaved receptor was performed. The C-terminal fragment of tie-1, which remains cell-associated after cytokine treatment,20 was isolated by affinity chromatography and subjected to N-terminal microsequencing. Fifteen N-terminal amino acids were identified with the sequence Ser-Arg-Ala-Ala-Glu-Glu-Gly-Leu-Asp-Gln-Gln-Leu-Ile-Leu-Ala. As shown in Table 1, these data indicated that tie-1 was cleaved between amino acids E749 and S750in the juxtamembrane domain. The same amino acid sequence was obtained from HUVECs treated with either TNFα or PMA or from stably transfected CHO-tie-1 cells treated with PMA. The sequence surrounding the tie-1 cleavage site was compared with the consensus cleavage sites of several proteases in the hopes of identifying the tie-1 protease. However, convincing homology between these sequences was not readily apparent. The closest match was to human neutrophil collagenase, which shows a threefold increase in protease activity when the P1amino acid is changed to Glu, yielding a consensus cleavage sequence of GPQE/IAGQ.25 Thus, identification of the tie-1 cleavage site did not unequivocally identify the tie-1 protease.
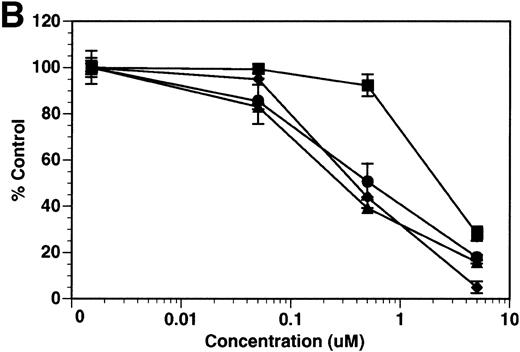
Although tie-1 protease activity is unaffected by serine-, cysteine-, aspartate-, or traditional metallo-protease inhibitors, activity is inhibited by EGTA.20 These data, along with recent evidence that PMA-stimulated shedding of TNF-R is inhibited by the hydroxamic acid derivative TAPI,26 suggested that the tie-1 protease might be a variant metalloprotease. Release of soluble tie from HUVECs was evaluated in the presence of BB-24, a hydroxamic acid-derived protease inhibitor.27 28 BB-24 inhibited the TNFα-mediated release of soluble tie-1 from HUVECs in a dose-dependent manner (Fig 6A). Similarly, BB-24 inhibited the low levels of basal tie-1 release from unstimulated cells. The IC50 for soluble tie-1 release was approximately 0.5 μmol/L for both TNF-dependent and basal release. BB-24 was also tested in endothelial cells stimulated with PMA, VEGF, IL-1β, or LPS+CD14 (Fig 6B). Inhibition of soluble tie-1 release was dose-dependent, but the extent of inhibition depended on the stimulus. The IC50s for VEGF-, IL-1β–, and LPS+CD14-mediated release were in the range of 0.3 to 0.5 μmol/L, similar to the IC50 for TNFα. However, the IC50 for PMA-mediated tie-1 release was approximately 2.5 μmol/L, which is about 10-fold higher. Interestingly, these results correlated with the extent of soluble tie-1 release in cells; PMA-mediated tie-1 release was approximately twice that seen with other stimuli, suggesting that PMA might also activate a second protease capable of cleaving tie-1. These data indicated that the release of soluble tie-1 was mediated by an endothelial cell metalloprotease sensitive to hydroxamic acid-derived inhibitors.
Inhibition of soluble tie-1 release by the hydroxamic acid-based metalloprotease inhibitor BB-24. HUVECs were preincubated with the indicated concentrations of BB-24 for 15 minutes before the stimulation of tie-1 release by (A) 10 ng/mL TNF (•) or (B) 10 ng/mL PMA (▪), 100 ng/mL VEGF (•), 10 ng/mL IL-1β (▴), or 10 ng/mL LPS+100 ng/mL soluble CD14 (⧫). After 1 hour of incubation, supernatants were collected and analyzed for soluble tie-1 release by sandwich ELISA as described in Materials and Methods. Basal soluble tie-1 release in the presence of media alone (○) was also inhibited by BB-24. Results are expressed as the percentage control (±SD), where 100% control represents soluble tie released by HUVECs in the absence of inhibitor.
Inhibition of soluble tie-1 release by the hydroxamic acid-based metalloprotease inhibitor BB-24. HUVECs were preincubated with the indicated concentrations of BB-24 for 15 minutes before the stimulation of tie-1 release by (A) 10 ng/mL TNF (•) or (B) 10 ng/mL PMA (▪), 100 ng/mL VEGF (•), 10 ng/mL IL-1β (▴), or 10 ng/mL LPS+100 ng/mL soluble CD14 (⧫). After 1 hour of incubation, supernatants were collected and analyzed for soluble tie-1 release by sandwich ELISA as described in Materials and Methods. Basal soluble tie-1 release in the presence of media alone (○) was also inhibited by BB-24. Results are expressed as the percentage control (±SD), where 100% control represents soluble tie released by HUVECs in the absence of inhibitor.
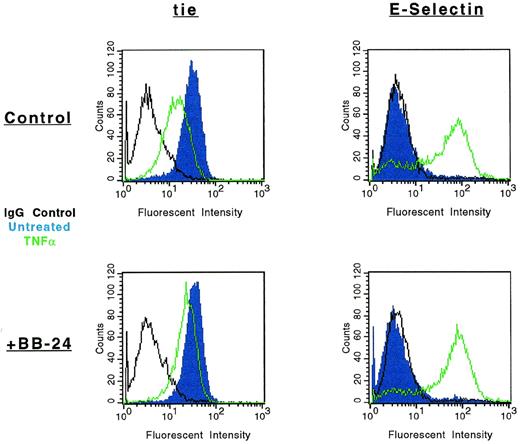
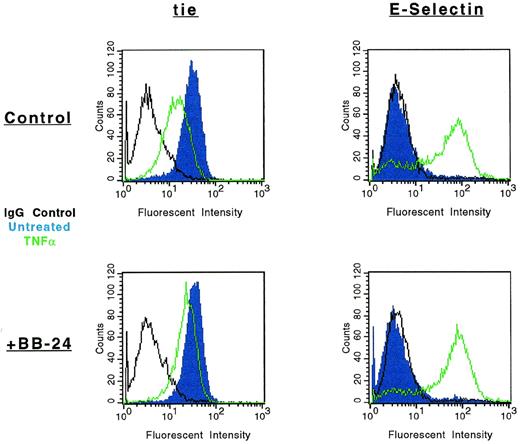
The modulation of tie-1 cell surface expression by BB-24 was also evaluated by FACS (Fig 7). As previously shown, cell surface expression of tie-1 decreased dramatically in HUVECs treated with TNFα. However, in the presence of BB-24, tie-1 expression was essentially unchanged, demonstrating that inhibition of soluble tie-1 release leads to the retention of full-length tie-1 on the cell surface. To rule out the possibility that BB-24 nonspecifically inhibited cytokine signaling, expression of E-selectin on HUVECs was evaluated in the presence and absence of BB-24. Although resting HUVECs did not express detectable levels of E-selectin (Fig 7), cell activation by TNFα significantly upregulated E-selectin expression. Moreover, upregulation of E-selectin expression by TNFα was unaffected by BB-24. These results indicated that cell surface levels of tie-1 remained stable in the presence of TNFα when shedding was inhibited by BB-24 and that BB-24 did not interfere with cytokine signaling in general. In addition, BB-24 did not exhibit any nonspecific cytotoxic effects on HUVECs (data not shown).
Modulation of tie-1, but not E-selectin, cell surface expression by BB-24. HUVECs were incubated in the presence or absence of 5 μmol/L BB-24 for 10 minutes before the addition of TNF (green) or DMSO (blue) for 1 hour. After harvesting with versine-EDTA, cells were stained on ice with Abs specific for tie-1 or CD62E (E-selectin). Control samples were stained with isotype matched Abs (black). FACS analysis was performed as described in Materials and Methods.
Modulation of tie-1, but not E-selectin, cell surface expression by BB-24. HUVECs were incubated in the presence or absence of 5 μmol/L BB-24 for 10 minutes before the addition of TNF (green) or DMSO (blue) for 1 hour. After harvesting with versine-EDTA, cells were stained on ice with Abs specific for tie-1 or CD62E (E-selectin). Control samples were stained with isotype matched Abs (black). FACS analysis was performed as described in Materials and Methods.
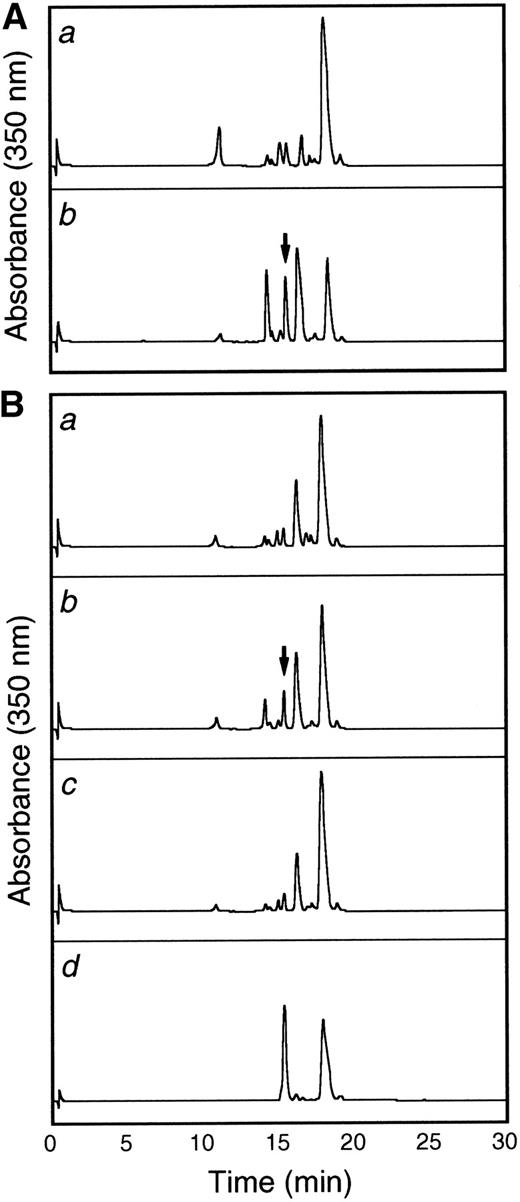
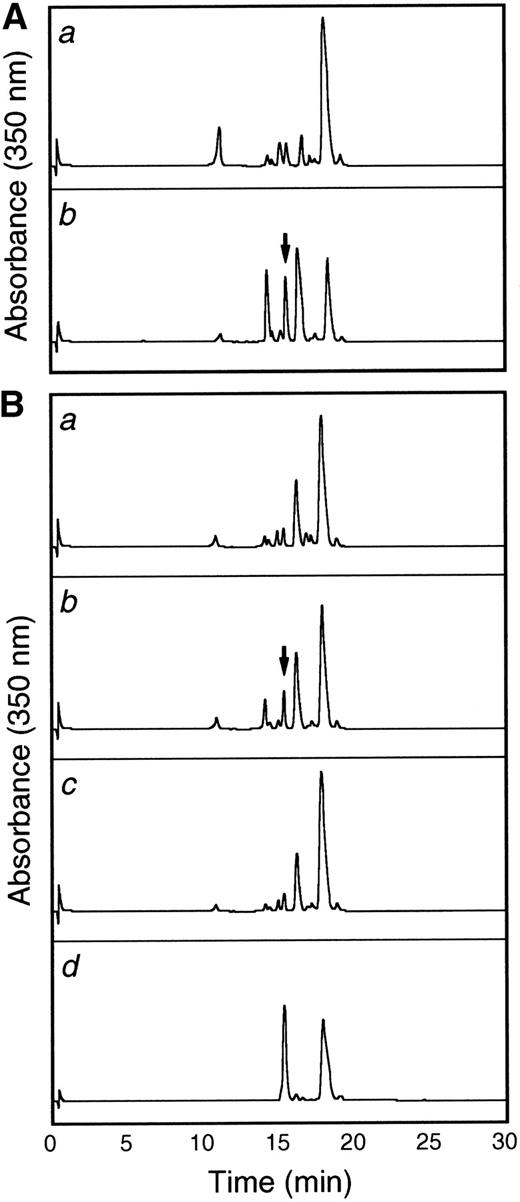
To more thoroughly characterize the endothelial cell tie-1 protease, a tie-1 peptide assay was developed. HUVEC plasma membranes were prepared by cell fractionation and differential centrifugation and incubated with a DNP-labeled peptide substrate spanning the tie-1 cleavage point. Cleavage of the substrate was determined by HPLC analysis and compared with a control intact peptide or a peptide truncated at the appropriate Glu residue. In Fig 8A, tie-1 protease activity was evaluated in the cytosol (a) and membrane fractions (b) of TNF-treated HUVECs. Protease activity was associated primarily with the membrane fraction. The low level of activity in the cytosol could be attributed to spillover or the presence of a second protease. As shown in Fig 8B, membranes prepared from untreated cells (a) did not cleave the intact peptide substrate into a product of the correct size and mobility. The appearance of a peak with intermediate mobility (slower than the truncated control peptide but faster than the intact control) suggested that HUVEC membranes may contain a second TNFα-insensitive protease. More significantly, when membranes from TNFα-treated cells were incubated with the intact substrate (b), a cleaved peptide of the correct size and mobility was generated. Furthermore, generation of the TNF-dependent cleavage product was inhibited in the presence of BB-24 (c). These data demonstrated that tie-1 protease activity was membrane-associated and specifically activated by TNFα. In addition, TNFα-dependent peptide cleavage was inhibited by BB-24, thus establishing specificity and correlating protease activity in intact endothelial cells with activity in cell membranes. These experiments also established a suitable assay system for the characterization and identification of the tie-1 protease.
The tie-1 protease is membrane-associated, activated specifically by TNF, and inhibited by BB-24. HUVECs incubated in the presence or absence of TNF were harvested, and cell fractionation was performed as described in Materials and Methods. (A) Cytosol (a) or membrane (b) fractions prepared from TNF-treated HUVECs were incubated with 500 μg/mL tie-1a peptide for 1 hour at 37°C. (B) HUVEC membrane fractions from untreated (a) or TNF-treated (b and c) cells were incubated with 500 μg/mL tie-1a peptide in the presence (c) or absence (b) of BB-24 for 1 hour at 37°C. Samples quenched with EDTA/EGTA were separated by HPLC and peptide elution followed by absorbance at 350 nm. The control full-length tie-1a and truncated tie-1b peptides are shown in the control chromatogram (d). The arrows indicate the peaks that correspond to peptide fragments with the same size and mobility as the truncated control peptide tie-1b.
The tie-1 protease is membrane-associated, activated specifically by TNF, and inhibited by BB-24. HUVECs incubated in the presence or absence of TNF were harvested, and cell fractionation was performed as described in Materials and Methods. (A) Cytosol (a) or membrane (b) fractions prepared from TNF-treated HUVECs were incubated with 500 μg/mL tie-1a peptide for 1 hour at 37°C. (B) HUVEC membrane fractions from untreated (a) or TNF-treated (b and c) cells were incubated with 500 μg/mL tie-1a peptide in the presence (c) or absence (b) of BB-24 for 1 hour at 37°C. Samples quenched with EDTA/EGTA were separated by HPLC and peptide elution followed by absorbance at 350 nm. The control full-length tie-1a and truncated tie-1b peptides are shown in the control chromatogram (d). The arrows indicate the peaks that correspond to peptide fragments with the same size and mobility as the truncated control peptide tie-1b.
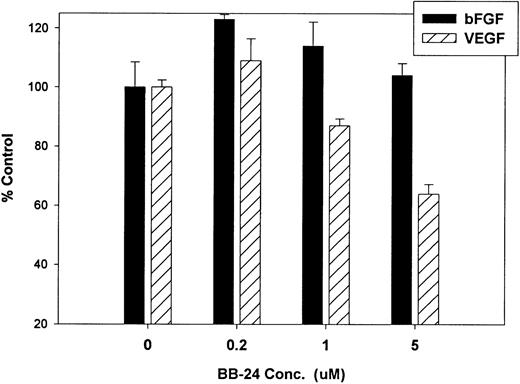
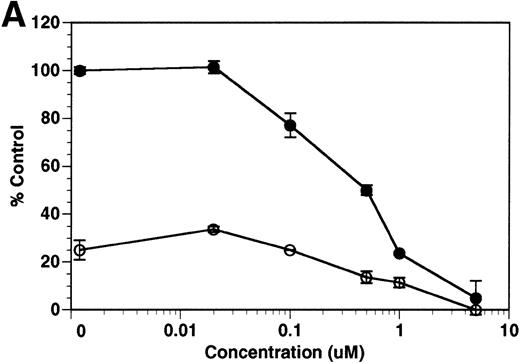
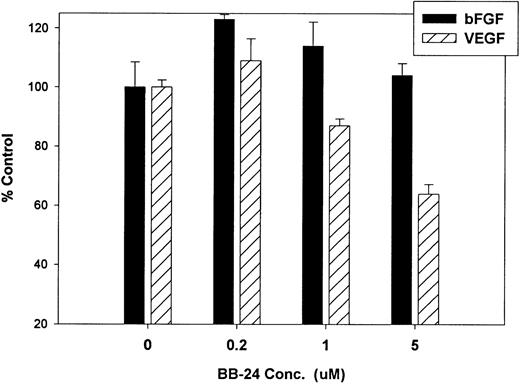
Finally, we investigated whether inhibition of soluble tie-1 release had an effect on endothelial cell proliferation. HUVECs were incubated with either bFGF or VEGF in the presence or absence of increasing concentrations of BB-24. As shown in Fig 9, BB-24 dose-dependently inhibited endothelial cell proliferation in response to VEGF, but not to bFGF. In fact, bFGF-mediated proliferation was modestly increased over controls in the presence of BB-24. Considering that VEGF, but not bFGF, stimulated soluble tie-1 release in cell culture (Fig 1A), these data suggested that release of soluble tie-1 is important in the regulation of endothelial cell proliferation. These data also provided further evidence that bFGF and VEGF use distinct signaling pathways in the modulation of endothelial cell proliferation.
BB-24 inhibits VEGF-mediated endothelial cell proliferation. HUVECs were incubated with increasing concentrations of BB-24 for 1 hour before the addition of 1 ng/mL bFGF or 20 ng/mL VEGF. After overnight incubation, BrdU was added for an additional 24 hours before measuring incorporation as described in Materials and Methods. Plates were read at 450 nm and results are expressed as mean percentage of control ± SD, where 100% control represents BrdU incorporation in the absence of inhibitor.
BB-24 inhibits VEGF-mediated endothelial cell proliferation. HUVECs were incubated with increasing concentrations of BB-24 for 1 hour before the addition of 1 ng/mL bFGF or 20 ng/mL VEGF. After overnight incubation, BrdU was added for an additional 24 hours before measuring incorporation as described in Materials and Methods. Plates were read at 450 nm and results are expressed as mean percentage of control ± SD, where 100% control represents BrdU incorporation in the absence of inhibitor.
DISCUSSION
The regulation of receptor expression in endothelial cells plays an important role in their ability to respond to external stimuli. Whether that response encompasses endothelial cell proliferation, as seen in angiogenesis, or activation, as seen in inflammation, receptor expression provides a mechanism to propagate and modulate alterations in endothelial cell function. In this study, we focused on the regulation of expression of tie-1, an orphan endothelial cell receptor likely to be involved in cell-cell interactions. Inflammatory cytokines and VEGF stimulated the release of soluble tie-1 from endothelial cells, effectively decreasing full-length receptor expression on the cell surface. Soluble tie-1 was not released from cells treated with bFGF, TGFα, or TGFβ. FACS analysis demonstrated the specificity of this response; the release of soluble receptors from endothelial cells was not a universal response to TNFα or PMA, and soluble tie-1 was not released from megakaryocytic UT-7 cells under any conditions. Furthermore, whereas activation of protein kinase C mediated PMA-dependent soluble tie-1 release, TNFα- and VEGF-dependent release signaled through an alternative pathway(s). Preliminary experiments have suggested that activation of p38 may play a role in soluble tie-1 release in response to TNFα and VEGF (data not shown). Although microsequencing demonstrated that tie-1 was cleaved between E749 and S750 in the sequence PVQESRAA, this information did not clarify the identity of the tie-1 protease. However, inhibition of soluble tie-1 release by BB-24 suggested that the protease was a metalloprotease sensitive to hydroxamic acid-based inhibitors. Cell fractionation studies indicated that the tie-1 protease was membrane-associated, inhibited by BB-24, and activated specifically in TNFα-treated cells. In addition, we found that inhibition of soluble tie-1 release inhibited endothelial cell proliferation in response to VEGF, but not bFGF. These results demonstrated that inflammatory cytokines and VEGF regulate tie-1 expression on endothelial cells through the activation of a membrane-associated metalloprotease that releases soluble tie from the cell surface.
The release of soluble forms of membrane-associated growth factors and receptors by PMA has been documented in many systems, including tumor cells, transfected CHO cells, and monocytes.29,30 However, PMA is such a pluripotent biological response modifier that investigators have questioned the specificity of the response as exemplified by Arribas et al,31 who showed that the PMA-dependent release of soluble L-selectin, TGFα, and IL-6Rα in transfected CHO cells occurs through a common proteolytic pathway. In contrast, shedding of the type II IL-1R in polymorphonuclear cells (PMNs) is stimulated by TNFα or GM-CSF, but not by several other cytokines, including IL-1β, even though TNFα and IL-1β share considerable similarity in their signal transduction pathways.32 Moreover, TNF does not induce shedding of class I MHC, FcγR, or β2 integrins in PMNs. These data suggest that the release of soluble receptors can be tightly regulated by signaling pathways that are cell-type specific. We have shown that the release of soluble tie-1 from endothelial cells is regulated by proinflammatory cytokines, including TNFα, IL-1β, and LPS, and the angiogenic growth factor VEGF. Interestingly, another angiogenic growth factor bFGF did not stimulate soluble tie-1 release, suggesting that activating endothelial cell mitogenesis per se is not sufficient to induce tie-1 shedding. This differential response to VEGF and bFGF was further underscored by the finding that inhibition of tie-1 shedding by BB-24 inhibited endothelial cell proliferation in response to VEGF, but not to bFGF. These results suggested that release of soluble tie-1 may play a role in VEGF-mediated proliferation.
Although the stimulation of tie-1 release by an inflammatory cytokine and an angiogenic growth factor, TNFα and VEGF, respectively, might appear to be unrelated, these factors do share the ability to increase vascular permeability. Because the ligand for tie-1 remains unidentified, direct examination of the role of tie-1 in vascular permeability is difficult. However, the death of tie-1 knock-out mice during embryogenesis due to extensive hemorrhaging suggests that tie-1 function is critical for vascular integrity.11,17 Recent data on the tie-2 receptor, and its ligands angiopoietin-1 and -2, strongly suggest a role for the tie receptor family in cell-cell interactions.16,18,33 Moreover, increased tie-1 expression has been associated with the leaky tumor vasculature in glioblastoma and melanoma, but not with the adjacent normal vasculature.12 13 These diverse pieces of evidence suggest that inflammatory cytokines and VEGF may modulate vascular permeability partly by regulating the expression of tie-1 on endothelial cells.
Although our results demonstrated that soluble tie-1 release was induced by a specific subset of cytokines and growth factors, it was unclear whether receptor shedding in response to cytokines was a general endothelial cell phenomenon. For example, LPS induces shedding of both TNFα and the p60 TNF-R from human peripheral blood monocytes.26 In this study, FACS analysis was used to demonstrate that loss of endothelial cell receptors was not a universal response to TNFα or PMA. The expression of CD31, a cell-cell adhesion molecule, was stable in the presence of both TNFα and PMA, as was the expression of several RTKs associated with endothelial cell proliferation, including the EGF-R and IGF-1R. In contrast, axl receptor expression decreased in the presence of PMA, but not TNFα. Loss of tie-1 expression in response to inflammatory cytokines or PMA was not observed in megakaryocytic UT-7 cells, even though these cells express tie-1, TNFα, and IL-1β receptors. Similarly, angiopoietin-2, the tie-2 receptor antagonist, stimulates receptor phosphorylation in transfected fibroblasts but not in endothelial cells expressing endogenous receptor.33 Thus, as yet undefined endothelial cell components may confer a level of specificity on these signaling pathways beyond receptor-ligand binding. These results demonstrated that activation of the tie-1 protease did not result in the universal cleavage of endothelial cell receptors and that soluble tie-1 release likely depends on endothelial cell-specific signal transduction pathways.
To further characterize the tie-1 protease, the cleavage site was determined by microsequencing. The amino acid sequence surrounding the cleavage site, PVQE/SRAA, did not match any known cleavage site consensus sequences and thus did not reveal the nature of the tie-1 protease. Comparison of the tie-1 cleavage site with the cleavage sites of other receptors or membrane-associated growth factors also failed to identify any significant homology.30 Currently, despite extensive research in this area, no consensus sequence for cleavage of these molecules has been established. Indeed, an alternative approach using mutagenized angiotensin-converting enzyme has generated evidence that cleavage may be a positional effect occurring at a minimum distance from both the membrane and the proximal extracellular domain.34 Clearly, the identification of the protease(s) involved in the release of soluble receptors and growth factors will answer many of these questions.
Initial attempts to categorize the tie-1 protease through the use of protease inhibitors were unsuccessful.20 Except for EGTA, none of the other inhibitors affected soluble tie-1 release, suggesting that the protease was Ca2+-dependent but little else. In this report, the hydroxamic acid-based inhibitor BB-24 was shown to dose-dependently inhibit soluble tie-1 release in response to inflammatory cytokines, VEGF and PMA, and inhibit cleavage of a tie-1 peptide by membranes prepared from TNFα-treated cells. The IC50 for TNFα-mediated release in the cell-based assay was approximately 500 nmol/L, similar to the activity of hydroxamic acid-based compounds in the inhibition of endotoxin-mediated release of TNFα from whole blood.35 These data suggested that the tie-1 protease was likely a metalloprotease, although the lack of inhibition by phosphoramidon20 and TIMP-2 (data not shown) suggested that it was not MMP-1, MMP-2, etc, or another well-characterized metalloprotease.35,36 However, the tie-1 protease may be a member of the disintegrin metalloprotease family that includes the TNFα convertase.27,37 These proteases are insensitive to phosphoramidon but susceptible to inhibition by hydroxamic acid-based compounds with IC50s in cell-based assays similar to that described for tie-1 proteolysis.21,35,36 Preliminary results demonstrated that the putative TNF convertase ADAM 10 did not cleave tie-1 in a peptide cleavage assay (D. Lyons, personal communication, June 1997) and expression of ADAM 10 was not detectable in HUVECs by immunoblot (data not shown). Furthermore, recent reports have suggested that the TNF convertase may be constitutively active,21,37 whereas in HUVECs, TNFα stimulation was necessary for protease activity. However, we cannot rule out the possibility that the tie-1 protease may be another member of the ADAM family of metalloproteases.38
The data presented in this study highlight an alternative pathway for the regulation of endothelial cell function. Although signaling can be directly initiated through receptor-ligand interactions, secondary control systems are clearly critical for fine tuning responses. Among these secondary pathways are the expression of endogenous antagonists such as angiopoietin-2,33 the downregulation of receptors through inhibition of transcription such as that reported for VEGF-R,39 and the levels of sequestering proteins in circulation such as the IGF-1 binding proteins.40 More recently, the regulation of receptor/growth factor activity through proteolysis of membrane-bound forms has emerged as an intriguing control mechanism. Even though the soluble receptors, eg, IL-2R or TNF-R, often retain binding activity, the change in localization has a dramatic effect on function. Thus, soluble IL-2R may act as a sink for IL-2 and suppress immune system activation,41 and soluble TNF-R may stabilize serum TNFα and act as a slow release reservoir exacerbating systemic inflammation in septic shock.42Similarly, the release of soluble tie-1 in response to inflammatory cytokines or VEGF could lead to the modulation of proliferation, inflammatory responses, or vascular permeability. The identification of the tie-1 ligand(s) and the tie-1 protease described in this report will help clarify these issues not only to illuminate the role of tie-1 in endothelial cell biology, but also to place the tie receptor family into the larger context of vascular physiology.
ACKNOWLEDGMENT
The authors thank D. Lyons, M. Rosendahl, D. Thomson, and R. Wahl for their excellent advice regarding ADAM proteases and inhibitors; the Amgen Protein Synthesis group for their expertise; and T. Burgess, S. Hu, B. Ratzkin, and K. Farrell for continuing support, interest, and critical review of the manuscript.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Rachel Yabkowitz, PhD, Hoechst Marion Roussel, Inc, Department of Oncology, M/S G203A, Route 202-206, PO Box 6800, Bridgewater, NJ 08807-0800; e-mail:rachel.yabkowitz@hmrag.com.