To the Editor:
The erythrocyte skeleton is a complex protein network responsible for the shape and the physical properties of red blood cells (RBCs), such as deformability and resistance to mechanical stress. Spectrin is the major constituent of this membrane skeleton and represents about 25% in weight of total RBC membrane proteins. Spectrin is composed of two elongated subunits, α and β chain (280 and 246 kD, respectively), which associate noncovalently in an antiparallel side-to-side orientation to form heterodimers (αβ). Spectrin heterodimers associate head-to-head to form tetramers (α2β2). These tetramers constitute the long flexible filaments of the network. Spectrin forms noncovalent associations with other proteins of the erythrocyte skeleton, such as ankyrin,1 band 4.1, actin, adducin, and tropomyosin.2 The α-spectrin chain consists largely of 20 homologous segments, which are about 106 amino acids long, whereas the β-spectrin chain contains 17 homologous segments.3
Here we would like to report an apparent sequencing error in the cDNA sequence of human erythroid α-spectrin (SPTA1) published by Sahr et al4 (Genbank accession no. M61877). We noted that the stretch of G’s at position 7379 of the published sequence consists of four G’s instead of three G’s. The presence of an additional G causes a change in reading frame which changes the last 31 residues of the original deduced amino acid sequence (Fig 1 A and B).
cDNA sequence and deduced amino acid sequence of C-terminus human -spectrin. (A) Published sequence. (B) Corrected sequence. The addition of G (underlined base) cause a change in reading frame resulting in a molecule 11 amino acids shorter (2418 residues instead of 2429) and different in amino acid sequence 2399-2418. (C) Comparison of C-terminus amino acid corrected sequence of human -spectrin (underlined sequence), human -fodrin, chicken -fodrin, drosophila -spectrin.
cDNA sequence and deduced amino acid sequence of C-terminus human -spectrin. (A) Published sequence. (B) Corrected sequence. The addition of G (underlined base) cause a change in reading frame resulting in a molecule 11 amino acids shorter (2418 residues instead of 2429) and different in amino acid sequence 2399-2418. (C) Comparison of C-terminus amino acid corrected sequence of human -spectrin (underlined sequence), human -fodrin, chicken -fodrin, drosophila -spectrin.
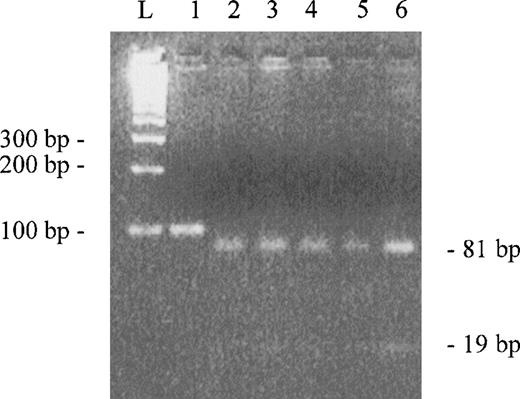
Our initial purpose was to clone the C-terminal domain of α-spectrin (repeat 22) as part of an effort to localize ubiquitination site(s) in this molecule. We used a healthy volunteer reticulocyte cDNA as template for amplification of the C-terminal coding region. To find a clone without errors in the sequence, we sequenced three clones selected at random. All clones sequenced showed the insertion of an extra G at position 7379 of the published sequence. In an attempt to verify if the insertion was a rare polymorphism or the correct wild-type sequence, we screened 16 healthy volunteers using a rapid method in which polymerase chain reaction (PCR) products, digested by a specific restriction enzyme and visualized in agarose gel, gave a clear pattern for the insertion of the additional G. PCR reactions were performed to amplify a region of 100 bp using genomic DNA as template (no introns are present in the amplified region). We designed an oligonucleotide primer to detect the G insertion, creating anScrFI restriction site when four G’s are present in the DNA template by introducing a mismatch base. The 100-bp region was amplified with the sense primer AGCAATATATGGACCCACG and antisense primer TCCACGAGGAGCTGCTTATT. The underlined base is the mismatch base. This mutagenic base in combination with the G at positions 7381 and 7382 (hypothetical) will create an ScrFI site (CCNGG). PCR products were digested with ScrFI and the digestion mixtures were analyzed in a 4% agarose gel (Fig2). All PCR products were cut in two fragments of 81 and 19 bp, indicating that the insertion of G is really present in wild-type α-spectrin gene and that the sequence published is incorrect. Furthermore, we compared the C-terminus amino acid sequences of human α-fodrin, chicken α-fodrin, and drosophila α-spectrin with our corrected C-terminus amino acid sequence of human α-spectrin (Fig 1C). The high homology found between these sequences are consistent with the modified sequence described here. Finally, we recently used automated DNA sequencing to reanalyze this region of a cDNA clone (gift from B. Forget, Yale University, New Haven, CT) which was initially sequenced using manual methods by Sahr et al4 to determine the published complete cDNA sequence. The automated sequence analysis of this original cDNA confirmed the four G’s at positions 7379-7382 and the predicted revised sequence described in Fig 1B.
Agarose gel electroforesis of PCR products digested byScrFI. Lane 1 is uncut control. Lanes 2 to 6 are digested PCR products (only 5 of the 16 samples are showed). L represents the ladder.
Agarose gel electroforesis of PCR products digested byScrFI. Lane 1 is uncut control. Lanes 2 to 6 are digested PCR products (only 5 of the 16 samples are showed). L represents the ladder.
In 1995, T.J. Gibson observed a difference in amino acids 2407 to 2418, due to a frameshift, compared with sequence published by Sahr et al4 (SwissProt databank accession no. P02549; Q15514). Now we confirm and extend these data by localizing the frameshift origin to the stretch of G’s at position 7379 and the consequent change in amino acid sequence from residue 2399 to 2418.
The corrected cDNA sequence and revised predicted amino acid sequence described here (Genbank accession no. AF060556) should be of interest to investigators when searching for spectrin mutations or when using cDNA from this region to express recombinant peptides for structure-function studies.